Abstract
Bosentan (Tracleer®) is an endothelin receptor antagonist prescribed for the treatment of pulmonary arterial hypertension (PAH). Its use is limited by drug-induced liver injury (DILI). To identify genetic markers of DILI, association analyses were performed on 56 Caucasian PAH patients receiving bosentan. Twelve functional polymorphisms in five genes (ABCB11, ABCC2, CYP2C9, SLCO1B1, SLCO1B3) implicated in bosentan pharmacokinetics were tested for associations with ALT, AST and DILI. After adjusting for BMI, CYP2C9*2 was the only polymorphism associated with ALT, AST and DILI (β = 2.16, P = 0.024; β = 1.92, P = 0.016; OR 95% CI = 2.29 - ∞, P = 0.003, respectively). Bosentan metabolism in vitro by CYP2C9*2 was significantly reduced compared to CYP2C9*1 and was comparable to CYP2C9*3. These results suggest that CYP2C9*2 is a potential genetic marker for prediction of bosentan-induced liver injury and warrants investigation for the optimization of bosentan treatment.
Keywords: bosentan, CYP2C9, pharmacogenetics
Introduction
Pulmonary arterial hypertension (PAH) is a progressive debilitating condition characterized by increased resistance in the pulmonary arterial circulation. Clinically, PAH is defined as a resting mean pulmonary artery pressure ≥ 25 mm Hg with a normal pulmonary venous pressure ≤ 15 mm Hg (1). PAH may have diverse etiology but with a common pathogenesis resulting in pulmonary vascular remodeling with proliferation and hypertrophy of pulmonary arterial smooth muscle cells, vasoconstriction, endothelial dysfunction and thrombosis in situ (1). Several vascular changes are associated with PAH, including down regulation of prostacyclin synthase (2) resulting in a relative deficiency of prostacyclin (PGI2) (3), overexpression of endothelin-1 (ET-1) (4) as well as down regulation of nitric oxide (NO) production with a decrease in intracellular cGMP expression (5).
Though prognosis for PAH still remains poor, major improvements have been made in disease management with the introduction of therapeutic agents targeting various affected pathways (6). Current treatments of PAH include parenteral prostanoids (epoprostenol, treprostinil and ilioprost), oral endothelin receptor antagonists (bosentan, sitaxentan, ambrisentan), and oral inhibitors of phosphodiesterase-5 (PDE5; sildenafil, tadalafil) (1). To date there is no compelling evidence for one class of drugs being superior over another, but it is generally accepted that prostanoids, which are difficult to administer and maintain, are used in patients with severe disease or in those who have failed to respond to other therapies. Therefore, oral endothelin receptor antagonists and PDE5 inhibitors often become a first-line treatment in mild to moderate PAH.
Bosentan is an oral dual endothelin receptor antagonist approved for treatment of PAH patients with WHO class II-IV symptoms. Bosentan is usually initiated at a dose of 62.5 mg twice daily and increased to 125 mg twice daily after 4 weeks of treatment. The pharmacokinetics of bosentan have been well described (7). After oral administration bosentan is absorbed with an absolute bioavailability of 50%, reaching peak plasma concentrations in about 3-4 hours. Bosentan is taken up from the blood into liver by OATP1B1 and OATP1B3 transporters, and metabolized by CYP3A4 and CYP2C9 to three major metabolites, one of which (Ro 48-5033) may contribute to up to 20% of total drug response. Bosentan is mainly cleared by hepatic metabolism with an elimination half-life of approximately 4-5 hours. Bosentan has been shown to significantly improve exercise ability and reduce the rate of clinical worsening in PAH patients (8, 9). Unfortunately, exposure-dependent liver toxicity manifesting in the form of increased activity of aminotransferases in 10-12% of treated patients (9, 10) limits bosentan use.
The possible mechanisms of bosentan-induced liver injury include a decrease in hepatic uptake of bosentan by OATP1B1/1B3 transporters and subsequent increase in systemic exposure (11, 12) as well as a decrease in activity of P450 enzymes and subsequent increase in both systemic and intrahepatic exposure (13, 14). There is also evidence that bosentan inhibits hepatic canalicular efflux transporters such as the bile salt export pump (BSEP) and multidrug resistance-associated protein 2 (MRP2), which are responsible for bile acid excretion, leading to intrahepatic bile acid accumulation and consequently liver damage (15-17). The latter mechanism might be independent or sequential to increased bosentan exposure. The majority of studies have focused on drug-drug interactions resulting in inhibition of liver uptake transporters or cytochrome P450 enzymes. To date no attempts have been made to study the influence of naturally occurring reduced function single nucleotide polymorphisms (SNPs) in genes encoding for the proteins involved in bosentan disposition (CYP2C9, CYP3A4, OATP1B1/1B3, BSEP, MRP2).
The current study was designed to examine associations between bosentan-induced liver injury and reduced function SNPs in genes encoding for CYP2C9, CYP3A4, OATP1B1, OATP1B3, BSEP, and MRP2. Putative genetic markers of bosentan-induced liver injury could be further investigated as biomarkers for stratification of PAH patients for appropriate treatment regimens.
Results
Cohort analysis
The original study cohort consisted of 92 ethnically diverse PAH patients. The majority of patients were Caucasians (61%), with smaller numbers of Asians (15%), Hispanics (15%) and African Americans (5%). There was no association of race with either change in ALT and AST levels or DILI (data not shown). Only the Caucasian group (n = 56) was large enough for genetic association analysis. Characteristics of the Caucasians included in the current analysis are shown in Table 1. The cohort consisted mostly of females (71%) with an average age of 51 ± 13 years and mean BMI of 30.8 ± 7.6 kg/m2. Less than 10% of the patients had a history of previous liver disease. The range of cumulative bosentan exposure was large, reflecting interpatient differences in the time of bosentan therapy. More than half of the patients (57.1%) were co-administered an OATP1B1 inhibitor (sildenafil in most cases). Co-medications that were known inhibitors of CYP2C9, CYP3A4 and MRP2 were used in 5.3%, 8.9% and 3.6% of patients, respectively.
Table 1.
Anthropometric, physiologic and clinical characteristics of Caucasian PAH study population.
| Number of patients | 56 |
| Age (yr) | 51 ± 131 |
| Sex M : F (%) | 29 : 71 |
| BMI (kg/m2) | 30.8 ± 7.6 |
| History of liver disease (%) | 8.9 |
| Bosentan exposure (g/m2) | 611 ± 677 |
| CYP2C9 inhibitors (%) | 5.3 |
| CYP3A4 inhibitors (%) | 8.9 |
| MRP2 inhibitors (%) | 3.6 |
| BSEP inhibitors (%) | 0 |
| OATP1B1 inhibitors (%) | 57.1 |
| OATP1B3 inhibitors (%) | 0 |
Data is presented as either mean ± S.D. or percent of total group.
Associations between candidate SNPs and increase in ALT and AST activity in Caucasian PAH patients treated with bosentan
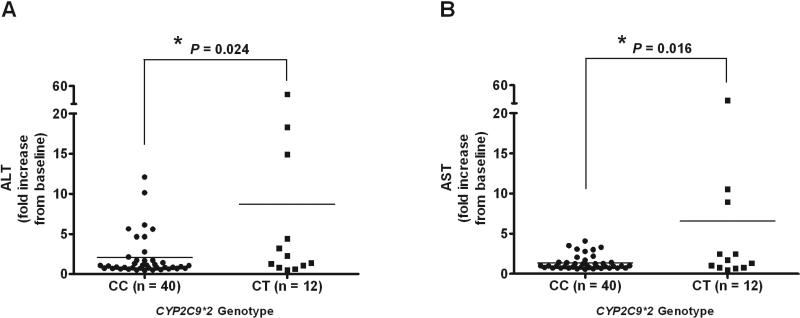
In a univariate linear regression analysis BMI showed a trend for negative correlation with ALT and AST increase (β = 0.96, P = 0.06; β = 0.98, P = 0.13, respectively). A single SNP in CYP2C9 (CYP2C9*2, rs1799853) showed a trend for association with ALT and AST increase in univariate analysis (Table 2) and was further tested in multivariate analysis. In a multivariate model adjusted for BMI, CYP2C9*2 remained significantly associated with the increase in ALT (β = 2.16, P = 0.024; Figure 1A) and AST (β = 1.92, P = 0.016; Figure 1B) activity.
Table 2.
Genetic association analysis for ALT and AST increase in 56 PAH patients treated with bosentan.
| ALT1 | AST1 | |||
|---|---|---|---|---|
| Fold Change2 | P | Fold Change2 | P | |
| Age | 1.01 | 0.53 | 1.01 | 0.28 |
| Sex | 1.15 | 0.67 | 1.12 | 0.68 |
| BMI | 0.96 | 0.06 | 0.98 | 0.13 |
| Hx liver disease | 0.87 | 0.78 | 1.07 | 0.87 |
| Bosentan exposure | 1.00 | 0.85 | 1.00 | 0.81 |
| Co-medications | 1.01 | 0.96 | 0.90 | 0.69 |
| ABCC2 | ||||
| rs717620 | 0.92 | 0.74 | 1.04 | 0.76 |
| rs2273697 | 0.92 | 0.78 | 0.81 | 0.34 |
| rs1885301 | 0.94 | 0.73 | 1.04 | 0.82 |
| rs7910642 | 1.49 | 0.33 | 1.08 | 0.82 |
| rs2804402 | 1.07 | 0.73 | 0.98 | 0.82 |
| ABCB11 | ||||
| rs2287622 | 1.19 | 0.39 | 1.01 | 0.92 |
| SLCO1B1 | ||||
| rs11045819 | 1.31 | 0.39 | 1.31 | 0.27 |
| rs2306283 | 1.16 | 0.53 | 1.01 | 0.99 |
| rs4149056 | 1.09 | 0.76 | 0.84 | 0.48 |
| SLCO1B3 | ||||
| rs7311358 | 1.28 | 0.38 | 1.15 | 0.52 |
| CYP2C9 | ||||
| *2/rs1799853 | 2.18 | 0.026 | 1.94 | 0.015 |
| *3/rs1057910 | 0.61 | 0.21 | 0.71 | 0.28 |
Univariate linear regression was performed and variants with a P ≤ 0.2 were carried into the multivariate regression analysis adjusted for BMI. Multivariate regression results are presented in the text.
Fold change in ALT and AST was not normally distributed and was natural log transformed for analysis. Coefficients are absolute fold change values. The regression coefficients and the standard errors from the transformed data are presented in Supplemental Table 3.
Figure 1.
Association between CYP2C9*2 and changes in (A) ALT and (B) AST levels in Caucasian PAH patients treated with bosentan. Data are presented as mean fold increase from baseline and were analyzed in a multivariate model corrected for body mass index (BMI); the P-value is indicated.
Associations between candidate SNPs and DILI in Caucasian PAH patients treated with bosentan
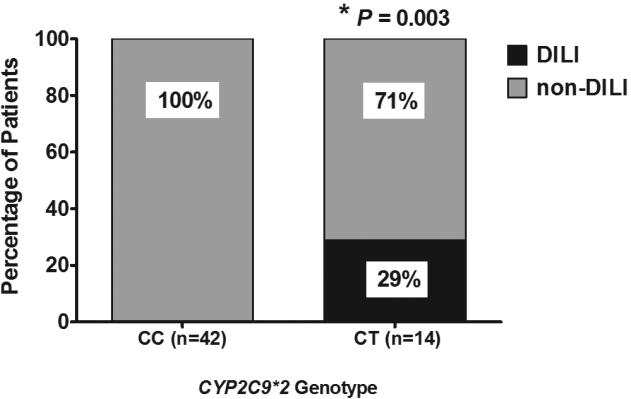
In univariate logistic regression (continuous variables) or by Fisher's exact test (dichotomous variables) none of the clinical covariates showed any trend for association with occurrence of DILI (Table 3) while a SNP in SLCO1B1 (rs11045819) showed a trend for association with bosentan-induced liver injury (Table 3). CYP2C9*2 was the only polymorphism significantly associated with occurrence of DILI (OR 95% CI 2.29 - ∞, P = 0.003) (Table 3 and Figure 2). Multivariate analysis was not performed due to the small size of the cohort and DILI group.
Table 3.
Genetic association analysis for drug-induced liver injury (DILI) in 56 PAH patients treated with bosentan.
| Odds ratio1 | OR 95% CI lower limit | OR 95% CI upper limit | P | |
|---|---|---|---|---|
| Age | 0.94 | 0.86 | 1.04 | 0.24 |
| Sex | 1.21 | 0.09 | 68.2 | 1.00 |
| BMI | 0.96 | 0.83 | 1.11 | 0.62 |
| Hx liver disease | 0.00 | 0.00 | 18.2 | 1.00 |
| Bosentan exposure | 0.99 | 0.99 | 1.00 | 0.39 |
| Co-medications | 0.58 | 0.04 | 8.61 | 0.62 |
| ABCC2 | ||||
| rs717620 | 1.59 | 0.11 | 23.5 | 0.64 |
| rs2273697 | 0.46 | 0.01 | 6.18 | 0.64 |
| rs1885301 | 1.86 | 0.14 | 103 | 1.00 |
| rs7910642 | 0.00 | 0.00 | 8.41 | 1.00 |
| rs2804402 | 0.28 | 0.02 | 4.21 | 0.23 |
| ABCB11 | ||||
| rs2287622 | 0.45 | 0.03 | 6.74 | 0.59 |
| SLCO1B1 | ||||
| rs11045819 | 4.60 | 0.30 | 71.5 | 0.17 |
| rs2306283 | 2.01 | 0.15 | 112 | 1.00 |
| rs4149056 | 0.54 | 0.01 | 7.26 | 1.00 |
| SLCO1B3 | ||||
| rs7311358 | 0.83 | 0.02 | 11.2 | 1.00 |
| CYP2C9 | ||||
| *2/ rs1799853 | ∞ | 2.29 | ∞ | 0.003 |
| *3/ rs1057910 | 0.00 | 0.00 | 8.41 | 1.00 |
Odds ratio was calculated by logistic regression for continuous variables and by a Fisher exact test for dichotomous variables.
Figure 2.
Association between CYP2C9*2 and drug induced liver injury (DILI) in Caucasian PAH patients treated with bosentan. Data are presented as percent of DILI and non-DILI patients in each allele group and were analyzed using Fisher's exact test; the P-value is indicated.
In vitro bosentan metabolite identification and functional characterization of CYP2C9*1, CYP2C9*2 and CYP2C9*3 enzymes in bosentan metabolism
Bosentan metabolic pathways were confirmed as described previously (7). Bosentan is metabolized to Ro 48-5033 through hydroxylation and to Ro 47-8634 through O-demethylation. Ro 47-8634 is further hydroxylated to Ro 64-1056 and Ro 48-5033 is subsequently metabolized through O-demethylation to form Ro 64-1056. In preliminary experiments it was confirmed that bosentan is metabolized 17%, 61% and 11% in 60 min by human liver microsomes, rCYP3A4 and rCYP2C9, respectively. All above mentioned metabolites were formed in human liver microsomes and CYP3A4 reactions, while Ro 48-5033 was the only metabolite formed in CYP2C9 reactions (Supplemental Table 1). Other tested rCYP enzymes (CYP1A2, CYP2B6, CYP2C8, CYP2C19, and CYP2D6) were not implicated in bosentan metabolism (Supplemental Table 1).
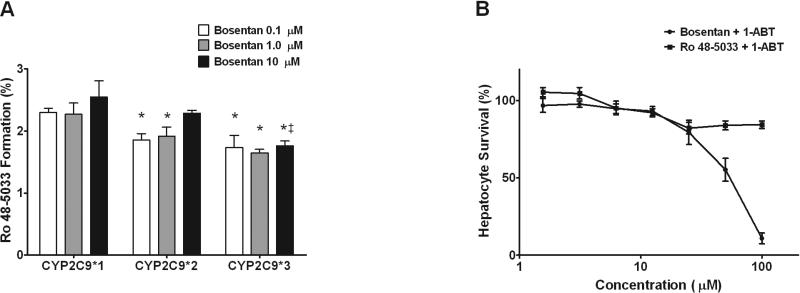
Functional activity of CYP2C9*1 (reference), CYP2C9*2 and CYP2C9*3 enzymes in bosentan metabolism to Ro 48-5033 was evaluated using recombinant enzymes. Both CYP2C9*2 and CYP2C9*3 variants had moderately reduced Ro 48-5033 formation compared to CYP2C9*1 at all three bosentan concentrations. In particular, ~2.3% of bosentan was converted to Ro 48-5033 by CYP2C9*2, ~1.8% by CYP2C9*3 and ~2.8% by CYP2C9*1 (Figure 3A).
Figure 3.
Bosentan metabolism by CYP2C9 variant proteins and CYP-dependent hepatotoxicity. (A) rCYP2C9*1 (reference), rCYP2C9*2 and rCYP2C9*3 were incubated with 0.1, 1 and 10 μM bosentan for 0 and 60 min and formation of Ro 48-5033 was quantified by LC/MS/MS. Data are presented as percent of bosentan converted to Ro 48-5033 ± S.D. (N=3). *P < 0.05 for difference between CYP2C9 reference and variants; ‡P < 0.05 for difference between CYP2C9*2 and CYP2C9*3. (B) Hepatocytes pre-incubated with 1-aminobenzotriazle were treated with increasing concentrations (0 – 100 μM) of bosentan or its metabolite Ro 48-5033 for 48 hours. Cell viability was assessed by measuring cellular ATP and GSH levels. Data are presented as percent of vehicle control. IC50 values were calculated using GraphPad Prism software.
Assessment of hepatotoxicity of bosentan and its major metabolite Ro 48-5033 was performed in human cultured hepatocytes in the presence and absence of 1-aminobenzotriazole (1-ABT), a pan-CYP450 inhibitor. In the absence of 1-ABT, bosentan and its major metabolite were quickly metabolized and no hepatocyte toxicity was observed (data not shown). In the presence of 1-ABT, bosentan exhibited hepatotoxicity with an estimated IC50 of ~ 54 μM, while its major metabolite had no significant effect on hepatocyte viability at the tested concentrations (Figure 3B).
Bosentan uptake by OATP1B1 reference and Pro155Thr (SLCO1B1 rs11045819) stably transfected HEK293 cells
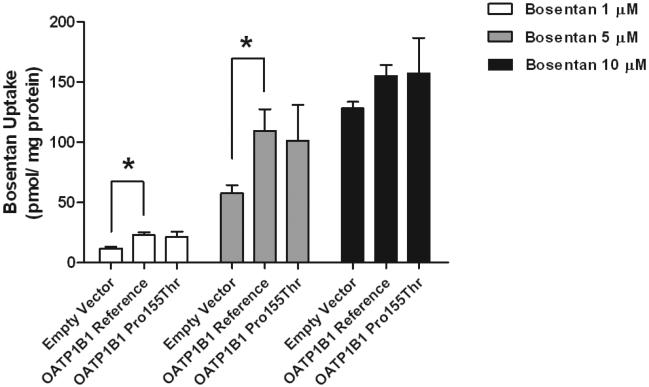
Bosentan uptake was significantly greater in HEK293 cells stably expressing the OATP1B1 reference transporter compared to cells expressing empty vector (Figure 4). This difference in uptake gradually disappeared with increasing bosentan concentrations from 10 μM to 100 μM (data shown in part in Figure 4). Bosentan transport by the OATP1B1 Pro155Thr variant was similar to that of the reference transporter (Figure 4).
Figure 4.
Bosentan transport by OATP1B1 reference and Pro155Thr variant transporters. Empty vector, OATP1B1 reference and OATP1B1 Pro155Thr expressing cells were incubated with 1, 5 and 10 μM bosentan for 5 min. Bosentan uptake was measured and normalized to protein. Data are presented as mean ± S.D. (N=3); P < 0.05.
Discussion
Bosentan significantly improves manageability and prognosis in PAH patients (1, 8, 9), but unfortunately, exposure dependent liver toxicity manifesting in the form of elevated aminotransferases in 10-12% of treated patients (9, 10) limits its use. The mechanisms of bosentan-induced liver injury have been intensively studied over the past decade. Bosentan-induced liver injury has been attributed at least in part to the accumulation of bile acids in hepatocytes as a result of inhibition of bile salt export pump (BSEP) by bosentan (16-18). BSEP is an ATP–dependent transmembrane transporter located on the canalicular membrane of hepatocytes. It pumps bile acids from hepatocytes into the bile, therefore inhibition of its function results in cholestasis (19). However, in in vitro studies bosentan produced inhibitory effects on BSEP at much higher (50 - 100 μM) (16-18) than clinically observed plasma concentrations (Cmax ~2 - 4 μM) (7). This suggests possible effects of additional pharmacokinetic factors which may lead to increased bosentan exposure. Another suggested mechanism of bosentan-induced liver injury involves alterations in multidrug resistance protein 2 (MRP2)–mediated bile salt formation. MRP2 is also an ATP–dependent transmembrane transporter located on the canalicular membrane of hepatocytes and is largely involved in the efflux of bilirubin, glutathione and glucuronide conjugated compounds into bile. It has been shown that bosentan stimulates MRP2 function (15, 17) causing an increase in bilirubin and glutathione secretion, which results in intermittent uncoupling of lipids from bile salt secretion (15). Phospholipids normally reduce the cytotoxicity of bile salts, thus uncoupling of lipids from bile salt secretion increases bile salt toxicity and may contribute to liver damage.
Based on the data implicating MRP2 and BSEP in bosentan disposition we hypothesized that decreased function variants in BSEP might increase a patient's risk to bosentan-induced liver damage, while reduced function MRP2 variants should afford protection against this toxicity. The common ABCB11 polymorphism rs2287622 encoding the V444A variant of BSEP was previously associated with reduced hepatic BSEP expression (20) and drug-induced cholestasis, but not drug-induced hepatocellular injury (21). ABCB11 variants may also have a role in intrahepatic cholestasis of pregnancy, although this is controversial (22, 23). In the current study, an association of rs2287622 with the development of bosentan-induced liver toxicity was not observed, possibly due to the small sample size. However, considering that bosentan-induced liver injury is more hepatocellular in nature (24), perhaps it is not surprising that ABCB11 variants did not contribute to risk of this toxicity.
The ABCC2 promoter variant rs717620 has been previously associated with diclofenac-induced hepatotoxicity, which is similar in presentation to bosentan liver injury (25). ABCC2 haplotypes containing rs717620 were also associated with hepatocellular injury from herbal compounds (26). Functional data on ABCC2 promoter variants suggests only modest effects on ABCC2 expression (27) and there is no consistent report of altered transport by the V417I MRP2 variant (rs2273697) (28, 29). In the current study ABCC2 variants were not implicated in bosentan-induced liver injury, suggesting involvement of other factors.
Bosentan-induced liver injury has also been attributed to mechanisms leading to increased exposure. Inhibition of proteins involved in metabolism of bosentan (CYP3A4 and CYP2C9) and/or its uptake into the liver (OATP1B1, OATP1B3) results in increased bosentan systemic exposure and a higher frequency of bosentan–associated adverse effects. Inhibitors of CYP3A4 (ketoconazole, itraconazole, ritonavir) and CYP2C9 (fluconazole, amiodarone) as well as inhibitors of OATP1B1/1B3 (cyclosporine A, sildenafil, ritonavir) increase bosentan Cmax and AUC in healthy volunteers (7, 14, 30). CYP3A4 accounts for 60% and CYP2C9 for 40% of bosentan metabolism in hepatic microsomes (7) and reduction in their functionality might be critical in the development of liver damage. In vitro assays indicated that hepatotoxicity is attributed to bosentan itself (IC50 ~ 54 μM) and not its metabolites (Figure 3B). The organic anion uptake transporters, OATP1B1/1B3, are expressed on the basolateral membrane of hepatocytes, and play a major role in transporting bosentan into hepatocytes (11, 18). Loss of their functionality increases systemic bosentan exposure and may influence non-hepatic adverse effects as well. Based on the data strongly supporting involvement of CYP3A4, CYP2C9, OATP1B1 and OATP1B3 in bosentan pharmacokinetics it was hypothesized that reduced function SNPs in these genes will confer higher risk of developing bosentan-induced liver injury.
CYP2C9*2 is a common polymorphism associated with a poor metabolizer phenotype. This allele encodes the R144C variant of CYP2C9 which has ~50% reduced function (31). CYP2C9*2 has been implicated in reduced clearance of warfarin (32), phenytoin (33), celecoxib (34) and losartan (35) and CYP2C9*2 genotyping is commonly used in determining initial warfarin dose (32). The current data suggests that it is the single most important factor influencing the risk of bosentan-induced hepatic injury. Additional studies will be required to validate its use as a biomarker for identifying patients at increased risk of liver toxicity from bosentan treatment. Considering the allele distribution in major populations, CYP2C9*2 will be most relevant in the Caucasian and Hispanic populations, where the minor allele frequency for this variant is 10-15%, compared to African American and Asian populations where it is rarely found (4% and 0% minor allele frequency, respectively) (36). A preliminary replication in the Hispanic population shows a trend toward association of this SNP with elevated ALT/AST levels (ALT: β = 4.01, P = 0.084; AST: β = 2.48, P = 0.154).
CYP2C9*3 encodes the I359L CYP2C9 variant, which is characterized by almost complete loss of function (31, 36). This polymorphism is less common, having a minor allele frequency of 7%, 2% and 3% in Caucasian, African American and Asian populations, respectively (36). It has also been implicated in reduced clearance of warfarin (32), phenytoin (33), celecoxib (34) and losartan (35). In this study, CYP2C9*3 did not show an association with bosentan-induced ALT or AST increase or bosentan-induced liver injury, although this might not be evident due to sample size limitations. Post hoc power calculations for each SNP confirm the limited power of detecting an association with CYP2C9*3 and other SNPs with relatively low minor allele frequencies (Supplemental Table 3). It is plausible that the lack of an association of bosentan liver injury with CYP2C9*3 could also be due to differential metabolism of bosentan by the variants, but this is not supported by the similar reductions in bosentan metabolism with CYP2C9*2 and CYP2C9*3 recombinant proteins. We also considered the possibility that CYP2C9*2 is not causative but is tagging a regulatory SNP in the genomic region. CYP2C9 promoter polymorphisms become of greater importance when induction of CYP2C9 plays a substantial role in a drug's pharmacokinetics (37). Bosentan induces CYP2C9 at therapeutic concentrations (7) through activation of PXR (38). Interestingly, CYP2C9*2 is in linkage disequilibrium (R2 = 0.57) with rs2185570 located downstream of CYP2C9. This SNP is located in a transcription factor binding region and DNaseI hypersensitivity cluster as confirmed by ChiP-Seq and DNase-Seq data from ENCODE and has been associated in a genome wide association study with dehydroepiandrosterone sulfate levels. There is no evidence that CYP2C9 is directly involved in dehydroepiandrosterone or dehydroepiandrosterone sulfate metabolism, however dehydroepiandrosterone is a known CYP2C9 inducer (39). CYP2C9*2 is also in linkage disequilibrium (R2 = 0.462) with an intronic SNP (rs4086116), which has been associated with warfarin (40) and acenocumarol dosing (41). Further studies will be required to determine if regulatory SNPs in the CYP2C9 genomic region might be contributing to bosentan-induced liver injury.
Several common polymorphisms in SLCO1B1 have been associated with reduced transporter function, altered pharmacokinetics and increased risk of substrate-induced toxicity (42). The SLCO1B1 polymorphism encoding for the OATP1B1 Pro155Thr variant (rs11045819) has been associated with gallstone formation (43) and enhanced lipid-lowering activity of fluvastatin in elderly hypercholesterolemic patients (44). In the present study, SLCO1B1 polymorphisms were not significantly associated with ALT or AST increase, although rs11045819 showed a trend toward an association with DILI (Table 3). Functional studies confirmed that bosentan is transported by OATP1B1 but there was no evidence for reduced function of this variant (Figure 4). At higher bosentan concentrations OATP1B1 transport was not efficient, suggesting that there might be another transporter(s) involved. Recent studies suggest that OATP1B3 has lower affinity but higher capacity for bosentan transport compared to OATP1B1 (11). Therefore, OATP1B3 might be responsible for this dose-dependent phenomenon in our cell model. Functional variants also exist for SLCO1B3 (45, 46) but the current analysis found no evidence that these are important for risk of bosentan-induced hepatic injury. The lack of associations with SLCO1B1 and SLCO1B3 variants is perhaps not surprising since reduced function variants would increase systemic exposure and more likely manifest as non-hepatic toxicities.
In summary, CYP2C9*2 was identified as a potential risk factor for bosentan-induced liver injury. Additional studies are needed to elucidate the molecular basis of this association since effects on bosentan metabolism are modest. The implications of this finding will require validation in additional patients. If validated, this pharmacogenetic marker might be useful in the identification of patients at increased risk of this serious toxicity and lead to alterations in treatment that would avoid this unnecessary risk.
Methods
Study population
Human investigations were approved by the local Committee on Human Research and all patients gave informed consent to participate. A total of 92 ethnically diverse PAH patients at the University of California San Francisco (UCSF) Medical Center Pulmonary Hypertension Clinic were recruited into the study during 2003-2008. Medical, social and laboratory data including age, sex, anthropometric parameters, smoking status, and history of liver disease were collected at the time of enrollment. A blood sample was collected for genotyping. Treatment with 125-250 mg bosentan daily was initiated and continued for variable periods of time depending on the time of enrollment and/or withdrawal from the study due to development of bosentan-induced liver toxicity. Co-medications included PDE5 inhibitors, prostanoids, calcium channel blockers, anticoagulants, diuretics, statins, antifungals and antimicrobial agents. Activities of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured before initiating bosentan treatment and then monthly to monitor liver function.
Clinical study design
This pharmacogenetic study was designed as a prospective cohort study. First, clinical covariates were defined, which included age, sex, body mass index (BMI), history of liver disease, bosentan exposure (cumulative dose in mg/m2 of body surface area) and co-medications which are known inhibitors of CYP3A4 (ritonavir, verapamil), CYP2C9 (fluconazole, metronidazole), OATP1B1 (sildenafil, statins), OATP1B3 (statins, valsartan), MRP2 (ritonavir, statins) and BSEP (rifampicin). Second, genetic covariates were selected. A literature search for reduced function SNPs was focused on six genes (CYP3A4, CYP2C9, SLCO1B1, SLCO1B3, ABCB11 and ABCC2) encoding proteins involved in bosentan pharmacokinetics. A total of 15 SNPs were selected (Supplemental Table 2).
Two types of outcomes for testing genetic associations were established. Outcome I was defined as the fold increase in ALT and AST activities from time of enrollment to the end of the study or to the time of bosentan discontinuation due to liver toxicity. Outcome I was expressed as a continuous variable, logarithmically transformed for analysis. Outcome II was classification of drug-induced liver injury (DILI), defined as a 3-fold increase of either ALT or AST above upper limit of norm (ULN) at any point in the study (n = 4). Outcome II was expressed as a binary variable. In addition to classifying drug-induced liver injury based on liver enzyme elevations, the data was evaluated with the Roussel Uclaf Causality Assessment Method (RUCAM) (47, 48). RUCAM was performed by two independent individuals and all Caucasian cases previously classified as DILI cases based on liver enzyme elevations received RUCAM scores of “possible (n = 3)” and “probable (n = 1)”, increasing confidence in the assignment of cases. It should be noted that data on re-challenges were largely absent. One clinical team cared for these pulmonary hypertension patients; initially two patients were re-challenged and both redeveloped an elevation in liver function test. Based on knowledge that drug-induced liver injury was a recognized complication of the drug, re-challenges were discontinued with the arrival of the clinical availability of a newer agent with reduced risk of liver injury.
Genotyping
DNA samples from 92 PAH patients were extracted and genotyped on the DMET chip (Affimetrix, Inc., DMET Early Access Program). The following SNPs were extracted for analysis: CYP2C9 (CYP2C9*2/rs1799853, CYP2C9*3/rs1057910), SLCO1B1 (rs11045819, rs2306283, rs4149056), SLCO1B3 (rs414917, rs7311358), ABCB11 (rs2287622), and ABCC2 (rs717620, rs2273697, rs8187692, rs17222723). Additionally, ABCC2 SNPs (rs1885301, rs7910642, rs2804402) not present on the chip were genotyped by direct sequencing at the UCSF genetics core facility.
Genetic association analysis
To avoid potential spurious associations as a result of population substructure and admixture, analysis was restricted to 56 Caucasian PAH patients. Selected SNPs have been checked for deviation from Hardy-Weinberg Equilibrium and filtered for minor allele frequency (MAF) greater than 5% and linkage disequilibrium (LD) lower than 0.8. After filtering, twelve SNPs in five genes: CYP2C9 (rs1799853, rs1057910), SLCO1B1 (rs11045819, rs2306283, rs4149056), SLCO1B3 (rs7311358), ABCB11 (rs2287622) and ABCC2 (rs717620, rs2273697, rs1885301, rs2804402, rs7910642) were analyzed for associations with changes in AST and ALT activity (Outcome I) and occurrence of DILI (Outcome II) using linear and logistic regression analyses or Fisher's exact test. An additive inheritance model was applied in the linear regression analysis and a dominant inheritance model was used for the Fisher exact test. Linear regression analysis was performed in two steps. First, each clinical covariate and each SNP were tested for associations in univariate analyses and those with P < 0.2 were carried into multivariate analyses. Multivariate models were adjusted for significant clinical covariates with inclusion of SNPs from step one. Associations with P < 0.05 in multivariate analyses were considered significant. A multivariate analysis for genetic associations with DILI was not performed due to the small size of the cohort and DILI group. Post hoc power calculations for the genetic associations with ALT and AST were conducted for effect sizes equal to the observed effect estimates using the power.t.test function in R v.3.0.1. Post hoc power for Fisher exact tests of the association of genetic carrier status with DILI were conducted using the power.fisher.test function from the statmod library in R v.3.0.1 with 10000 simulated datasets.
Hepatocyte Toxicity Assay
The assay was performed as described previously (49). Briefly, human hepatocytes were plated at 0.3 × 105 cells/well in collagen I-coated plates. Four hours after plating, cells were pre-incubated for 16 hr with 0.5 mM 1-aminobenzotriazole (1-ABT) (Sigma-Aldrich, St Louis, MO). Subsequently, medium was replaced with fresh medium containing bosentan, its metabolite Ro 48-5033 or chlorpromazine as a positive control at varying concentrations (0 -100 μM), and incubated for an additional 48 hr. DMSO was used as the negative control. At the end of incubation, cell viability was assessed by measuring ATP levels using CellTiter-Glo™ and cellular GSH levels using GSH-Glo following the manufacturer's protocols (Promega; Madison, WI). Data are presented as mean percent of control ± SD of triplicate experiments. IC50 values were calculated using Graph Pad Prism (version 5).
Enzyme kinetic studies with human liver microsomes (HLM) and recombinant CYP2C9*1, CYP2C9*2 and CYP2C9*3 enzymes
Bosentan was purchased from Waterstone Technology (Carmel, IN, USA). Human liver microsomes and rCYP450 reference enzymes were purchased from BD Biosciences (San Jose, CA, USA). The CYP2C9*1, CYP2C9*2 and CYP2C9*3 enzymes for functional comparison experiments were purchased from Cypex Limited (Dundee, Scotland, UK). Enzyme activities were measured by parent drug disappearance and metabolite formation in incubations with 0.1 μM, 1 μM and 10 μM bosentan. All incubations were performed in 100 mM phosphate buffer (pH 7.4). The final protein concentration was 0.5 mg/mL in human liver microsome incubations, and the enzyme concentration was 40 pmol/mL in incubations with recombinant CYP2C9 enzymes. All incubations were performed at 37°C for 60 min. Reactions were initiated by adding NADPH (10 mM final concentration) and were terminated by adding a 3X volume of acetonitrile. Samples were vortexed for 5 min prior to centrifugation at 2690g for 10 min. The supernatant was transferred to new tubes and dried down at room temperature with a Labconco Centrivap DNA Concentrator. It was then reconstituted with a water:acetonitrile mixture (2:1), followed by vortexing for 5 min and centrifugation at 11,000g for 10 min. The supernatant was analyzed by LC-MS/MS. Negative controls consisting of incubation matrix without substrate were incubated under identical conditions. Data is presented as mean ± SD of triplicate experiments. Statistical analysis was performed using two-way ANOVA; P < 0.05 was considered significant.
Functional assay of bosentan uptake by SLCO1B1 reference and variant proteins
Stable cell lines expressing the OATP1B1 reference and variant transporters have been described elsewhere (50). Uptake assays were performed in triplicate on poly-D-lysine-coated 48-well plates (BD Biosciences, CA). Empty vector, OATP1B1 reference and the Pro155Thr variant (rs11045819, C463A) expressing cells were seeded at 1.5 × 105 cells/well in complete DMEM supplemented with 10% FBS. On the following day medium was supplemented with 5 mM sodium butyrate and cells were incubated for another 24 hr. Twenty four hours later the cells were washed with warm Krebs-Henseleit buffer (KHB) and incubated with 1, 5, 10, 25, 50 or 100 μM bosentan in KHB for 5 min at 37°C. After accumulation, cells were washed twice with ice-cold KHB and were lysed with a 10% sodium dodecyl sulfate and 1 N NaOH solution. Bosentan was extracted with two volumes of acetonitrile. An aliquot of the cell lysate was used to determine protein concentration with a BCA TM protein assay kit (Pierce Biotechnology, IL) and bosentan levels were normalized to total protein concentrations. Data are presented as mean ± SD of triplicate experiments. Statistical analysis was performed using two-way ANOVA; P < 0.05 was considered significant.
Chromatographic and mass spectrometric conditions for measuring bosentan and its metabolites
LC-MS analysis was conducted using a Thermo LTQ XL coupled to an Ultra High Pressure Liquid Chromatograph (UPLC, Accela Pump), an Accela autosampler, and an Accela PDA (San Jose, CA, USA). The mass spectrometric conditions were set as follows: positive mode ESI, capillary voltage at 16 V, source voltage at 4.5 kV, tube lens voltage at 95 V, capillary temperature at 350°C, sheath gas flow at 57 units, aux gas flow at 24 units, sweep gas flow at 5 units using full scan, MSn, and targeted MSn scan modes. The HPLC column used for both systems was a Thermo Hypersil Gold C18 (100 × 2.1 mm, 1.9 μm; Thermo Scientific, Pittsburgh, PA, USA). The solvent system consisted of solvent A (0.1% formic acid (FA) in water) and solvent B (0.1% FA in acetonitrile). Solvent B was delivered initially at 5%, held for 1 min and increased to 95% via a 25 min gradient, then decreased back to 5% at 26 min and equilibrated for 4 min at a flow rate of 400 μL/min and a total run time of 30 min. The injection volume was 20 μL.
Supplementary Material
Study highlights.
What is the current knowledge on the topic? Bosentan (Tracleer®) is a dual endothelin receptor antagonist prescribed as first-line treatment in pulmonary arterial hypertension (PAH) patients. Drug induced liver injury (DILI; 10-12% incidence) is the most serious adverse effect of bosentan.
What question this study addressed? This study was designed to identify genetic markers associated with bosentan-induced liver injury in PAH patients. CYP2C9*2 was strongly associated with ALT and AST increase and with occurrence of drug-induced liver injury (β = 2.16, P = 0.024; β = 1.92, P = 0.016; and OR 95% CI 2.29 - ∞, P = 0.003, respectively).
What this study adds to our knowledge? Bosentan metabolism in vitro by CYP2C9*2 was moderately reduced compared to CYP2C9*1 and was comparable to CYP2C9*3. CYP2C9*2 is a potential genetic marker for prediction of bosentan-induced liver injury.
How this might change clinical pharmacology and therapeutics? This genetic information may be of use in guiding endothelin receptor antagonist therapy in pulmonary hypertension and prevention of bosentan-induced liver injury.
Acknowledgments
This work was funded by NIH Grants GM61390 and GM32165, the Foundation for Cardiac Research, and the Affymetrix, Inc., Early Access Program.
Footnotes
Conflict of interests
There are no conflicts of interest to report.
Author Contributions
Svetlana Markova and Deanna Kroetz wrote the manuscript. Teresa DeMarco, Jason Halladay, Allan E. Rettie, Cyrus Khojasteh, Dana McGlothlin, Alan Wu, and Janice Schwartz designed the research. Svetlana Markova, Erin Kobashigawa, Hoa Le, Jasleen Sodhi, and Chenghong Zhang performed the research. Svetlana Markova, Nasrine Bendjilali , Joel Mefford, Hoa Le, Jasleen Sodhi, and Chenghong Zhang, Jason Halladay, Wen-Chi Hsueh, John Witte, and Deanna Kroetz analyzed the data.
References
- 1.McLaughlin VV, Archer SL, Badesch DB, Barst RJ, Farber HW, Lindner JR, et al. ACCF/AHA 2009 expert consensus document on pulmonary hypertension a report of the American College of Cardiology Foundation Task Force on Expert Consensus Documents and the American Heart Association developed in collaboration with the American College of Chest Physicians; American Thoracic Society, Inc.; and the Pulmonary Hypertension Association. J Am Coll Cardiol. 2009;53:1573–619. doi: 10.1016/j.jacc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Tuder RM, Cool CD, Geraci MW, Wang J, Abman SH, Wright L, et al. Prostacyclin synthase expression is decreased in lungs from patients with severe pulmonary hypertension. Am J Respir Crit Care Med. 1999;159:1925–32. doi: 10.1164/ajrccm.159.6.9804054. [DOI] [PubMed] [Google Scholar]
- 3.Christman BW, McPherson CD, Newman JH, King GA, Bernard GR, Groves BM, et al. An imbalance between the excretion of thromboxane and prostacyclin metabolites in pulmonary hypertension. N Engl J Med. 1992;327:70–5. doi: 10.1056/NEJM199207093270202. [DOI] [PubMed] [Google Scholar]
- 4.Stewart DJ, Levy RD, Cernacek P, Langleben D. Increased plasma endothelin-1 in pulmonary hypertension: marker or mediator of disease? Ann Intern Med. 1991;114:464–9. doi: 10.7326/0003-4819-114-6-464. [DOI] [PubMed] [Google Scholar]
- 5.Giaid A, Saleh D. Reduced expression of endothelial nitric oxide synthase in the lungs of patients with pulmonary hypertension. N Engl J Med. 1995;333:214–21. doi: 10.1056/NEJM199507273330403. [DOI] [PubMed] [Google Scholar]
- 6.Thenappan T, Shah SJ, Rich S, Gomberg-Maitland M. A USA-based registry for pulmonary arterial hypertension: 1982-2006. Eur Respir J. 2007;30:1103–10. doi: 10.1183/09031936.00042107. [DOI] [PubMed] [Google Scholar]
- 7.Dingemanse J, van Giersbergen PL. Clinical pharmacology of bosentan, a dual endothelin receptor antagonist. Clin Pharmacokinet. 2004;43:1089–115. doi: 10.2165/00003088-200443150-00003. [DOI] [PubMed] [Google Scholar]
- 8.Channick RN, Simonneau G, Sitbon O, Robbins IM, Frost A, Tapson VF, et al. Effects of the dual endothelin-receptor antagonist bosentan in patients with pulmonary hypertension: a randomised placebo-controlled study. Lancet. 2001;358:1119–23. doi: 10.1016/S0140-6736(01)06250-X. [DOI] [PubMed] [Google Scholar]
- 9.Rubin LJ, Badesch DB, Barst RJ, Galie N, Black CM, Keogh A, et al. Bosentan therapy for pulmonary arterial hypertension. N Engl J Med. 2002;346:896–903. doi: 10.1056/NEJMoa012212. [DOI] [PubMed] [Google Scholar]
- 10.Denton CP, Pope JE, Peter HH, Gabrielli A, Boonstra A, van den Hoogen FH, et al. Long-term effects of bosentan on quality of life, survival, safety and tolerability in pulmonary arterial hypertension related to connective tissue diseases. Ann Rheum Dis. 2008;67:1222–8. doi: 10.1136/ard.2007.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treiber A, Schneiter R, Hausler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug Metab Dispos. 2007;35:1400–7. doi: 10.1124/dmd.106.013615. [DOI] [PubMed] [Google Scholar]
- 12.Burgess G, Hoogkamer H, Collings L, Dingemanse J. Mutual pharmacokinetic interactions between steady-state bosentan and sildenafil. Eur J Clin Pharmacol. 2008;64:43–50. doi: 10.1007/s00228-007-0408-z. [DOI] [PubMed] [Google Scholar]
- 13.van Giersbergen PL, Halabi A, Dingemanse J. Single- and multiple-dose pharmacokinetics of bosentan and its interaction with ketoconazole. Br J Clin Pharmacol. 2002;53:589–95. doi: 10.1046/j.1365-2125.2002.01608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingemanse J, van Giersbergen PL, Patat A, Nilsson PN. Mutual pharmacokinetic interactions between bosentan and lopinavir/ritonavir in healthy participants. Antivir Ther. 15:157–63. doi: 10.3851/IMP1506. [DOI] [PubMed] [Google Scholar]
- 15.Fouassier L, Kinnman N, Lefevre G, Lasnier E, Rey C, Poupon R, et al. Contribution of mrp2 in alterations of canalicular bile formation by the endothelin antagonist bosentan. J Hepatol. 2002;37:184–91. doi: 10.1016/s0168-8278(02)00107-1. [DOI] [PubMed] [Google Scholar]
- 16.Fattinger K, Funk C, Pantze M, Weber C, Reichen J, Stieger B, et al. The endothelin antagonist bosentan inhibits the canalicular bile salt export pump: a potential mechanism for hepatic adverse reactions. Clin Pharmacol Ther. 2001;69:223–31. doi: 10.1067/mcp.2001.114667. [DOI] [PubMed] [Google Scholar]
- 17.Mano Y, Usui T, Kamimura H. Effects of bosentan, an endothelin receptor antagonist, on bile salt export pump and multidrug resistance-associated protein 2. Biopharm Drug Dispos. 2007;28:13–8. doi: 10.1002/bdd.527. [DOI] [PubMed] [Google Scholar]
- 18.Hartman JC, Brouwer K, Mandagere A, Melvin L, Gorczynski R. Evaluation of the endothelin receptor antagonists ambrisentan, darusentan, bosentan, and sitaxsentan as substrates and inhibitors of hepatobiliary transporters in sandwich-cultured human hepatocytes. Can J Physiol Pharmacol. 88:682–91. doi: 10.1139/Y10-060. [DOI] [PubMed] [Google Scholar]
- 19.Stieger B, Meier Y, Meier PJ. The bile salt export pump. Pflugers Arch. 2007;453:611–20. doi: 10.1007/s00424-006-0152-8. [DOI] [PubMed] [Google Scholar]
- 20.Ho RH, Leake BF, Kilkenny DM, Meyer Zu Schwabedissen HE, Glaeser H, Kroetz DL, et al. Polymorphic variants in the human bile salt export pump (BSEP; ABCB11): functional characterization and interindividual variability. Pharmacogenet Genomics. 20:45–57. doi: 10.1097/FPC.0b013e3283349eb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang C, Meier Y, Stieger B, Beuers U, Lang T, Kerb R, et al. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- 22.Pauli-Magnus C, Lang T, Meier Y, Zodan-Marin T, Jung D, Breymann C, et al. Sequence analysis of bile salt export pump (ABCB11) and multidrug resistance p-glycoprotein 3 (ABCB4, MDR3) in patients with intrahepatic cholestasis of pregnancy. Pharmacogenetics. 2004;14:91–102. doi: 10.1097/00008571-200402000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Dixon PH, van Mil SW, Chambers J, Strautnieks S, Thompson RJ, Lammert F, et al. Contribution of variant alleles of ABCB11 to susceptibility to intrahepatic cholestasis of pregnancy. Gut. 2009;58:537–44. doi: 10.1136/gut.2008.159541. [DOI] [PubMed] [Google Scholar]
- 24.Humbert M, Segal ES, Kiely DG, Carlsen J, Schwierin B, Hoeper MM. Results of European post-marketing surveillance of bosentan in pulmonary hypertension. Eur Respir J. 2007;30:338–44. doi: 10.1183/09031936.00138706. [DOI] [PubMed] [Google Scholar]
- 25.Daly AK, Aithal GP, Leathart JB, Swainsbury RA, Dang TS, Day CP. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 2007;132:272–81. doi: 10.1053/j.gastro.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Choi JH, Ahn BM, Yi J, Lee JH, Nam SW, Chon CY, et al. MRP2 haplotypes confer differential susceptibility to toxic liver injury. Pharmacogenet Genomics. 2007;17:403–15. doi: 10.1097/01.fpc.0000236337.41799.b3. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen TD, Markova S, Liu W, Gow JM, Baldwin RM, Habashian M, et al. Functional characterization of ABCC2 promoter polymorphisms and allele-specific expression. Pharmacogenomics J. doi: 10.1038/tpj.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirouchi M, Suzuki H, Itoda M, Ozawa S, Sawada J, Ieiri I, et al. Characterization of the cellular localization, expression level, and function of SNP variants of MRP2/ABCC2. Pharm Res. 2004;21:742–8. doi: 10.1023/b:pham.0000026422.06207.33. [DOI] [PubMed] [Google Scholar]
- 29.Haenisch S, May K, Wegner D, Caliebe A, Cascorbi I, Siegmund W. Influence of genetic polymorphisms on intestinal expression and rifampicin-type induction of ABCC2 and on bioavailability of talinolol. Pharmacogenet Genomics. 2008;18:357–65. doi: 10.1097/FPC.0b013e3282f974b7. [DOI] [PubMed] [Google Scholar]
- 30.Venitz J, Zack J, Gillies H, Allard M, Regnault J, Dufton C. Clinical Pharmacokinetics and Drug-Drug Interactions of Endothelin Receptor Antagonists in Pulmonary Arterial Hypertension. J Clin Pharmacol. doi: 10.1177/0091270011423662. [DOI] [PubMed] [Google Scholar]
- 31.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–63. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 32.Cavallari LH, Shin J, Perera MA. Role of pharmacogenomics in the management of traditional and novel oral anticoagulants. Pharmacotherapy. 31:1192–207. doi: 10.1592/phco.31.12.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Weide J, Steijns LS, van Weelden MJ, de Haan K. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics. 2001;11:287–91. doi: 10.1097/00008571-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Tang C, Shou M, Rushmore TH, Mei Q, Sandhu P, Woolf EJ, et al. Invitro metabolism of celecoxib, a cyclooxygenase-2 inhibitor, by allelic variant forms of human liver microsomal cytochrome P450 2C9: correlation with CYP2C9 genotype and in-vivo pharmacokinetics. Pharmacogenetics. 2001;11:223–35. doi: 10.1097/00008571-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 35.Lee CR, Pieper JA, Frye RF, Hinderliter AL, Blaisdell JA, Goldstein JA. Tolbutamide, flurbiprofen, and losartan as probes of CYP2C9 activity in humans. J Clin Pharmacol. 2003;43:84–91. doi: 10.1177/0091270002239710. [DOI] [PubMed] [Google Scholar]
- 36.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhry AS, Urban TJ, Lamba JK, Birnbaum AK, Remmel RP, Subramanian M, et al. CYP2C9*1B promoter polymorphisms, in linkage with CYP2C19*2, affect phenytoin autoinduction of clearance and maintenance dose. J Pharmacol Exp Ther. 332:599–611. doi: 10.1124/jpet.109.161026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss J, Theile D, Ruppell MA, Speck T, Spalwisz A, Haefeli WE. Interaction profile of macitentan, a new non-selective endothelin-1 receptor antagonist, in vitro. Eur J Pharmacol. 701:168–75. doi: 10.1016/j.ejphar.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Belic A, Temesvari M, Kohalmy K, Vrzal R, Dvorak Z, Rozman D, et al. Investigation of the CYP2C9 induction profile in human hepatocytes by combining experimental and modelling approaches. Curr Drug Metab. 2009;10:1066–74. doi: 10.2174/138920009790820147. [DOI] [PubMed] [Google Scholar]
- 40.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teichert M, Eijgelsheim M, Rivadeneira F, Uitterlinden AG, van Schaik RH, Hofman A, et al. A genome-wide association study of acenocoumarol maintenance dosage. Hum Mol Genet. 2009;18:3758–68. doi: 10.1093/hmg/ddp309. [DOI] [PubMed] [Google Scholar]
- 42.Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 63:157–81. doi: 10.1124/pr.110.002857. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava A, Srivastava N, Choudhuri G, Mittal B. Organic anion transporter 1B1 (SLCO1B1) polymorphism and gallstone formation: High incidence of Exon4 CA genotype in female patients in North India. Hepatol Res. 41:71–8. doi: 10.1111/j.1872-034X.2010.00736.x. [DOI] [PubMed] [Google Scholar]
- 44.Couvert P, Chapman MJ, Carrie A. Impact of genetic variation in the SLCO1B1 gene on statin efficacy in low-density lipoprotein cholesterol-lowering therapy. Pharmacogenomics. 12:137–9. doi: 10.2217/pgs.10.214. [DOI] [PubMed] [Google Scholar]
- 45.Geng F, Jiao Z, Dao YJ, Qiu XY, Ding JJ, Shi XJ, et al. The association of the UGT1A8, SLCO1B3 and ABCC2/ABCG2 genetic polymorphisms with the pharmacokinetics of mycophenolic acid and its phenolic glucuronide metabolite in Chinese individuals. Clin Chim Acta. 413:683–90. doi: 10.1016/j.cca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz UI, Meyer zu Schwabedissen HE, Tirona RG, Suzuki A, Leake BF, Mokrab Y, et al. Identification of novel functional organic anion-transporting polypeptide 1B3 polymorphisms and assessment of substrate specificity. Pharmacogenet Genomics. 21:103–14. doi: 10.1097/FPC.0b013e328342f5b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benichou C, Danan G, Flahault A. Causality assessment of adverse reactions to drugs--II. An original model for validation of drug causality assessment methods: case reports with positive rechallenge. J Clin Epidemiol. 1993;46:1331–6. doi: 10.1016/0895-4356(93)90102-7. [DOI] [PubMed] [Google Scholar]
- 48.Danan G, Benichou C. Causality assessment of adverse reactions to drugs--I. A novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol. 1993;46:1323–30. doi: 10.1016/0895-4356(93)90101-6. [DOI] [PubMed] [Google Scholar]
- 49.Pai R, French D, Ma N, Hotzel K, Plise E, Salphati L, et al. Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicol Sci. 126:446–56. doi: 10.1093/toxsci/kfs011. [DOI] [PubMed] [Google Scholar]
- 50.Tamraz B, Fukushima H, Wolfe AR, Kaspera R, Totah RR, Floyd JS, Ma B, Chu C, Marciante KD, Heckbert SR, Psaty BM, Kroetz DL, Kwok P-Y. OATP1B1 related drug-drug and drug-gene interactions as potential risk factors for cerivastatin induced rhabdomyolysis. Pharmacogenet Genomics. 2013 doi: 10.1097/FPC.0b013e3283620c3b. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.