Abstract
The Shank3 gene encodes a scaffolding protein that anchors multiple elements of the postsynaptic density at the synapse. Previous attempts to delete the Shank3 gene have not resulted in a complete loss of the predominant naturally occurring Shank3 isoforms. We have now characterized a homozygous Shank3 mutation in mice that deletes exon 21, including the Homer binding domain. In the homozygous state, deletion of exon 21 results in loss of the major naturally occurring Shank3 protein bands detected by C-terminal and N-terminal antibodies, allowing us to more definitively examine the role of Shank3 in synaptic function and behavior. This loss of Shank3 leads to an increased localization of mGluR5 to both synaptosome and postsynaptic density-enriched fractions in the hippocampus. These mice exhibit a decrease in NMDA/AMPA excitatory postsynaptic current ratio in area CA1 of the hippocampus, reduced long-term potentiation in area CA1, and deficits in hippocampus-dependent spatial learning and memory. In addition, these mice also exhibit motor-coordination deficits, hypersensitivity to heat, novelty avoidance, altered locomotor response to novelty, and minimal social abnormalities. These data suggest that Shank3 isoforms are required for normal synaptic transmission/plasticity in the hippocampus, as well as hippocampus-dependent spatial learning and memory.
Introduction
Autism is characterized by differences in key behavioral domains: social behavior, language, and restricted and repetitive behaviors (Schreibman, 1988). Deletions and other loss-of-function mutations of the gene encoding the synaptic scaffolding protein SHANK3 have been strongly implicated in human autism (Durand et al., 2007; Moessner et al., 2007; Gauthier et al., 2009; Boccuto et al., 2012). Furthermore, there are hundreds of children with Phelan–McDermid syndrome (22q13 deletion syndrome, intellectual disability with autism or autistic features) in which SHANK3 is strongly implicated in the autistic features and broader neurodevelopmental phenotype (Bonaglia et al., 2001, 2006; Wilson et al., 2003; Dhar et al., 2010; Boccuto et al., 2012), making hemizygous SHANK3 deletion the most common SHANK3 mutation associated with autistic features. Thus, a complete understanding of SHANK3 function in the CNS is critical to understand a subset of autism spectrum disorders caused by SHANK3 deletion or mutation.
Shank3 is a member of the Shank family of postsynaptic scaffolding proteins enriched in postsynaptic densities (PSDs) and was discovered in yeast two-hybrid screens as a binding partner of guanylate kinase-associated protein (GKAP) and postsynaptic density protein 95 (PSD-95; Naisbitt et al., 1999). Shank3 binds to the integral machinery of PSDs through its several functional domains. The ankyrin repeat domain of Shank3 mainly interacts with cytoskeletal proteins (Böckers et al., 2001). Its PSD protein/Drosophila disc large tumor suppressor/zonula occludens-1 protein (PDZ) domain interacts with ionotropic glutamate receptors either directly or indirectly via GKAP and PSD-95 (Garner et al., 2000; Uchino et al., 2006). The Homer binding domain of Shank3 binds to Homer, which then binds to the group 1 metabotropic glutamate receptors, such as mGluR1/5 (Tu et al., 1999).
Initial attempts to create mouse models lacking all Shank3 isoforms were unsuccessful, although they added important information of potential relevance to autism caused by SHANK3 mutations. Exon 4–9 or 4–7 deletion mouse models, coding for the ankyrin repeat domain, led to loss of only one of three major protein isoforms of Shank3 (Shank3α) by Western blot analysis (Bozdagi et al., 2010; Peça et al., 2011; Wang et al., 2011). A deletion model encompassing exons 13–16 (coding for the PDZ domain) led to loss of only two of the three major protein isoforms of Shank3 (Shank3α and Shank3β; Peça et al., 2011) using a single antibody.
Interestingly, an exon 21 deletion (coding for the Homer binding domain) mouse model results in loss of the predominant naturally occurring isoforms of Shank3 in the homozygous state, providing an excellent model in which to understand the effects of loss of naturally occurring Shank3 isoforms. This exon 21 deletion mouse model is based on a particular autism-associated mutation in humans that involves a guanine nucleotide insertion in exon 21, creating a frame shift and premature stop codon near the Homer binding domain. In the hemizygous state (Shank3+/ΔC), this model results in only partial loss of the major naturally occurring Shank3 proteins (data not shown). In the homozygous state (Shank3ΔC/ΔC), this model results in loss of major naturally occurring isoforms of Shank3 detected by N-terminal and C-terminal antibodies. Thus, examining the effects of homozygous mutation is informative of the function of Shank3, although only hemizygous mutation in exon 21 of the Shank3 gene has been linked to autism.
In this study, we examine the biochemical, behavioral, and electrophysiological consequences of homozygous loss of major naturally occurring Shank3 isoforms in the exon 21 deletion mouse model. We find that Shank3ΔC/ΔC mice show deficits in spatial learning and memory, motor coordination, sensitivity to sensory stimuli, and responses to novelty. To begin to understand the underlying mechanisms of these deficits, we examine synaptic physiology in area CA1 of the hippocampus of these mice and find impairments in hippocampal synaptic transmission and plasticity. These findings highlight the importance of Shank3 in normal synaptic function and behavior and provide additional evidence of potential treatment targets for autism and intellectual disability associated with Shank3 deletion/mutation.
Materials and Methods
Generation of Shank3ΔC/ΔC mice.
The Shank3 targeting construct was designed to delete exon 21 with Cre-mediated excision. To “flox” exon 21, Shank3 bacterial artificial chromosome DNA clone (Geneservice) was modified using standard recombineering technology. The final targeting construct had two homology arms of 6.0 and 1.7 kb, respectively. To identify targeted ES cells by PCR screen, a PCR control vector was constructed, which retains the Neo cassette and the short homology arm present in the targeting vector and additional Shank3 genomic sequence contiguous to the short arm. The targeting construct was electroporated into ES cells (129 s6 SvEv Tac background) and ES clones were selected for G418 resistance. The ES clones with targeted homologous recombination were identified by PCR with two sets of primers (forward, 50-TCGCCTTCTTGACGAGTTCT; reverse, 50-ACACGCTTTGGACACTTCTC). The authenticity of desired homologous recombination in the ES clones was confirmed by sequencing the PCR products. The positive ES clones were then injected into blastocysts (C57BL/6 strain) to generate chimeras at the Transgenic Facility of Johns Hopkins University School of Medicine. The chimeric mice were bred with C57BL/6 mice to confirm germ-line transmission of floxed Shank3, which was identified by PCR with primers as follows: forward, 50-ACTTCGTATAATGTATGCTATACGAAG; reverse, 50-GGCCATTGAATGGCTTCTCTGG. The floxed Shank3 mice were then mated with mice expressing actin–cre to excise exon 21. The resulting progeny were genotyped using a combination of three primers. The primer sequences were as follows: 50-TCCTGTGTCCCCTCATTGATGTT, 50-CTCTGCCACCTTCTGCCTACAAA, and 50-TGTCCTGTTGCAGGTAGGGAGAG. After confirming excision, the Shank3+/ΔC mice were mated with wild-type (WT) C57BL/6 mice to cross out the cre allele, and the progeny were further backcrossed with C57BL/6 mice for at least five generations. All mice tested were sex-matched, littermate progeny of matings between heterozygous Shank3 mutant mice. Overall, the Shank3ΔC/ΔC mutant mice appeared healthy except for their smaller body weight (measured at ∼17 weeks of age; Shank3ΔC/ΔC, 24.57 ± 0.13 g; WT, 31.95 ± 0.15 g; p < 0.001; Table 1).
Table 1.
Statistical analysis of behavioral studies
| Parameter | Comparison | Results |
|---|---|---|
| Body weight (n = 19) | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 25.47, p < 0.00001; main effect of genotype: F(1,34) = 14.60, p < 0.001; sex × genotype interaction: F(1,34) = 0.17, p = 0.67 |
| Open field (n = 19) | ||
| Time in center | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.33, p = 0.56; main effect of genotype: F(1,34) = 0.10, p = 0.74; sex × genotype interaction: F(1,34) = 4.26, p < 0.4 |
| Frequency in center | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.05, p = 0.82; main effect of genotype: F(1,34) = 5.59, p < 0.03; sex × genotype interaction: F(1,34) = 2.47, p = 0.12 |
| Time in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.99, p = 0.32; main effect of genotype: F(1,34) = 4.44, p < 0.05; sex × genotype interaction: F(1,34) = 0.13, p = 0.71 |
| Frequency in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: p < 0.0004, F(1,34) = 0.86, p = 0.36; main effect of genotype: F(1,34) = 15.83; sex × genotype interaction: F(1,34) = 1.19, p = 0.28 |
| Time in center/time in periphery | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.14, p = 0.70; main effect of genotype: F(1,34) = 0.003, p = 0.95; sex × genotype interaction: F(1,34) = 3.34, p = 0.07 |
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.02, p = 0.88; main effect of genotype: F(1,34) = 23.54, p < 0.00003; sex × genotype interaction: F(1,34) = 0.04, p = 0.83 |
| Velocity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.02, p = 0.88; main effect of genotype: F(1,34) = 23.42, p < 0.00003; sex × genotype interaction: F(1,34) = 0.04, p = 0.84 |
| Dark/light box (n = 19) | ||
| Total activity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.16, p = 0.68; main effect of genotype: F(1,34) = 35.60, p < 0.000002; sex × genotype interaction: F(1,34) = 0.0001, p = 0.99 |
| Time in light side | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 2.32, p = 0.13; main effect of genotype: F(1,34) = 29.06, p < 0.00006; sex × genotype interaction: F(1,34) = 3.47, p = 0.071 |
| Crosses | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 1.55, p = 0.22; main effect of genotype: F(1,34) = 26.83, p < 0.00002; sex × genotype interaction: F(1,34) = 0.71, p = 0.40 |
| Latency to enter light side | Sex and genotype; parametric | 2-way ANOVA; main effect of sex: F(1,34) = 0.002, p = 0.96; main effect of genotype: F(1,34) = 21.08, p < 0.00006; sex × genotype interaction: F(1,34) = 0.006, p = 0.93 |
| Time in dark side | Sex and genotype; nonparametric | 2-way ANOVA; main effect of sex: F(1,34) = 2.32, p = 0.14; main effect of genotype: F(1,34) = 29.06, p < 0.00006; sex × genotype interaction: F(1,34) = 3.47, p = 0.07 |
| Elevated plus maze (n = 18) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.01, p = 0.90; main effect of genotype: F(1,32) = 0.05, p = 0.82; sex × genotype interaction: F(1,32) = 0.01, p = 0.91 |
| Velocity | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.14, p = 0.70; main effect of genotype: F(1,32) = 0.48, p = 0.49; sex × genotype interaction: F(1,32) = 0.14, p = 0.70 |
| Time in open arms/time in both arms | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.50, p = 0.48; main effect of genotype: F(1,32) = 0.12, p = 0.73; sex × genotype interaction: F(1,32) = 0.06, p = 0.79 |
| Entries in open/entries in both | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 3.78, p = 0.06; main effect of genotype: F(1,32) = 3.07, p = 0.08; sex × genotype interaction: F(1,32) = 0.48, p = 0.49 |
| Morris water maze–initial training (n = 18) | ||
| Latency to reach platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.35, p = 0.55; main effect of genotype: F(1,32) = 13.55, p < 0.001; main effect of day: F(7,224) = 36.47, p < 0.000001; sex × genotype interaction: F(1,32) = 0.12, p = 0.72; sex × day interaction: F(7,224) = 1.58, p = 0.14; genotype × day interaction: F(7,224) = 2.54, p < 0.05; sex × genotype × day interaction: F(7,224) = 0.53, p = 0.806 |
| Percentage thigmotaxis | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.49, p = 0.49; main effect of genotype: F(1,32) = 7.62, p < 0.01; main effect of day: F(7,224) = 75.09, p < 0.000001; sex × genotype interaction: F(1,32) = 0.07, p = 0.79; sex × day interaction: F(7,224) = 5.09, p < 0.0001; genotype × day interaction: F(7,224) = 2.41, p < 0.05; sex × genotype × day interaction: F(7,224) = 2.10, p < 0.05 |
| Distance traveled to platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.83, p = 0.36; main effect of genotype: F(1,32) = 17.41, p < 0.001; main effect of day: F(7,224) = 41.61, p < 0.000001; sex × genotype interaction: F(1,32) = 2.39, p = 0.13; sex × day interaction: F(7,224) = 1.02, p = 0.41; genotype × day interaction: F(7,224) = 3.25, p < 0.005; sex × genotype × day interaction: F(7,224) = 0.72, p = 0.65 |
| Average swim speed | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 3.51, p = 0.07; main effect of genotype: F(1,32) = 0.5, p = 0.48; main effect of day: F(7,224) = 2.00, p = 0.05; sex × genotype interaction: F(1,32) = 5.48, p < 0.02; sex × day interaction: F(7,224) = 0.59, p = 0.75; genotype × day interaction: F(7,224) = 0.92, p = 0.48; sex × genotype × day interaction: F(7,224) = 1.12, p = 0.34 |
| WT versus KO | Planned comparisons: females (WT vs KO): p < 0.05; Males (WT vs KO): p = 0.22 | |
| Males versus females | Planned comparisons: WT (M vs F): p = 0.74 KO (M vs F): p < 0.0006 | |
| Morris water maze–probe trial (n = 18) | ||
| Percentage time in quadrant | Sex, genotype, and quadrant | 3-way rmANOVA; main effect of sex: F(1,32) = 0.91, p = 0.34; main effect of genotype: F(1,32) = 0.22, p = 0.63; main effect of quadrant: F(3,96) = 33.49, p < 0.000001; sex × genotype interaction: F(1,32) = 3.64, p = 0.06; sex × quadrant interaction: F(3,96) = 0.51, p = 0.67; genotype × quadrant interaction: F(3,96) = 4.21, p < 0.01; sex × genotype × quadrant interaction: F(3,96) = 0.63, p = 0.59 |
| WT versus KO within quadrants | Planned comparisons: TargetNW quad: p < 0.01; RightNE quad: p = 0.87; LeftSW quad: p = 0.40; OppositeSE quad: p < 0.01 | |
| Target versus other quads within WT | Planned comparisons: TargetNW versus RightNE: p < 0.00001; TargetNW versus LeftSW: p < 0.00001; TargetNW versus OppositeSE: p < 0.00001 | |
| Target versus other platforms KO | Planned comparisons: TargetNW versus RightNE: p < 0.02; TargetNW versus LeftSW: p < 0.02; TargetNW versus OppositeSE: p < 0.0001 | |
| Number of platform crossings | Sex, genotype, and platform | 3-way rmANOVA; main effect of sex: F(1,32) = 1.59, p = 0.21; main effect of genotype: F(1,32) = 0.01, p = 0.91; main effect of quadrant: F(3,96) = 13.00, p < 0.000001; sex × genotype interaction: F(1,32) = 8.39, p < 0.01; sex × quadrant interaction: F(3,96) = 1.30, p = 0.27; genotype × quadrant interaction: F(3,96) = 2.13, p = 0.10; sex × genotype × quadrant interaction: F(3,96) = 0.47, p = 0.70 |
| WT versus KO within platforms | Planned Comparisons: TargetNW quad: p = 0.27; RightNE quad: p = 0.69; LeftSW quad: p = 0.83; OppositeSE quad: p < 0.01 | |
| Target versus other platforms WT | Planned comparisons: TargetNW versus RightNE: p < 0.001; TargetNW versus LeftSW: p < 0.01; TargetNW versus OppositeSE: p < 0.0001 | |
| Target versus other platforms KO | Planned comparisons: TargetNW versus RightNE: p = 0.08; TargetNW versus LeftSW: p = 0.08; TargetNW versus OppositeSE: p = 0.052 | |
| WT versus KO | Planned comparisons: females (WT v sKO): p = 0.05; males (WT v sKO): p < 0.05 | |
| Males versus females | Planned comparisons: WT (M vs F): p = 0.25; KO (M vs F): p < 0.007 | |
| Average swim speed | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 2.51, p = 0.12; main effect of genotype: F(1,32) = 0.03, p = 0.84; sex × genotype interaction: F(1,32) = 5.17, p < 0.01 |
| WT versus KO | Planned comparisons: females (WT vs KO): p = 0.10; males (WT vs KO): p = 0.12 | |
| Males versus females | Planned comparisons: WT (M vs F): p = 0.62; KO (M vs F): p < 0.05 | |
| Percentage thigmotaxis | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 1.42, p = 0.24; main effect of genotype: F(1,32) = 5.45, p < 0.01; sex × genotype interaction: F(1,32) = 0.03, p = 0.84 |
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 2.52, p = 0.12; main effect of genotype: F(1,32) = 0.03, p = 0.86; sex × genotype interaction: F(1,32) = 5.12, p < 0.05 |
| WT versus KO | Planned comparisons: females (WT vs KO): p = 0.11; males (WT vs KO): p = 0.12 | |
| Males versus females | Planned comparisons: WT (M vs F): p = 0.63; KO (M vs F): p < 0.02 | |
| Morris water maze–reversal training (n = 18) | ||
| Latency to reach platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.0000041, p = 0.99; main effect of genotype: F(1,32) = 2.63, p = 0.11; main effect of day: F(4,128) = 8.05, p < 0.0009; sex × genotype interaction: F(1,32) = 1.21, p = 0.27; sex × day interaction: F(4,128) = 0.64, p = 0.63; genotype × day interaction: F(4,128) = 1.69, p = 0.15; sex × genotype × day interaction: F(4,128) = 0.27, p = 0.89 |
| Percentage thigmotaxis | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.50, p = 0.48; main effect of genotype: F(1,32) = 2.29, p = 0.13; main effect of day: F(4,128) = 1.50, p = 0.20; sex × genotype interaction: F(1,32) = 0.39, p = 0.53; sex × day interaction: F(4,128) = 1.22, p = 0.30; genotype × day interaction: F(4,128) = 0.57, p = 0.68; sex × genotype × day interaction: F(4,128) = 0.77, p = 0.54 |
| Distance traveled to platform | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 0.44, p = 0.51; main effect of genotype: F(1,32) = 2.08, p = 0.15; main effect of day: F(4,128) = 4.99, p < 0.0009; sex × genotype interaction: F(1,32) = 0.01, p = 0.89; sex × day interaction: F(4,128) = 0.91, p = 0.45; genotype × day interaction: F(4,128) = 2.64, p < 0.05; sex × genotype × day interaction: F(4,128) = 0.65, p = 0.62 |
| Average swim speed | Sex, genotype, and day | 3-way rmANOVA; main effect of sex: F(1,32) = 2.48, p = 0.12; main effect of genotype: F(1,32) = 0.006, p = 0.93; main effect of day: F(4,128) = 1.08, p = 0.36; sex × genotype interaction: F(1,32) = 3.05, p = 0.09; sex × day interaction: F(4,128) = 1.21, p = 0.30; genotype × day interaction: F(4,128) = 0.68, p = 0.60; sex × genotype × day interaction: F(4,128) = 1.84, p = 0.12 |
| Morris water maze–reversal probe (n = 18) | ||
| Percentage time in quadrant | Sex, genotype, and quadrant | 3-way rmANOVA; main effect of sex: F(1,32) = 0.71, p = 0.40; main effect of genotype: F(1,32) = 0.83, p = 0.36; main effect of quadrant: F(3,96) = 5.87, p < 0.01; sex × genotype interaction: F(1,32) = 0.76, p = 0.38; sex × quadrant interaction: F(3,96) = 1.76, p = 0.15; genotype × quadrant interaction: F(3,96) = 6.51, p < 0.001; sex × genotype × quadrant interaction: F(3,96) = 0.59, p = 0.61 |
| WT versus KO within platforms | Planned comparisons: TargetSE quad: p < 0.001; RightNE quad: p = 0.08; LeftSW quad: p = 0.76; OppositeNW quad: p < 0.01 | |
| Target versus other platforms WT | Planned comparisons: TargetSE versus RightNE: p < 0.001; TargetSE versus LeftSW: p < 0.001; TargetSE versus OppositeNW: p < 0.0001 | |
| Target versus other platforms KO | Planned comparisons: TargetSE versus RightNE: p = 0.55; TargetSE versus LeftSW: p = 0.32; TargetSE versus OppositeNW: p = 0.60 | |
| Number of platform crossings | Sex, genotype, and platform | 3-way rmANOVA; main effect of sex: F(1,32) = 0.003, p = 0.95; main effect of genotype: F(1,32) = 6.11, p < 0.02; main effect of quadrant: F(3,96) = 4.93, p < 0.01; sex × genotype interaction: F(1,32) = 0.63, p = 0.43; sex × quadrant interaction: F(3,96) = 0.35, p = 0.78; genotype × quadrant interaction: F(3,96) = 1.98, p = 0.12; sex × genotype × quadrant interaction: F(3,96) = 0.04, p = 0.98 |
| WT versus KO within platforms | Planned comparisons: TargetSE quad: p < 0.05; RightNE quad: p = 0.39; Left SW quad: p = 0.19; OppositeNW quad: p = 0.86 | |
| Target versus other platforms WT | Planned comparisons: TargetSE versus RightNE: p < 0.05; TargetSE versus LeftSW: p < 0.01; TargetSE versus OppositeNW: p < 0.01 | |
| Target versus other platforms KO | Planned comparisons: TargetSE versus RightNE: p = 0.66; TargetSE versus LeftSW: p = 0.20; TargetSE versus OppositeNW: p = 0.68 | |
| Average swim speed | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 1.67, p = 0.20; main effect of genotype: F(1,32) = 1.16, p = 0.28; sex × genotype interaction: F(1,32) = 3.13, p = 0.08 |
| Percentage thigmotaxis | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.49, p = 0.48; main effect of genotype: F(1,32) = 1.32, p = 0.25; sex × genotype interaction: F(1,32) =1.04, p = 0.31 |
| Distance traveled | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 1.72, p = 0.19; main effect of genotype: F(1,32) = 1.12, p = 0.29; sex × genotype interaction: F(1,32) = 3.23, p = 0.08 |
| Visible water maze (n = 18) | ||
| Latency to platform | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.15, p = 0.69; main effect of genotype: F(1,32) = 2.93, p = 0.09; sex × genotype: F(1,32) = 0.07, p = 0.78 |
| Percentage thigmotaxis | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.0008, p = 0.97; main effect of genotype: F(1,32) = 0.98, p = 0.32; sex × genotype: F(1,32) = 0.04, p = 0.82 |
| Distance traveled to platform | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.82, p = 0.36; main effect of genotype: F(1,32) = 3.34, p = 0.07; sex × genotype: F(1,32) = 0.02, p = 0.86 |
| Average swim speed | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.89, p = 0.35; main effect of genotype: F(1,32) = 0.34, p = 0.56; sex × genotype: F(1,32) = 0.12, p = 0.72 |
| Hotplate (n = 18) | ||
| Time to lick shake hindpaw | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.68, p = 0.41; main effect of genotype: F(1,32) = 4.38, p < 0.05; sex × genotype: F(1,32) = 0.97, p = 0.33 |
| Nesting behavior (n = 19) | ||
| Increase in nest height | Sex, genotype, and time | 3-way rmANOVA; main effect of sex: F(1,34) = 6.004, p < 0.02; main effect of genotype: F(1,34) = 17.78, p < 0.001; main effect of trial: F(2,68) = 17.44, p < 0.0001; sex × genotype interaction: F(1,34) = 7.57, p < 0.01; sex × trial interaction: F(2,68) = 1.78, p = 0.17; genotype × trial interaction: F(2,68) = 4.73, p < 0.02; sex × genotype × trial interaction: F(4,128) = 0.55, p = 0.57 |
| Increase in nest width | Sex, genotype, and time | 3-way rmANOVA; main effect of sex: F(1,34) = 3.77, p = 0.06; main effect of genotype: F(1,34) = 17.06, p < 0.001; main effect of trial: F(2,68) = 7.60, p < 0.01; sex × genotype interaction: F(1,34) = 3.55, p = 0.06; sex × trial interaction: F(2,68) = 1.84, p = 0.16; genotype × trial interaction: F(2,68) = 6.00, p < 0.004; sex × genotype × trial interaction: F(4,128) = 1.48, p = 0.23 |
| Three-choice interaction test– baseline (n = 17) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 0.52, p = 0.47; main effect of genotype: F(1,30) = 2.70, p = 0.11; sex × genotype interaction: F(1,30) = 0.0004, p = 0.98 |
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 0.43, p = 0.51; main effect of genotype: F(1,30) = 2.36, p = 0.13; sex × genotype interaction: F(1,30) = 0.0023, p = 0.96 |
| Time spent sniffing | Sex, genotype, and interaction target (front vs back) | 3-way rmANOVA: main effect of sex: F(1,30) = 0.03, p = 0.86; main effect of genotype: F(1,30) = 1.23, p = 0.27; main effect of target: F(1,30) = 0.01, p = 0.90; sex × genotype interaction: F(1,30) = 4.01, p = 0.05; sex × target interaction: F(1,30) = 1.00, p = 0.32; genotype × target interaction: F(1,30) = 0.06, p = 0.80; sex × genotype × target interaction: F(1,30) = 0.20, p = 0.65 |
| Effect of target (within each genotype) | Planned comparisons; A: p = 0.92; B: p = 0.79 | |
| Three-choice interaction test– social preference (n = 17) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 3.57, p = 0.06; main effect of genotype: F(1,30) = 2.68, p = 0.11; sex × genotype interaction: F(1,30) = 0.14, p = 0.70 |
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 1.30, p = 0.26; main effect of genotype: F(1,30) = 1.53, p = 0.22; sex × genotype interaction: F(1,30) = 0.55, p = 0.46 |
| Interaction time | Sex, genotype, and interaction target (inanimate vs social) | 3-way rmANOVA: main effect of sex: F(1,30) = 0.23, p = 0.63; main effect of genotype: F(1,30) = 2.95, p = 0.09; main effect of target: F(1,30) = 20.81, p < 0.0001; sex × genotype interaction: F(1,30) = 0.002, p = 0.96; sex × target interaction: F(1,30) = 1.54, p = 0.22; genotype × target interaction: F(1,30) = 0.36, p = 0.54; sex × genotype × target interaction: F(1,30) = 0.55, p = 0.46 |
| Effect of target within each genotype | Planned comparisons; A: p < 0.01; B: p < 0.001 | |
| Effect of gentoype (WT vs KO) within each target | Planned comparisons; social target: p = 0.60; inanimate: p < 0.01 | |
| Three-choice interaction test– social novelty (n = 17) | ||
| Distance traveled | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 14.80, p < 0.0005; main effect of genotype: F(1,30) = 0.87, p = 0.35; sex × genotype interaction: F(1,30) = 0.13, p = 0.71 |
| Velocity | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,30) = 15.90, p < 0.0003; main effect of genotype: F(1,30) = 0.45, p = 0.50; sex × genotype interaction: F(1,30) = 0.007, p = 0.93 |
| Interaction time | Genotype and interaction target (familiar vs stranger) | 3-way rmANOVA: main effect of sex: F(1,30) = 0.004, p = 0.94; main effect of genotype: F(1,30) = 0.56, p = 0.45; main effect of target: F(1,30) = 4.16, p = 0.05; sex × genotype interaction: F(1,30) = 0.10, p = 0.74; sex × target interaction: F(1,30) = 3.35, p = 0.07; genotype × target interaction: F(1,30) = 1.68, p = 0.20; sex × genotype × target interaction: F(1,30) = 0.17, p = 0.67 |
| Effect of target within each genotype | Planned comparisons; A: p < 0.05; B: p = 0.60 | |
| Effect of gentoype (WT vs KO) within each target | Planned comparisons; familiar target: p = 0.87; novel: p = 0.23 | |
| Social interaction with juvenile (n = 18) | ||
| Interaction time | Sex, genotype, and trial | 3-way rmANOVA; main effect of sex: F(1,32) = 1.46, p = 0.23; main effect of genotype: F(1,32) = 0.502, p = 0.48; main effect of trial: F(1,32) = 138.76, p < 0.000001; sex × genotype: F(1,32) = 1.00, p = 0.32; sex × trial interaction: F(1,32) = 0.008, p = 0.92; genotype × trial interaction: F(1,32) = 3.88, p = 0.05; sex × genotype × trial interaction: F(1,32) = 1.175, p = 0.28 |
| Trial (initial vs recognition) within each genotype | Planned comparisons; A: p < 0.00001; B: p < 0.00001 | |
| Locomotor habituation (n = 19) | ||
| Total beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,34) = 0.32, p = 0.57; main effect of genotype: F(1,34) = 0.49, p = 0.48; main effect of trial: F(7,782) = 41.30, p < 0.000001; sex × genotype interaction: F(1,34) = 0.06, p = 0.80; sex × trial interaction: F(7,782) = 1.09, p = 0.34; genotype × trial: F(7,782) = 1.21, p = 0.22; sex × genotype × trial interaction: F(7,782) = 0.88, p = 0.61 |
| First bin only | Genotype and sex | 2-way ANOVA; main effect of sex: F(1,34) = 2.28, p = 0.14; main effect of genotype: F(1,34) = 4.43, p < 0.05; sex × genotype interaction: F = 0.09, p = 0.76 |
| Stereodypy beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,34) = 0.45, p = 0.50; main effect of genotype: F(1,34) = 1.99, p = 0.16; main effect of trial: F(7,782) = 21.07, p < 0.000001; sex × genotype interaction: F(1,34) = 1.07, p = 0.30; sex × trial interaction: F(7,782) = 1.16, p = 0.27; genotype × trial: F(7,782) = 0.72, p = 0.81; sex × genotype × trial interaction: F(7,782) = 0.72, p = 0.82 |
| Ambulatory beam breaks | Sex, genotype, and bin | 3-way rmANOVA; main effect of sex: F(1,34) = 1.03, p = 0.31; main effect of genotype: F(1,34) = 2.07, p = 0.15; main effect of trial: F(7,782) = 39.29, p < 0.000001; sex × genotype interaction: F(1,34) = 0.001, p = 0.96; sex × trial interaction: F(7,782) = 1.07, p = 0.36; genotype × trial: F(7,782) = 1.42, p = 0.08; sex × genotype × trial interaction: F(7,782) = 0.89, p = 0.60 |
| Rotarod (n = 19) | ||
| Time to fall off | Sex, genotype, and trial | 3-way rmANOVA; main effect of sex: F(1,34) = 7.17, p < 0.02; main effect of genotype: F(1,34) = 6.95, p < 0.02; main effect of trial: F(7,238) = 11.71, p < 0.000001; sex × genotype interaction: F(1,34) = 0.45, p = 0.50; sex × trial interaction: F(7,238) = 0.51, p = 0.82; genotype × trial: F(7,238) = 1.60, p = 0.13; sex × genotype × trial interaction: F(7,238) = 1.88, p = 0.07 |
| WT versus KO within each gender | Planned comparisons: within females: p < 0.05; within males: p = 0.162 | |
| Males versus females within each genotype | Planned comparisons: within WT: p < 0.05; within KO: p = 0.164 | |
| Prepulse inhibition (n = 18) | ||
| Initial startle response | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.15, p = 0.69; main effect of genotype: F(1,32) = 0.04, p = 0.83; sex × genotype interaction: F(1,32) = 0.12, p = 0.72 |
| Second set startle response | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.01, p = 0.91; main effect of genotype: F(1,32) = 0.01, p = 0.89; sex × genotype interaction: F(1,32) = 0.08, p = 0.77 |
| Third set startle response | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,32) = 0.002, p = 0.95; main effect of genotype: F(1,32) = 0.008, p = 0.92; sex × genotype interaction: F(1,32) = 0.0, p = 0.99 |
| Startle habituation | Sex, genotype, and trial | 3-way rmANOVA; main effect of sex: F(1,32) = 0.02, p = 0.87; main effect of genotype: F(1,32) = 0.02, p = 0.88; main effect of trial: F(2,64) = 6.16, p < 0.03; sex × genotype interaction: F(1,32) = 0.05, p = 0.81; sex × decibel interaction: F(2,64) = 1.13, p = 0.32; genotype × decibel interaction: F(2,64) = 0.09, p = 0.90; sex × genotype × decibel interaction: F(2,64) = 0.81, p = 0.44 |
| Prepulse inhibition | Sex, genotype, and decibel | 3-way rmANOVA; main effect of sex: F(1,34) = 5.85, p < 0.03; main effect of genotype: F(1,34) = 3.06, p = 0.08; main effect of trial: F(2,64) = 54.86, p < 0.0001; sex × genotype interaction: F(1,34) = 5.50, p < 0.03; sex × trial interaction: F(2,64) = 0.03, p = 0.96; genotype × trial interaction: F(2,64) = 0.20, p = 0.81; sex × genotype × trial interaction: F(2,64) = 1.14, p = 0.32 |
| WT versus KO | Planned comparisons: females (WT vs KO): p < 0.01; males (WT vs KO): p = 0.65 | |
| Males versus females | Planned comparisons: WT (M vs F): p = 0.95 KO (M vs F): p < 0.01 | |
| Startle threshold (n = 18) | ||
| Startle response | Sex, genotype, and decibel | 3-way rmANOVA; main effect of sex: F(1,32) = 0.19, p = 0.65; main effect of genotype: F(1,32) = 0.008, p = 0.92; main effect of trial: F(5,160) = 34.95, p < 0.0001; sex × genotype interaction: F(1,32) = 0.47, p = 0.49; sex × decibel interaction: F(5,160) = 0.25 p = 0.93; genotype × decibel interaction: F(5,160) = 0.03, p = 0.99; sex × genotype × decibel interaction: F(5,160) = 0.34, p = 0.88 |
| Marble burying (n = 19) | ||
| Number of marbles buried | Sex and genotype | 2-way ANOVA; main effect of sex: F(1,34) = 0.01, p = 0.90; main effect of genotype: F(1,34) = 58.20, p < 0.000001; sex × genotype interaction: F(1,34) = 0.19, p = 0.66 |
| Grooming (9–18 weeks old) (n = 19) | ||
| Time spent grooming | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,34) = 2.04, p = 0.16; main effect of genotype: F(1,34) = 0.36, p = 0.54; sex × genotype interaction: F(1,34) = 0.03, p = 0.85 |
| Number of bouts | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,34) = 0.17, p = 0.67; main effect of genotype: F(1,34) = 1.32, p = 0.25; sex × genotype interaction: F(1,34) = 0.25, p = 0.62 |
| Time per bout | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,34) = 1.92, p = 0.17; main effect of genotype: F(1,34) = 0.44, p = 0.50; sex × genotype interaction: F(1,34) = 0.88, p = 0.35 |
| Grooming (10–13 months old) (n = 16) | ||
| Time spent grooming | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,28) = 0.62, p = 0.43; main effect of genotype: F(1,28) = 4.69, p < 0.04; sex × genotype interaction: F(1,28) =1.61, p = 0.21 |
| Number of bouts | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,28) = 1.13, p = 0.29; main effect of genotype: F(1,28) = 1.86, p = 0.18; sex × genotype interaction: F(1,28) = 0.53, p = 0.47 |
| Time per bout | Sex and genotype | 2-way ANOVA: main effect of sex: F(1,28) = 1.60, p = 0.21; main effect of genotype: F(1,28) = 6.00, p < 0.03; sex × genotype interaction: F(1,28) =2.44, p = 0.12 |
| USVs (10–13 months old) (n = 8) | ||
| Latency to call | Genotype | 1-way ANOVA; main effect of genotype: F(1,14) = 0.14, p = 0.71 |
| Number of calls | Genotype | 1-way ANOVA; main effect of genotype: F(1,14) = 0.40, p = 0.52 |
For ANOVAs, between-subjects factors are generally sex and genotype, with repeated measures (day, time, or trial). F(x,y), F ratio statistic is used to determine whether the variances in two independent samples are equal; x, y are degrees of freedom. Degrees of freedom is a measure of the number of independent pieces of information on which the precision of a parameter estimate is based. x, Number of groups − 1; y, number of animals per group − 1, multiplied by the number of groups.
Western blot.
Synaptic protein levels from seven pairs (WT/Shank3ΔC/ΔC, 5–6 months old) were determined by immunoblotting whole hippocampus dissected in ice-cold artificial CSF [ACSF; 124 mm NaCl, 5 mm KCl, 1.25 mm NaH2PO4, 1 mm MgCl2, 26 mm NaHCO3, 10 mm dextrose, 2 mm CaCl2, 5 mm EDTA, and 1× Halt protease and phosphatase inhibitor mixture (Thermo Fisher Scientific)] and then homogenized in 1% SDS, 50 mm NaF, 5 mm EDTA, and 1× Halt protease and phosphatase inhibitor mixture (Thermo Fisher Scientific). Ten micrograms of protein were loaded per lane and blotted with antibodies for synaptic proteins and internal loading controls (β-actin). An Image Works film processor was used to develop films, and the chemiluminescence signals were quantified, normalized, and statistically analyzed using NIH ImageJ, Image Studio, and Microsoft Excel.
Synaptosome preparation.
All steps were performed at 4°C or on ice; all buffers contained Halt protease and phosphatase inhibitors (Thermo Fisher Scientific). Mice were killed by live decapitation, and rapidly dissected hippocampi were homogenized in Syn-PER synaptic protein extraction reagent (Thermo Fisher Scientific), ∼1 ml/100 mg tissue. Samples were centrifuged at 1200 × g for 10 min. The resulting supernatant was then centrifuged at 15,000 × g for 20 min. Pellets (synaptosomes) were resuspended in buffer B (3 mm sucrose in 6 mm Tris, pH 8.0) with 1% SDS, briefly sonicated, and flash frozen in liquid nitrogen. Samples were stored at −80°C, and protein concentration was quantified by DC Protein Assay (Bio-Rad).
PSDII preparation.
Synaptosome pellets were isolated as described above, resuspended in 1 ml of buffer B, and homogenized. Homogenates were gently layered onto sucrose density gradients (1.15, 1, and 0.85 m sucrose) and centrifuged at 82,500 × g. The fraction between 1.15 and 1 m sucrose layer was isolated, resuspended in buffer C (6 mm Tris, pH 8.0, 1% Triton X-100), and incubated for 15 min before centrifugation at 32,800 × g for 20 min. The pellets were resuspended in buffer D (6 mm Tris, pH 8.0, 0.5% Triton X-100) and incubated for 15 min. After centrifugation for 1 h at 201,800 × g, the resulting pellets (PSD-II) were resuspended in a minimal volume of buffer D and flash frozen in liquid nitrogen.
Behavioral overview.
Behavioral tests were performed on a cohort of 9 female and 10 male, age- and sex-matched littermate pairs (n = 19 WT and n = 19 homozygous mutant) during the light cycle of the mice. All mice were born within 10 weeks of each other. Behaviors were tested at 2–6 months of age (unless otherwise noted) by an experimenter blind to genotype in the following order: elevated plus maze, dark/light, open field, locomotor, grooming, three-box social interaction test, marble burying, rotarod, social interaction with a juvenile, nesting behavior, Morris water maze, visible water maze, paired-pulse inhibition, startle threshold, footshock sensitivity, and hotplate sensitivity. One littermate pair was excluded from the analysis of elevated plus maze and social interaction with a juvenile as one littermate jumped out of the apparatus. Similarly, two littermate pairs were excluded from the three-box social interaction test as the mice climbed out and began circling the outer edge of the apparatus. Also, one mouse was found dead in its home cage on the morning after nesting behavior was performed, so its littermate pair was excluded from the study after nesting behavior. Ultrasonic vocalizations (USVs) emitted by male mice in the presence of a free-roaming estrous female were recorded in eight male age–matched littermate pairs at 10–13 months of age. Afterward, grooming behavior was repeated in a cohort of seven male and an additional nine female littermate pairs (n = 16 WT and n = 16 homozygous mutant).
Behavioral results are not described in the order they were tested in an effort to ease presentation and interpretation of the data. All statistical analyses of behavioral data were conducted using Statistica software (version 5.5; Statsoft) using either two-way ANOVAs or three-way repeated-measures ANOVA (rmANOVA) using genotype and sex as the main variables and trial as the repeated measure when applicable. Post hoc planned comparisons were applied for significant effects and interactions. For detailed information and numerical statistical results, see Table 1.
Morris water maze.
The Morris water maze task was conducted essentially as described previously (Powell et al., 2004; Tabuchi et al., 2007). Briefly, a white, circular pool 1.2 m in diameter was filled with water (22 ± 1οC) made opaque with nontoxic, “gothic white” liquid tempera paint, and a circular platform (10 cm in diameter) was submerged ∼1 cm beneath the surface of the water. The testing room was well lit and filled with a number of extramaze cues. Training was conducted over 9 consecutive days with four trials/d using an intertrial interval of 1–1.5 min. Mice were placed pseudorandomly into each of four starting locations for each of four daily training trials. In each trial, mice swam until they found the hidden platform or were guided to it by the experimenter if not found within 60 s. Mice remained on the platform for 15 s before being removed to the home cage. Daily data were averaged across the four trials. A probe trial was conducted on day 10; the hidden platform was removed, and mice were placed in the pool and allowed to swim for 60 s. For reversal water maze training, training resumed the day after the probe trial for 5 d with the platform in the opposite quadrant of the maze; on day 6 after reversal, a second probe trial was administered. Data were analyzed using three-way rmANOVA with genotype and sex as between-subject factors and trial days as a within-subject factor for training. For probe trials, quadrant or platform location was used as the within-subject factor.
Accelerating rotarod.
Coordination and motor learning were tested using a rotarod essentially as described previously (Powell et al., 2004). Mice were placed on a stationary rotarod (IITC Life Sciences) in a well lit room that was then activated and accelerated from 0 to 45 revolutions/min over 5 min. The latency for mice to fall off the rod or take one revolution was measured. Trials were repeated four times with intertrial intervals of 30 min over a single day. Data were analyzed using three-way rmANOVA with genotype and sex as between-subject factors and trials as a within-subject factor.
Hotplate sensitivity.
This test was performed as described previously (Powell et al., 2004; Blundell et al., 2010b). Mice were placed on a black, anodized plate that was held at a constant temperature of 52°C (IITC Life Sciences model 39 hotplate) covered with a Plexiglas enclosure. Mice were removed after the first hindpaw lick or after 30 s if no response was elicited. The plate was cleaned with water between mice and allowed to return to temperature. Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Nesting.
Nesting behavior was performed in a well lit (∼80 lux) room by first habituating the mouse to a novel home cage with ∼1.5 cm of bedding for 15 min, and then a cotton nestlet (5.5 × 5.5 × 0.5 cm) was put in the cage. Height and width of the nests were measured at 30, 60, and 90 min (Etherton et al., 2009). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Marble burying.
As described previously (Blundell et al., 2010b), 20 marbles were evenly placed around the edges of a novel home cage with 5 cm of bedding, and mice were given 30 min in the cage. After 30 min, the number of marbles buried was recorded. A marble was defined as buried when <25% of the marble was visible. The test room was well lit (∼80 lux). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Dark/light.
The dark/light test was performed as described previously (Powell et al., 2004). Briefly, the dark/light apparatus consisted of two chambers (each chamber was 25 × 26 cm), one brightly lit (∼2900 lux) and the other kept dark with a small door (7 × 7 cm) separating the two. Mice were habituated for 2 min in the dark side, the door was opened, and then mice were allowed to move freely between the two sides for 10 min. Time spent in and the number of entries into each side were measured along with locomotor activity using photobeams monitored by Med PC IV data acquisition software. Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Elevated plus maze.
Mice were placed in the center of a white Plexiglas elevated plus maze (each arm was 33 cm long and 5 cm wide, with 15-cm-high black Plexiglas walls on closed arms) and allowed to explore for 5 min (Powell et al., 2004). The test was conducted in dim white light (∼7 lux). Mice were monitored using CleverSys TopScan Software, and time spent in and entries into the open and closed arms were measured. Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Open field.
The open-field test was performed as described previously (Powell et al., 2004; Blundell et al., 2009), with the exception of the data acquisition software. Mice were monitored using CleverSys TopScan Software after being placed in a white plastic arena (48 × 48 × 48 cm) for 10 min. Time spent in and number of entries into the center of the arena (15 × 15 cm) were recorded in addition to locomotor activity. The test was conducted in dim white light (∼7 lux). Data were analyzed using two-way ANOVA with genotype and sex as between-subject factors.
Locomotor.
Locomotor activity was tested by placing the mice in a fresh home cage with minimal bedding and monitoring their activity for 2 h using photobeams linked to computer data acquisition software (San Diego Instruments; Powell et al., 2004). The test was conducted in the dark. Three-way rmANOVA was used to analyze the data with genotype and sex as between-subject factors and time as a within-subject factor.
Three-chambered social approach.
Social versus inanimate object preference and preference for social novelty analyses were performed in a three-chambered box with small openings connecting the chambers as described previously (Blundell et al., 2009) and based to a large extent on the original descriptions (Moy et al., 2004; Nadler et al., 2004). The test was conducted in dim white light (∼7 lux). The mouse behavior was monitored using CleverSys TopScan Software. This test consisted of three 10-min trials. During the first trial, the mouse was allowed to explore the entire apparatus with empty cages in each end-chamber. In the second trial, the mouse was free to choose between an inanimate cage and a caged, social target. For the third trial, the mouse was free to choose between a caged, novel social target versus a caged, familiar social target. Locations of empty cages and social targets were counterbalanced, and mice were placed back into the home cage for very brief intervals between trials. Data were analyzed using three-way mixed ANOVA with genotype and sex as between-subject factors and target as a within-subject factor.
Social learning.
Social interaction with a juvenile and social learning were performed as described previously (Kwon et al., 2006). Mice were first habituated to a dimly lit testing room (∼7 lux) for 20 min. After the habituation period, an experimental adult mouse and a juvenile target were placed together in a fresh home cage with no bedding. Time spent interacting with the juvenile was recorded live by an observer blind to genotype for 2 min. After 3 d, the above procedure was repeated to assess social learning. Data were analyzed using three-way mixed ANOVA with genotype and sex as between-subject factors and test session as a within-subject factor.
Prepulse inhibition and startle.
As described previously (Blundell et al., 2010a), startle chambers (San Diego Instruments) were modified for mice and mounted atop a piezoelectric accelerometer that detected and transduced animal movements. Acoustic stimuli were delivered by high-frequency speakers mounted 33 cm above the cylinders. Animal movements were digitized and stored using computer software supplied by San Diego Instruments. From the onset of startle stimuli, 65 1-ms readings were recorded, and the amplitude of the startle responses was obtained in arbitrary units. Chambers were calibrated before each set of mice, and sound levels were monitored using a sound meter (Tandy). For prepulse inhibition (PPI), mice were subjected to five trial types in a 22 min session: pulse alone (40 ms, 120 dB, white noise pulse), three different prepulse/pulse trials (20 ms prepulse of 4, 8, or 16 dB above background noise level of 70 dB preceded the 120 dB pulse by 100 ms onset–onset interval), and no stimulus. All trials were presented pseudorandomly with an average of 15 s (7–23 s) between the 62 trials. Testing began with a 5 min acclimation to the cylinders, followed by four blocks of test trials. The first and last blocks consisted of six pulse-alone trials. Blocks 2 and 3 each contained six pulse-alone trials, five of each level of prepulse/pulse trials, and five no-stimulus trials. Data were analyzed for baseline startle amplitude (initial pulse-alone trials) and PPI (percentage decrease in startle amplitude for prepulse/pulse trials compared with pulse-alone trials).
For the startle reactivity test, eight presentations of six trial types were given in pseudorandom order: no stimulus, 80, 90, 100, 110, or 120 dB pulses. Mean startle amplitudes for each condition were calculated. Data were analyzed using three-way mixed ANOVA with genotype and sex as between-subject factors and trial as a within-subject factor.
Grooming.
Mice were placed in a novel home cage without the bedding, and time spent grooming the face, head, or body was measured for 10 min. Number of grooming bouts that lasted >1 s was also recorded. Time per bout was calculated by dividing the total time spent grooming by the number of grooming bouts initiated. Each grooming parameter described above was then analyzed using a two-way ANOVA with genotype and sex as between-subject factors.
USV.
The male adult mouse was placed in a sound-attenuated (11.5 × 11.5 × 11.5 inches) chamber with an estrous female mouse and allowed to freely interact for 5 min while recording USV calls (Scattoni et al., 2010). The chamber was cleaned between trials. The Ultrasound Microphone (Avisoft UltraSoundGate condenser microphone CM16/CMPA; Avisoft Bioacoustics) was set up to record from a range of 10–200 kHz. The microphone was placed ∼5 inches above the floor through a hole in the back wall of the chamber. The recording software was set at default settings with a 250 kHz sampling rate, 16 bit format with a 32 ms buffer. To analyze the calls, the sound file was then converted to a spectrogram using the Avisoft SASLab Pro software. To reduce background noise, 25 kHz was set as a lower cutoff frequency. The analysis involved counting of calls and finding latency to first call. Data were analyzed using one-way ANOVA with genotype as a between-subject factor.
Electrophysiology.
Male mice were briefly anesthetized with isoflurane (Baxter Healthcare Corporation) and rapidly decapitated. Brains were quickly removed and submerged in ice-cold modified ACSF containing in the following (in mm): 75 sucrose, 87 NaCl, 3 KCl, 1.25 NaH2PO4, 7 MgSO4, 26 NaHCO3, 20 dextrose, and 0.5 CaCl2. Acute hippocampal slices 350–400 μm thick were made using a vibrating microtome (Vibratome). A cut was made between CA3 and CA1 to reduce recurrent excitation of CA3 neurons. Slices were allowed to recover at 33°C for 30 min in normal ACSF (in mm: 124 NaCl, 5 KCl, 1.25 NaH2PO4, 1 MgCl2, 26 NaHCO3, 10 dextrose, and 2 CaCl2) and slowly cooled to room temperature over a 45 min period before recording. All solutions were pH 7.4 and saturated with 95% O2/5% CO2.
All recordings were performed at 33 ± 0.5°C, and all data were collected using Clampex (pClamp software suite version 10.2; Molecular Devices). Recordings were filtered at 1 kHz and digitized at 10 kHz. CA3–CA1 synapses were stimulated by a 100 μs biphasic pulse through a monopolar tungsten microelectrode (FHC) placed 400–500 μm laterally from the recording electrode. The distance between the recording electrode and the stimulating electrode was kept constant within these bounds. For extracellular electrophysiology. the recording electrode (1–3 MΩ) was filled with normal ACSF and placed in the stratum radiatium using a SZX7 dissecting microscope (Olympus) at 35× magnification. For whole-cell electrophysiology, the recording electrode (4–6 MΩ) contained the following (in mm): 110 Cs-methanesulfonate, 15 CsCl, 8 NaCl, 10 tetraethylammonium-Cl, 2 EGTA, 10 HEPES, 3 N-(2,6-dimethylphenylcarbamoylmethyl)triethylammonium chloride (QX-314), 2 ATP, and 0.3 GTP. CA1 neurons were visually selected under 80× magnification using an AxioExaminer D1 differential interference contrast microscope (Zeiss).
The majority of experiments were performed on postnatal day 13 (P13) to P16 male mice, except long-term plasticity [long-term potentiation (LTP) and mGluR–long-term depression (LTD)] experiments, which were evaluated in young adult male mice (3–4 or 6–8 weeks, as noted). Sample size for extracellular field recordings [input/output (I/O) curve, paired-pulse ratio (PPR), long-term plasticity] represents number of slices tested with one to two slices used per mouse. Response size was determined by fitting a straight line to the initial slope (10–40%) of the field EPSP (fEPSP) using automated analysis in Clampfit (pClamp software suite version 10.2; Molecular Devices). For studies of LTP and PPR, stimulus intensity was set to generate ∼50% of the maximum fEPSP, as determined by the I/O curve. Stimulus intensity was set at 75–85% of the maximum fEPSP for mGluR–LTD experiments. I/O curves were performed in each slice immediately preceding each field recording, and stimulus intensity remained unchanged thereafter for the duration of the experiment.
Whole-cell patch-clamp recordings were performed in the presence of 100 μm picrotoxin to block fast inhibitory transmission and began 5–10 min (NMDA/AMPA ratio) or 10–15 min [miniature EPSCs (mEPSCs)] after successful break-in. The NMDA/AMPA ratio was measured at 0.1 Hz and elicited no short-term plasticity. For primarily AMPA receptor (AMPAR)-mediated EPSCs, holding potential was −70 mV, and peak amplitude was measured 10–15 ms after stimulus onset. For primarily NMDAR-mediated EPSCs, holding potential was +40 mV, and peak amplitude was measured 40–45 ms after stimulus onset. mEPSCs were recorded for 5 min at a holding potential of −65 mV in the presence of 1 μm octahydro-12-(hydroxymethyl)-2-imino-5,9:7,10a-dimethano-10aH[1,3]dioxocino[6,5-d]pyrimidine-4,7,10,11,12-pentol tetrodotoxin [tetrodotoxin (TTX)] to block evoked transmission. Recordings were rejected if holding current or series resistance varied >25% during the recording. Sample size indicates total number of cells from no less than four mice per group. Raw data were analyzed using Clampfit (pClamp software suite version 10.2; Molecular Devices). GraphPad Prism was used for statistical analysis and graphing. Means are expressed as mean ± SEM.
Histology.
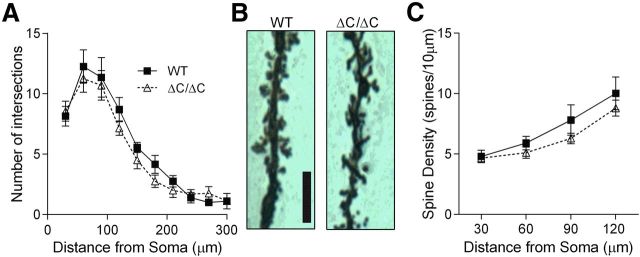
For histological studies, brains were dissected from 6- to 8-week-old mice and processed for Golgi–Cox staining with the FD Rapid GolgiStain Kit (FD Neurotechnologies). After live decapitation, brains were quickly removed and rinsed in double-distilled water and then immersed into impregnation solution, which was then replaced with fresh impregnation solution after 24 h and stored in the dark at room temperature (22–25°C). After 2 weeks, the brains were transferred to solution C and then shipped to FD Neurotechnologies within 48 h. Serial 100 μm cryostat sections were cut coronally through the brain containing the hippocampus and mounted on glass slides. Twenty CA1 neurons from the dorsal hippocampus were selected from five mice per genotype (WT and Shank3ΔC/ΔC). Neurons were traced using NeuroLucida 3D neuron tracing software (MicroBrightField) at 100× magnification by an experimenter blind to experimental conditions. Sholl analysis was conducted using NeuroLucida 3D software to study dendritic branching by drawing concentric circles 30 μm apart starting at 30 μm from the center of the cell body. For spine density experiments, 30 μm apical dendrite segments that did not have any interfering crossings were chosen at defined distances from the cell body (0–30, 30–60, 60–90, and 90–120 μm). Data from the four neurons from each mouse were averaged together before statistical analysis. Data were analyzed using two-way mixed ANOVA with genotype as between-subject factor and distance from the cell body as a within-subject factor.
Drugs.
TTX, picrotoxin, QX-314, and (RS)-3,5-dihydroxyphenylglycine (DHPG) were obtained from Tocris Bioscience. Cs-methanesulfonate and CsCl were obtained from Sigma-Aldrich. All other reagents were obtained from Thermo Fisher Scientific.
Results
Homozygous Shank3 mutation results in loss of major Shank3 isoforms
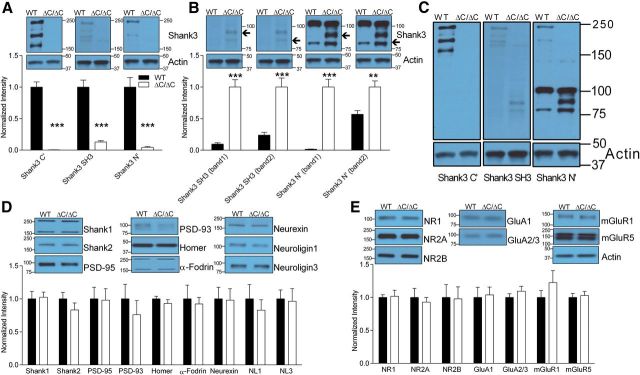
To quantify the loss of Shank3 in Shank3ΔC/ΔC mice compared with WT littermate controls, we performed quantitative Western blot analysis using antibodies directed against Shank3 C terminus (C′), N terminus (N′), and the SH3 domain (SH3) on hippocampal lysates from 5- to 6-month-old mice (n = 7). Western blot analysis reveals that Shank3ΔC/ΔC mice have lost major Shank3 isoforms detected by the C-terminal, SH3 domain, and N-terminal antibodies [Fig. 1A; Shank3(C′): t(12) = 11.418, p < 0.0001; Shank3(SH3): t(12) = 8.524, p < 0.0001; Shank3(N′): t(12) = 5.244, p < 0.001]. Interestingly, we also observe the appearance of lower-molecular-weight (<100 kDa) isoforms of Shank3 detected by the SH3 domain and the N-terminal antibodies in Shank3ΔC/ΔC hippocampus that are not substantially present in the WT controls [Fig. 1B; Shank3(SH3 band 1): t(12) = 8.199, p < 0.0001; Shank3(SH3 band 2): t(12) = 5.818, p < 0.0001; Shank3(N′ band 1): t(12) = 11.303, p < 0.0001; Shank3(N′ band 2): t(12) = 5.494, p < 0.001]. Figure 1C depicts longer representative Western blots with each of the three antibodies for reference.
Figure 1.
Loss of major naturally occurring Shank3 proteins in Shank3ΔC/ΔC mice. A, Quantification and representative Western blots of hippocampal lysates showing loss of the major isoforms of Shank3 with C′ (P. Worley, JH3025), SH3 (Abcam), and N′ (P. Worley) antibodies. B, Quantification and representative Western blots of lower-molecular-weight bands that appear or increase in intensity in the Shank3ΔC/ΔC hippocampal lysates using SH3 domain and N′ antibodies. C, Larger representative Western blots showing the comparison of bands detected by the three Shank3 antibodies. D, E, Quantification and representative blots of whole hippocampus lysates with antibodies against PSD proteins and receptors that interact directly or indirectly with Shank3. In A, D, and E, data are normalized to β-actin control and then to average of WT levels. In B, data are normalized to β-actin control and then to average of Shank3ΔC/ΔC. Error bars represent SEM, **p < 0.01, ***p < 0.001.
In addition to Shank3, we also probed for Shank1, Shank2, and multiple other PSD proteins and receptors that bind directly or indirectly to Shank3 to identify any compensatory effects of Shank3 loss from hippocampal lysates. We did not observe any changes in levels of PSD proteins or synaptic receptor subunits in whole hippocampal homogenates (Fig. 1D,E).
Homozygous Shank3 mutants exhibit increased mGluR5 in synaptic fractions
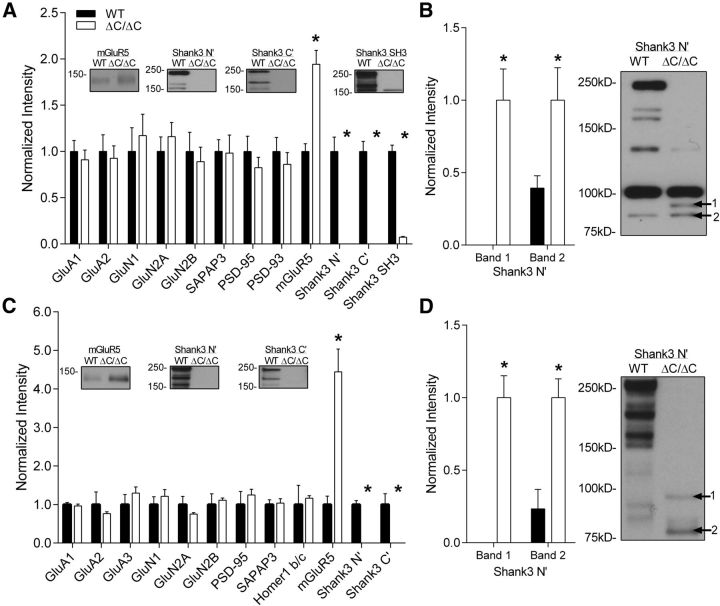
To determine the effects of a complete loss of Shank3 on the subcellular distribution of synaptic proteins, we isolated the synaptosome and postsynaptic density (PSDII) fractions of the hippocampus of Shank3ΔC/ΔC mice compared with WT littermate controls following established methods with modifications (Cohen et al., 1977). As in the whole hippocampal lysates, we found dramatic loss of all three Shank3 isoforms in the synaptosome fractions [Fig. 2A; Shank3(N′): t(12) = 5.514, p < 0.001; Shank3(C′): t(12) = 7.929, p < 0.001; Shank3(SH3): t(12) = 11.552, p < 0.001]. We also detected the presence of lower-molecular-weight (<100 kDa) isoforms of Shank3 in synaptosomes using the N-terminal antibody [Fig. 2B; Shank3(N′ band 1): t(12) = 5.417, p < 0.001; Shank3(N′ band 2): t(12) = 2.81256, p = 0.016]. Similar loss of Shank3 isoforms was observed in the PSDII fractions [Fig. 2C; Shank3(N′): t(4) = 9.82, p < 0.001; Shank3(C′): t(4) = 3.575, p = 0.023] along with the presence of lower-molecular-weight isoforms of Shank3 in PSDs [Fig. 2D; Shank3(N′ band 1): t(4) = 6.594, p = 0.003; Shank3(N′ band 2): t(4) = 4.114, p = 0.015] using the N-terminal antibody, suggesting that these lower-molecular-weight isoforms are present at hippocampal synapses.
Figure 2.
Increased mGluR5 protein levels in hippocampal synaptosome and PSDII fractions of Shank3ΔC/ΔC mice. A, Quantification of PSD proteins in synaptosome fractions of the hippocampus shows a complete loss of the major isoforms of Shank3 using the C terminus, N terminus, and SH3 domain antibodies against Shank3 in Shank3ΔC/ΔC mice (*p < 0.0001) as well as an increase in mGluR5 (*p < 0.0001) compared with WT (WT, n = 8; ΔC/ΔC, n = 6). Representative blots are shown inset for proteins showing significant differences. B, Quantification of the <100 kDa bands that appear or increase in the hippocampal synaptosomes of Shank3ΔC/ΔC mice. C, Quantification of PSD proteins in PSDII fractions of the hippocampus shows a complete loss of Shank3 using the C-terminal antibody (*p < 0.05) as well as the N-terminal antibody (*p < 0.001) of Shank3 in Shank3ΔC/ΔC mice as well as a robust increase in mGluR5 (*p < 0.01) compared with WT (for each group, n = 3 sets of hippocampi pooled from 2 mice each). D, Quantification and representative Western blot of the <100 kDa bands that appear or increase in the hippocampal PSDII fraction of Shank3ΔC/ΔC mice. For A–D, data were normalized to β-actin levels and then to the average of WT (A, C) or ΔC/ΔC (B, D). Data shown as average ± SEM. Representative blots are shown inset for proteins showing significant differences.
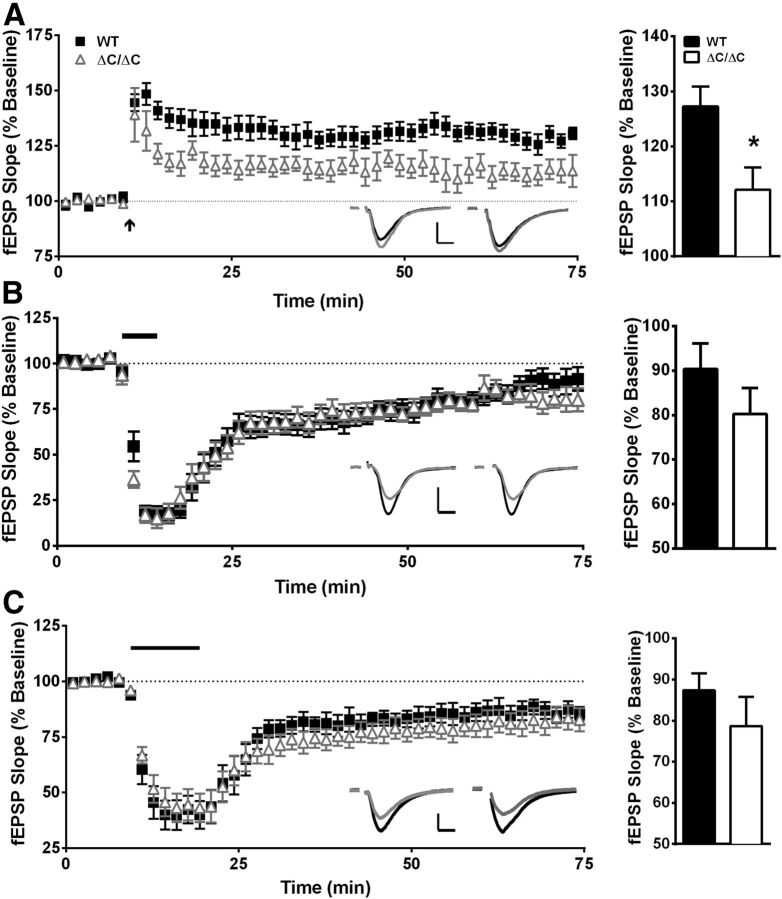
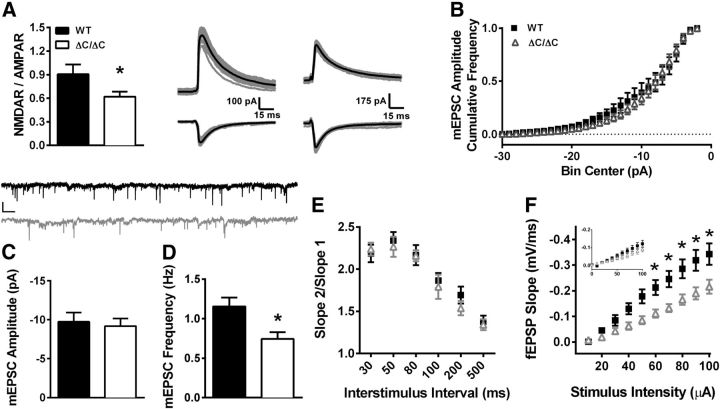
In addition to Shank3, we analyzed the levels of synaptic proteins and receptors that are either directly or indirectly bound to Shank3 in the hippocampus. Strikingly, a significant increase in mGluR5 was seen in the synaptosome fraction (Fig. 2A; t(12) = 5.867, p < 0.001) and to an even greater extent in the PSDII fraction (Fig. 2C; t(4) = 5.465, p = 0.005), suggesting enhanced mGluR5 localization to hippocampal synapses. We found no significant differences in ionotropic glutamate receptor subunits (NMDA or AMPA) or in other scaffolding and linker proteins (Fig. 2A,C) in the hippocampus of Shank3ΔC/ΔC mice.
Homozygous Shank3 mutants are impaired in spatial learning
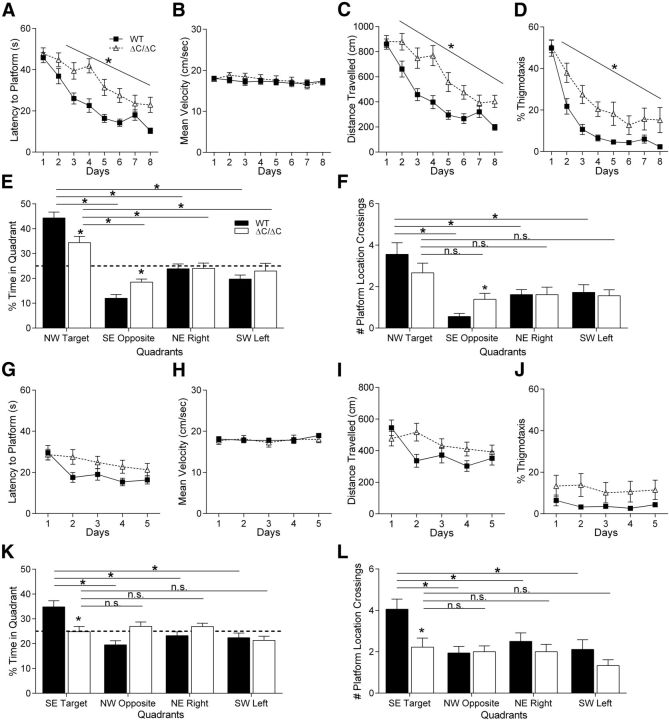
Because 40–50% of autism patients exhibit intellectual disability (Wingate et al., 2012) and a majority of patients with SHANK3 mutation/deletion exhibit some degree of intellectual disability (Gong et al., 2012), we tested the Shank3ΔC/ΔC mice in the Morris water maze task, a test of spatial learning and memory. Shank3ΔC/ΔC mice were significantly impaired in spatial learning compared with WT littermate controls using latency to reach the hidden platform as a measure (Fig. 3A; three-way rmANOVA; main effect of genotype: F(1,32) = 13.55, p < 0.001; main effect of day: F(7,224) = 36.47, p < 0.000001; genotype × day interaction: F(7,224) = 2.54, p < 0.05; for complete statistical results for all experiments, see Table 1). The increased latency to reach the hidden platform in Shank3ΔC/ΔC mice was not accounted for by overall differences in swim speed during training (Fig. 3B; three-way rmANOVA; main effect of genotype: F(1,32) = 0.5, p = 0.48; main effect of day: F(7,224) = 2.00, p = 0.05; genotype × day interaction: F(7,224) = 0.92, p = 0.48). Interestingly, there was a significant genotype × sex interaction in swim speed (Table 1). ANOVA results revealed that the Shank3ΔC/ΔC female group had significantly higher swim speed during training compared with WT females (genotype × sex interaction; Table 1).
Figure 3.
Shank3ΔC/ΔC mice exhibit impaired spatial learning. A–D, Training days for the Morris water maze task. For each day of training, data were averaged across four daily trials. A, Latency to reach hidden platform on successive water maze days. Shank3ΔC/ΔC mice take longer to reach the submerged platform. B, Swim speed on successive water maze training days. The average swim speed was unchanged in Shank3ΔC/ΔC mice. C, Distance traveled before reaching the hidden platform on successive water maze training days. Shank3ΔC/ΔC mice travel a more circuitous route (longer distance) before reaching the submerged platform. D, Percentage time spent in thigmotaxis on successive water maze training days. E, Time spent in target quadrant and other quadrants during probe trial in which target platform is removed. Shank3ΔC/ΔC mice spend more time in the target quadrant versus other quadrants but less time in target quadrant compared with littermate controls. F, Number of target location crossings and corresponding phantom platform location crossings in other quadrants during the probe trial. Shank3ΔC/ΔC mice fail to show a preference for the target platform location. G–J, Training trials for the Morris water maze reversal task. No differences were observed during training for the Morris water maze reversal learning task in latency to platform (G), mean swim speed (H), distance traveled (I), or thigmotaxis (J). K, On the probe trial for the Morris water maze reversal task, Shank3ΔC/ΔC mice failed to show preference for the target quadrant and spent equal time in all four quadrants. L, On the probe trial for the Morris water maze reversal task, Shank3ΔC/ΔC mice did not show preference for the target platform location (n = 18 in all panels, data depicted as average ± SEM, *p < 0.05).
Not surprisingly, Shank3ΔC/ΔC mice also exhibited significantly decreased learning using distance traveled to reach the platform, a measure that eliminates swim speed as a confound (Fig. 3C; three-way rmANOVA; main effect of genotype: F(1,32) = 17.41, p < 0.001; main effect of day: F(7,224) = 41.61, p < 0.000001; genotype × day interaction: F(7,224) = 3.25, p < 0.005). On the initial trials, Shank3ΔC/ΔC and WT littermates spent the same amount of time in thigmotaxis (swimming near the maze walls). As the WT mice learned, they progressed to alternative search strategies, whereas the Shank3ΔC/ΔC mice were slower to shift to alternative search strategies from the initial thigmotaxic strategy (Fig. 3D; three-way rmANOVA; main effect of genotype: F(1,32) = 7.62, p < 0.01; main effect of day: F(7,224) = 75.09, p < 0.000001; genotype × day interaction: F(7,224) = 2.41, p < 0.05). These findings are consistent with a significant decrease in spatial learning in the Shank3ΔC/ΔC mice.
Although Shank3ΔC/ΔC mice were slower to learn the water maze task, their spatial memory performance was only somewhat affected during the initial probe trial. Both WT and Shank3ΔC/ΔC mice showed a significant preference for the target quadrant compared with all three other quadrants [Fig. 3E; three-way rmANOVA; main effect of genotype: F(1,32) = 0.22, p = 0.63; main effect of quadrant: F(3,96) = 33.49, p < 0.000001; genotype × quadrant interaction: F(3,96) = 4.21, p < 0.01; planned comparisons for WT mice: TargetNW vs RightNE: p < 0.00001; TargetNW vs LeftSW: p < 0.00001; TargetNW vs OppositeSE: p < 0.00001; planned comparisons for Shank3ΔC/ΔC mice: TargetNW vs RightNE: p < 0.02; TargetNW vs LeftSW: p < 0.02; TargetNW vs OppositeSE: p < 0.0001 (SE indicates southeast, SW indicates southwest, NE indicates northeast, and NW indicates northwest)], indicating that both groups were able to recall a previously learned spatial strategy. However, there was a significant decrease in the percentage time Shank3ΔC/ΔC mice spent in the target quadrant and a significant increase in the percentage time spent in the opposite quadrant compared with WT littermates (planned comparisons for WT vs Shank3ΔC/ΔC mice: TargetNW: p < 0.01; OppositeSE: p < 0.01), suggesting that Shank3ΔC/ΔC mutants may not have learned as efficient a spatial strategy as their WT littermate counterparts. To examine this further, we analyzed the number of times the mice crossed the exact target platform location and corresponding locations in the other three quadrants. Using this measure, only the WT group demonstrated a significant preference for the target platform location compared with the three other corresponding locations (Fig. 3F; three-way rmANOVA; main effect of genotype: F(1,32) = 0.01, p = 0.91; main effect of quadrant: F(3,96) = 13.00, p < 0.000001; genotype × quadrant interaction: F(3,96) = 2.13, p = 0.10; planned comparisons for WT mice: TargetNW vs RightNE, p < 0.001; TargetNW vs LeftSW, p < 0.01; TargetNW vs OppositeSE, p < 0.0001; planned comparisons for Shank3ΔC/ΔC mice: TargetNW vs RightNE, p = 0.08; TargetNW vs LeftSW, p = 0.08; TargetNW vs OppositeSE, p = 0.052). Statistical analysis also demonstrated a genotype × sex interaction that, on closer analysis, suggests that the inability to show a preference for target platform location can be attributed to the Shank3ΔC/ΔC mutant males because, during the probe trial, they show a decrease in the number of platform location crosses (data not shown). Together, these data suggest that Shank3ΔC/ΔC mice are slower to learn a spatial strategy in the water maze compared with WT littermate controls.
To examine for both cognitive flexibility (Ozonoff and Jensen, 1999) and insistence on sameness (Schreibman, 1988; Greaves et al., 2006; Richler et al., 2007) as well as an additional test of spatial learning/memory, we performed a reversal learning experiment using the water maze, identifying additional evidence of significantly decreased spatial memory or cognitive inflexibility in Shank3ΔC/ΔC mice. The day after the first probe trial, mice were trained for 5 additional days with the hidden platform in the opposite quadrant. Interestingly, Shank3ΔC/ΔC mice showed no difference in acquisition of the reversal task using latency to reach the platform (Fig. 3G; three-way rmANOVA; main effect of genotype: F(1,32) = 2.63, p = 0.11; main effect of day: F(4,128) = 8.05, p < 0.0009; genotype × day interaction: F(4,128) = 1.69, p = 0.15). Likewise, no differences were observed in swim speed (Fig. 3H; three-way rmANOVA; main effect of genotype: F(1,32) = 0.006, p = 0.93; main effect of day: F(4,128) = 1.08, p = 0.36; sex × genotype interaction: F(1,32) = 3.05, p = 0.09; genotype × day interaction: F(4,128) = 0.68, p = 0.60), distance traveled to reach the platform (Fig. 3I; three-way rmANOVA; main effect of genotype: F(1,32) = 2.08, p = 0.15; main effect of day: F(4,128) = 4.99, p < 0.0009; genotype × day interaction: F(4,128) = 2.64, p < 0.05), or the time spent in thigmotaxis (Fig. 3J; three-way rmANOVA; main effect of genotype: F(1,32) = 2.29, p = 0.13; main effect of day: F(4,128) = 1.50, p = 0.20; genotype × day interaction: F(4,128) = 0.57, p = 0.68).
However, on the probe trial, Shank3ΔC/ΔC mice failed to show any preference for the new target location, whereas WT littermates showed a clear preference for the target quadrant compared with all other quadrants (Fig. 3K; three-way rmANOVA; main effect of genotype: F(1,32) = 0.83, p = 0.36; main effect of quadrant: F(3,96) = 5.87, p < 0.01; genotype × quadrant interaction: F(3,96) = 6.51, p < 0.001; planned comparisons for WT mice: TargetSE vs RightNE, p < 0.001; TargetSE vs LeftSW, p < 0.001; TargetSE vs OppositeNW, p < 0.0001; planned comparisons for Shank3ΔC/ΔC mice: TargetSE vs RightNE, p = 0.55; TargetSE vs LeftSW, p = 0.32; TargetSE vs OppositeNW, p = 0.60). Decreased spatial memory in the reversal learning task was confirmed using number of platform location crossings as a measure, with WT littermates demonstrating a clear preference for the target location over all other locations and the Shank3ΔC/ΔC mice showing no preference (Fig. 3L; three-way rmANOVA; main effect of genotype: F(1,32) = 6.11, p < 0.02; main effect of quadrant: F(3,96) = 4.93, p < 0.01; genotype × quadrant interaction: F(3,96) = 1.98, p = 0.12; planned comparisons for WT mice: TargetSE vs RightNE, p < 0.05; TargetSE vs LeftSW, p < 0.01; TargetSE vs OppositeNW, p < 0.01; planned comparisons for Shank3ΔC/ΔC mice: TargetSE vs RightNE, p = 0.66; TargetSE vs LeftSW, p = 0.20; TargetSE vs OppositeNW, p = 0.68). These data are consistent with decreased spatial learning and memory and may or may not represent additional difficulties with cognitive flexibility or behavioral inflexibility. To test basic visual function and comprehension of the task, we measured latency and distance traveled to reach a visible cue atop the platform in the water maze. No significant differences were observed, but there was a trend toward increased latency and distance traveled to reach the visible cue.
Homozygous Shank3 mutants have impaired motor coordination
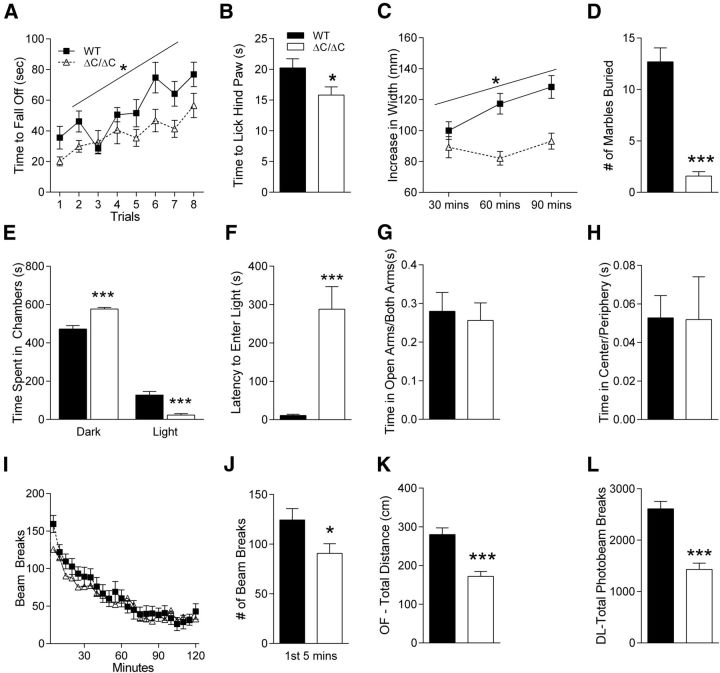
Shank3ΔC/ΔC mice exhibit impaired coordination on the accelerating rotarod. Shank3ΔC/ΔC mice had consistently decreased latencies to fall from the rotarod apparatus, although they demonstrated the same rate of motor learning indicated by improvement in their ability over subsequent trials (Fig. 4A; three-way rmANOVA; main effect of sex: F(1,34) = 7.17, p < 0.02; main effect of genotype: F(1,34) = 6.95, p < 0.02; main effect of trial: F(7,238) = 11.71, p < 0.000001; sex × genotype interaction: F(1,34) = 0.45, p = 0.50; sex × trial interaction: F(7,238) = 0.51, p = 0.82; genotype × trial: F(7,238) = 1.60, p = 0.13; sex × genotype × trial interaction: F(7,238) = 1.88, p = 0.07). Interestingly, in addition to the main effect of genotype, there was also a gender difference in performance that appeared to be attributable to the WT females because they were able to stay on the rotarod longer than the WT males (planned comparisons: females, WT vs Shank3ΔC/ΔC, p < 0.05; males, WT vs Shank3ΔC/ΔC, p = 0.162; WT, males vs females, p < 0.05; Shank3ΔC/ΔC, males vs females, p = 0.164).
Figure 4.
Shank3ΔC/ΔC mice exhibit impairments in other behavioral tasks. A, Latency to fall from or to go one full revolution on the rotarod task. Shank3ΔC/ΔC mice exhibit motor coordination impairments in eight trials of rotarod test conducted over 2 d (n = 19). Legend in A applies to C and I. B, Latency to lick hindpaw on the hotplate task. Shank3ΔC/ΔC mice show hypersensitivity to heat on a hotplate (n = 18). Legend in B applies to D–H and J–L. C, Width of nest built as a function of time in a nest-building task. Shank3ΔC/ΔC mice exhibit impairments in nest-building behavior over a 90 min period (n = 19). D, Number of marbles buried during a 30 min marble-burying task. Shank3ΔC/ΔC mice show impaired marble burying behavior (n = 19). E, Time spent in dark and light chambers during dark/light task. Shank3ΔC/ΔC mice spend more time in the dark than littermate controls (n = 19). F, Latency to enter the light chamber in the dark/light task. Shank3ΔC/ΔC mice exhibit dramatically increased latency to enter the light side (n = 19). G, Fraction of time in the open arms versus time in other arms in the elevated plus maze task. Shank3ΔC/ΔC mice spend the same time in open versus closed arms when compared with littermate controls (n = 18). H, Ratio of time spent in the center to time spent in the periphery in an open-field task. Shank3ΔC/ΔC mice behave the same as littermate controls (n = 19). I, Locomotor activity as measured by number of photobeam breaks during successive 5 min intervals over a 2 h period. Shank3ΔC/ΔC mice exhibit normal locomotor habituation over the full 2 h period (n = 19). J, Number of photobeam breaks during the initial 5 min of the locomotor task shown in I. Shank3ΔC/ΔC mice show decreased activity, initially suggesting abnormal locomotor response to novelty (n = 19). K, Total distance traveled during the 10 min open-field task. Shank3ΔC/ΔC mice have decreased locomotor activity in the open field (n = 19). L, Number of photobeam breaks during the 10 min dark/light task. Shank3ΔC/ΔC mice have decreased locomotor activity in dark/light (n = 19). Error bars represent SEM, *p < 0.05.
Shank3 mutant mice are hypersensitive in the hotplate task
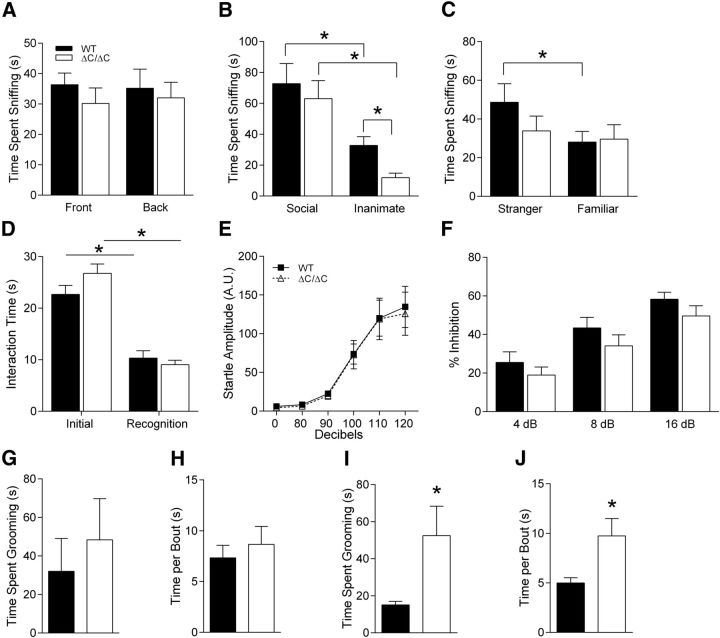
Some patients with Phelan–McDermid syndrome or 22q13 deletion syndrome are anecdotally said to have increased pain tolerance. Therefore, we tested the sensitivity of Shank3ΔC/ΔC mice to pain by testing the latency to lick the hindpaw on the hotplate task and found that it was decreased, suggesting a hypersensitivity to heat (Fig. 4B; two-way ANOVA; main effect of genotype: F(1,32) = 4.38, p < 0.05).
Shank3 mutant mice exhibit an avoidance phenotype toward inanimate objects
In attempts to measure nest building, marble burying, and other tasks, we uncovered an interesting avoidance of inanimate objects in the Shank3ΔC/ΔC mice. When we measured nest building by adding a nestlet to a novel cage after a habituation period, we found that Shank3ΔC/ΔC mice showed essentially no change in their nestlets, whereas WT littermates readily made nests from the material (Fig. 4C; three-way rmANOVA; main effect of genotype: F(1,34) = 17.06, p < 0.001; main effect of time: F(2,68) = 7.60, p < 0.01; genotype × time interaction: F(2,68) = 6.00, p < 0.004). At first, we interpreted this as deficient nest building, but later tasks revealed a potential alternative explanation.
Surprisingly, similar avoidance behavior was observed in the marble-burying task. Shank3ΔC/ΔC mice showed little to no interest in burying marbles (Fig. 4D; two-way ANOVA; main effect of genotype: F(1,34) = 58.20, p < 0.000001). In many instances, it appeared as if Shank3ΔC/ΔC mice did not even touch or walk over the marbles because they appeared completely undisturbed. A potentially related phenotype was observed in the three-chamber social interaction task wherein the Shank3ΔC/ΔC mice interacted significantly less with the inanimate object than WT littermate controls (see below and Fig. 5B).
Figure 5.
Shank3ΔC/ΔC mice exhibit minimal social interaction deficits and normal startle reactivity and PPI. A, Time spent in chambers with empty cages. For the first trial of three-chambered social interaction test, Shank3ΔC/ΔC mice were allowed to explore a three-chambered apparatus and showed no initial preference for either end of the box (n = 17). Legend in A applies to B–D and F. B, In the second trial when given a choice between social or inanimate target, both WT and Shank3ΔC/ΔC mice show a preference for a caged social target versus inanimate object. However, Shank3ΔC/ΔC mice avoided the inanimate object and spend less time sniffing it than the WT group (n = 17). C, In the third trial, when given a choice between novel social target versus a familiar social target, Shank3ΔC/ΔC mice failed to show a preference for the novel social target, unlike their WT littermate pairs (n = 17). D, Shank3ΔC/ΔC mice show normal social interaction with a juvenile conspecific mouse and, when presented with the same mouse 3 d later, exhibit normal social memory (n = 18). Shank3ΔC/ΔC mice exhibit normal response to startle (E) and show no deficits in PPI (E; n = 18). Shank3ΔC/ΔC mice show no change in total time spent in repetitive grooming behavior (G) or in time spent grooming per bout (H; n = 19). However, when tested at an older age, Shank3ΔC/ΔC mice show a significant increase in overall time spent grooming (I) and time spent grooming per bout (J; n = 16). Error bars represent SEM, *p < 0.05.
We also tested Shank3ΔC/ΔC mice in anxiety behaviors and found that these mice show a remarkable increase in avoidance of the brightly lit chamber in the dark/light task. Shank3ΔC/ΔC mice spent almost the entire 10 min of this task in the dark chamber, virtually completely avoiding the brightly lit chamber (Fig. 4E; time spent in dark side, two-way ANOVA; main effect of genotype: F(1,34) = 29.06, p < 0.00006; time spent in light side, two-way ANOVA; main effect of genotype: F(1,34) = 29.06, p < 0.00006). This avoidance behavior is also evident by the increased latency to enter the brightly lit side of the box (Fig. 4F; two-way ANOVA; main effect of genotype: F(1,34) = 21.08, p < 0.00006). Such behavior in this task is typically interpreted as an increase in anxiety-like behavior. To differentiate between increased anxiety-like behavior versus avoidance behavior, we tested Shank3ΔC/ΔC mice in two other standard tests for anxiety: elevated plus maze and open field. We did not observe an increase in anxiety-related behavior in the elevated plus maze because Shank3ΔC/ΔC mice and their WT littermate counterparts spent equal time in the open arms (Fig. 4G; two-way ANOVA; main effect of genotype: F(1,32) = 0.12, p = 0.73). Similarly, no anxiety-like phenotype was observed in the open-field task because the Shank3ΔC/ΔC mice spent the same amount of time in the center as their WT littermates (Fig. 4H; two-way ANOVA; main effect of genotype: F(1,34) = 0.003, p = 0.95). Thus, it appears that Shank3ΔC/ΔC mice do not have an anxiety phenotype but demonstrate avoidance behavior in multiple tasks, including avoiding the brightly lit chamber in the dark/light box test.
Shank3 mutant mice exhibit aberrant locomotor activity in response to novelty
Curiously, in some tasks, but not others, Shank3ΔC/ΔC mice demonstrated decreased locomotor activity that can be interpreted as a transiently decreased locomotor response to novel situations. At first glance, when examining locomotor activity over 2 h in a novel home cage, Shank3ΔC/ΔC mice demonstrated completely normal locomotor activity (Fig. 4I; three-way rmANOVA; main effect of genotype: F(1,34) = 0.49, p = 0.48; main effect of trial: F(7,782) = 41.30, p < 0.000001; genotype × trial: F(7,782) = 1.21, p = 0.22). For the first 5 min in this novel home cage situation, however, the Shank3ΔC/ΔC mice showed a significant decrease in locomotor activity that reverted to the WT level rapidly thereafter (Fig. 4J; two-way ANOVA; main effect of genotype: F(1,34) = 4.43, p < 0.05). Along the same lines, distance traveled in the novel open-field arena over 10 min was decreased in Shank3ΔC/ΔC mice (Fig. 4K; two-way ANOVA; main effect of genotype: F(1,34) = 23.54, p < 0.00003), and total number of photobeams interrupted during the 10 min in the dark/light chamber was decreased in Shank3ΔC/ΔC mice (Fig. 4L; two-way ANOVA; main effect of genotype: F(1,34) = 35.60, p < 0.000002). We interpret these data overall as a decrease in locomotor response to novel environments that reverts rapidly to WT levels.
Shank3 mutant mice exhibit minimal social interaction deficits
Social interaction deficits are one of the three characteristic features of autism (Schreibman, 1988; Mahjouri and Lord, 2012). Therefore, we tested social behaviors in Shank3ΔC/ΔC mice in two separate tasks. Much like their WT littermate pairs, Shank3ΔC/ΔC mice showed no a priori preference for either side in the three-chambered social interaction box before introduction of a social target (Fig. 5A; three-way rmANOVA: main effect of genotype: F(1,30) = 1.23, p = 0.27; main effect of target: F(1,30) = 0.01, p = 0.90; genotype × target interaction: F(1,30) = 0.06, p = 0.80). Similarly, in the test for social versus inanimate preference, both WT and Shank3ΔC/ΔC spent more time interacting with the social target (Fig. 5B; three-way rmANOVA: main effect of genotype: F(1,30) = 2.95, p = 0.09; main effect of target: F(1,30) = 20.81, p < 0.0001; genotype × target interaction: F(1,30) = 0.36, p = 0.54; planned comparisons, social vs inanimate: WT, p < 0.01; Shank3ΔC/ΔC, p < 0.001). However, in the test of preference for social novelty versus familiarity, Shank3ΔC/ΔC mice showed no preference for social novelty, unlike their WT littermate pairs (Fig. 5C; three-way rmANOVA: main effect of genotype: F(1,30) = 0.56, p = 0.45; main effect of target: F(1,30) = 4.16, p = 0.05; genotype × target interaction: F(1,30) = 1.68, p = 0.20; planned comparisons for WT mice: social vs inanimate, p < 0.05; planned comparisons for Shank3ΔC/ΔC mice: social vs inanimate, p = 0.23).
No differences were observed in either social interaction or social learning during reciprocal social interaction with a juvenile (Fig. 5D; three-way rmANOVA; main effect of genotype: F(1,32) = 0.502, p = 0.48; main effect of trial: F(1,32) = 138.76, p < 0.000001; genotype × trial interaction: F(1,32) = 3.88, p = 0.05; planned comparisons, initial vs recognition: WT, p < 0.00001; Shank3ΔC/ΔC, p < 0.00001).
Shank3 mutant mice exhibit normal startle response and PPI
SHANK3 and other autism genes are implicated in schizophrenia (Burbach and van der Zwaag, 2009; Gauthier et al., 2009, 2010; Verpelli et al., 2012). Thus, we tested Shank3ΔC/ΔC mice for PPI deficits and auditory startle responses. We observed no differences in the auditory startle response at the tested sound levels (Fig. 5E; three-way rmANOVA; main effect of genotype: F(1,32) = 0.008, p = 0.92; main effect of trial: F(5,160) = 34.95, p < 0.0001; genotype × decibel interaction: F(5,160) = 0.03, p = 0.99). Similarly, there was no difference in PPI between the WT and Shank3ΔC/ΔC mice (Fig. 5F; three-way rmANOVA; main effect of genotype: F(1,34) = 3.06, p = 0.08; main effect of trial: F(2,64) = 54.86, p < 0.0001; genotype × trial interaction: F(2,64) = 0.20, p = 0.81). We did observe a significant interaction between sex and genotype, and additional analysis revealed that the Shank3ΔC/ΔC female mice show less PPI than Shank3ΔC/ΔC males or WT females (data not shown; for detailed statistics, see Table 1).
Older, but not younger, Shank3 mutant mice exhibit increased grooming
We characterized grooming behavior in Shank3ΔC/ΔC, as a measure of the repetitive stereotyped behavior core symptom domain of autism. When tested at a younger age (9–18 weeks old), Shank3ΔC/ΔC mice did not exhibit an increase in total time spent grooming (Fig. 5G; two-way ANOVA; main effect of genotype: F(1,34) = 0.36, p = 0.54). Similarly, no difference was observed in the time spent per grooming bout (Fig. 5H; two-way ANOVA; main effect of genotype: F(1,34) = 0.44, p = 0.50).
However, when grooming was tested in older mice (10–13 months old), Shank3ΔC/ΔC mutants exhibited a significant increase in grooming when compared with WT (Fig. 5I; two-way ANOVA; main effect of genotype: F(1,28) = 4.69, p < 0.04). The older Shank3ΔC/ΔC mutants also showed an increase in time spent per grooming bout (Fig. 5J; two-way ANOVA; main effect of genotype: F(1,28) = 6.00, p < 0.03).
Shank3 mutant mice do not exhibit communication deficits
Communication deficits are also a hallmark of autism. To test communication, adult Shank3ΔC/ΔC mutant males were allowed to mate with free-roaming estrous females, and their USVs were recorded. No differences were observed in the latency to emit the first call or the total number of calls emitted in a 5 min recording period (data not shown; for statistical analysis, see Table 1).
Shank3 mutant mice are impaired in hippocampal synaptic transmission and plasticity
Because Shank3ΔC/ΔC mice exhibited significant deficits in hippocampus-dependent spatial learning, we examined long-term synaptic plasticity in the hippocampus. Extracellular “field” recordings made in the CA1 region of the hippocampus revealed a deficit in long-term plasticity (Fig. 6) that is consistent with the spatial learning deficits in Shank3ΔC/ΔC mice. LTP was significantly impaired in Shank3ΔC/ΔC mice compared with WT mice at 55–60 min after a 100 Hz, 1 s conditioning stimulus (Fig. 6A; WT, 127.2 ± 3.64%, n = 8 vs Shank3ΔC/ΔC, 112.1 ± 4.08%, n = 6; t(12) = 2.753, p = 0.018).
Figure 6.
Synaptic plasticity at hippocampal CA3–CA1 synapses is altered in Shank3ΔC/ΔC mice. A, LTP is decreased in Shank3ΔC/ΔC mice (n = 6) compared with WT controls (n = 8). Arrow indicates onset of 100 Hz train for 1 s. Inset, Average of 15 consecutive traces immediately before (black) and 60 min after (gray) 100 Hz tetanus. Calibration: 0.3 mV (WT) or 0.55 mV (Shank3ΔC/ΔC), 5 ms. Legends in A also apply to B and C. B, mGluR–LTD from 6- to 8-week-old mice is not significantly affected by exon 21 deletion (WT, n = 9; Shank3ΔC/ΔC, n = 7). Bar indicates 5 min bath application of DHPG. Inset, Average of 15 consecutive traces immediately before DHPG wash-in (black) and 60 min after the start of DHPG washout (gray). Calibration: 0.3 mV, 5 ms. C, There no significant difference in mGluR–LTD from 3- to 4-week-old Shank3ΔC/ΔC mice (n = 11) compared with WT (n = 9). Bar indicates 10 min bath application of DHPG. Inset, Average of 15 consecutive traces immediately before DHPG wash-in (black) and 60 min after the start of DHPG washout (gray). Calibration: 0.3 mV (WT) or 0.22 mV (Shank3ΔC/ΔC), 5 ms. Error bars represent SEM, *p < 0.05.
Shank3 also binds indirectly to group 1 mGluRs through interactions with Homer at the C-terminus proline-rich region (Tu et al., 1999). Specifically, Shank3 has been shown to regulate mGluR5 signaling in hippocampal neuronal cultures, and knockdown of Shank3 in cultured neurons causes a decrease in mGluR5 signaling and mEPSC frequency (Verpelli et al., 2011). However, in our Shank3ΔC/ΔC homozygous mice, we identified a significant increase in mGluR5 in synaptosome and PSDII fractions (Fig. 2A,C).
To determine whether mGluR5 signaling is altered in Shank3ΔC/ΔC mice, we first induced mGluR–LTD using a 5 min bath application of the group I mGluR agonist DHPG (100 μm) in 6- to 8-week-old mice. DHPG was used no more than 10 d after stock was prepared, and LTD experiments were performed with the experimenter blind to genotype and in an interleaved manner (alternating WT and mutant experiments) with no more than two slices per mouse. At 55–60 min after washout began (Fig. 6B), we found that modest LTD was induced in WT mice (90.36 ± 5.73%, n = 9) and in Shank3ΔC/ΔC mice (80.24 ± 5.84%, n = 7), but no difference in percentage LTD between WT and Shank3ΔC/ΔC mice was observed (t(18) = 0.996, p = 0.243). To optimize expression of mGluR–LTD, we repeated the experiment in 3- to 4-week-old mice with a 10 min bath application of DHPG (Fig. 6C). With a larger sample size and doubling of DHPG application time, we again found no difference in mGluR–LTD between Shank3ΔC/ΔC mice and WT controls (WT, 87.38 ± 4.14%, n = 9 vs Shank3ΔC/ΔC, 78.67 ± 7.12%, n = 11; t(14) = 1.219, p = 0.332).
Because long-term plasticity is dependent on proper function of glutamatergic AMPARs and NMDARs and because Shank3 is known to interact indirectly with both types of receptors (Naisbitt et al., 1999; Uchino et al., 2006; Arons et al., 2012), we hypothesized that the relative contributions of AMPARs and NMDARs to the EPSC would be altered in Shank3ΔC/ΔC mice. As predicted, there was a significant decrease in the NMDA/AMPA ratio in Shank3ΔC/ΔC compared with WT mice (Fig. 7A; WT, 0.91 ± 0.12, n = 22 vs Shank3ΔC/ΔC, 0.62 ± 0.06, n = 25; t(45) = 2.129, p = 0.039).
Figure 7.
Synaptic transmission is altered at hippocampal CA3–CA1 synapses in Shank3ΔC/ΔC mice. A, The NMDA/AMPA ratio is decreased in Shank3ΔC/ΔC mice (n = 25) compared with WT (n = 22). Fifteen consecutive traces (gray) and average trace (black) at −70 mV (bottom) and at +40 mV (top) from WT mice (left) and Shank3ΔC/ΔC mice (right). Legend in A applies to C and D. Cumulative frequency of mEPSC amplitude (B) and mean mEPSC amplitude (C) were unchanged, but mEPSC frequency (D) was significantly decreased in Shank3ΔC/ΔC compared with WT (WT, n = 15; Shank3ΔC/ΔC, n = 22). Inset, One minute raw traces from a WT CA1 neuron (black) and a Shank3ΔC/ΔC CA1 neuron (gray). Calibration: 10 pA, 2.5 s. Legend in B also applies to E and F. E, PPR is not different between WT and Shank3ΔC/ΔC mice at interstimulus intervals of 30–500 ms. n = 10 for each genotype. F, The relationship of stimulus intensity to fEPSP slope is decreased in Shank3ΔC/ΔC mice. Inset, Relationship of fiber volley to fEPSP slope is similar between WT and Shank3ΔC/ΔC mice. n = 10 for each genotype. Error bars represent SEM, *p < 0.05.
In an effort to determine whether the decreased NMDA/AMPA ratio was attributable to decreased NMDAR-mediated synaptic transmission or to increased AMPAR-mediated transmission, we examined the amplitude of spontaneous mEPSCs that primarily reflect AMPAR-mediated responses at individual synapses. Consistent with the NMDA/AMPA ratio decrease being attributable to decreased NMDAR-mediated responses, cumulative frequency of mEPSC amplitudes was not different in Shank3ΔC/ΔC mice compared with WT (Fig. 7B; Kolmagorov–Smirnov two-sample test, p > 0.1). Similarly, mean mEPSC amplitude was not affected by Shank3 deletion (Fig. 7C; WT, −9.74 ± 1.19 pA, n = 15 vs Shank3ΔC/ΔC, −9.19 ± 0.97 pA, n = 22; t(35) = 0.364, p = 0.718).
However, the frequency of mEPSCs was significantly decreased in Shank3ΔC/ΔC mice compared with WT (Fig. 7D; WT, 1.16 ± 0.11 Hz, n = 15 vs Shank3ΔC/ΔC, 0.74 ± 0.09 Hz, n = 22; t(35) = 2.971, p = 0.005). A decrease in mEPSC frequency can be attributable to a decrease in presynaptic evoked release probability (Pr), decreased synapse number, decreased synaptic release sites, or a selective decrease in spontaneous Pr. To distinguish among these possibilities, we examined both paired pulse ratios (PPRs) and baseline synaptic strength via input/output (I/O) curves. Alterations in PPRs often accompany changes in presynaptic Pr (Zucker and Regehr, 2002; Lauri et al., 2007; Regehr, 2012), but we identified no changes in PPR using a broad range of interstimulus intervals (50–500 ms) in Shank3ΔC/ΔC mice compared with WT littermate controls (Fig. 7E; two-way rmANOVA; genotype: F(1,107) = 0.748, p = 0.389; interstimulus interval: F(5,107) = 29.43, p < 0.0001; genotype × interstimulus interval: F(5,107) = 0.254, p = 0.937; WT, n = 10; Shank3ΔC/ΔC, n = 10). This finding is consistent with there being no change in evoked synaptic Pr.
The I/O relationship of stimulus intensity to slope of the fEPSP was decreased in Shank3ΔC/ΔC mice (Fig. 7F; two-way rmANOVA; genotype: F(1,18) = 8.969, p = 0.008; intensity: F(10,180) = 93.93, p < 0.0001; intensity × genotype: F(10,180) = 6.155, p < 0.0001; WT, n = 10; Shank3ΔC/ΔC, n = 10). Bonferroni's multiple comparisons indicate significant differences between WT and Shank3ΔC/ΔC mice at stimulus intensities >50 μA, with a maximum fEPSP slope 36.9% greater in WT compared with Shank3ΔC/ΔC mice (WT, −0.34 ± 0.04 mV/ms; Shank3ΔC/ΔC, −0.22 ± 0.03 mV/ms; Bonferroni's multiple comparisons, p < 0.0001). No difference was found between WT and Shank3ΔC/ΔC mice in the relationship between stimulus intensity and fiber volley amplitude (inset), suggesting no change in presynaptic axonal excitability (two-way rmANOVA; intensity: F(10,280) = 65.97, p < 0.0001; genotype: F(2,28) = 0.427, p = 0.656; intensity × genotype: F(20,280) = 0.936, p = 0.542). These alterations in spontaneous and evoked synaptic transmission without changes in mEPSC amplitude suggest a possible decrease in the number of functional CA3–CA1 synapses to account for decreased LTP in Shank3ΔC/ΔC mice.
To determine whether a change in the number of structural synaptic spines is contributing to functional deficits in synaptic transmission and plasticity, we used Golgi–Cox staining and histology to see whether CA1 pyramidal neuron morphology is altered in the Shank3ΔC/ΔC mice. No differences were observed in dendritic complexity of the WT and Shank3ΔC/ΔC mice (Fig. 8A; two-way rmANOVA; main effect of genotype: F(1,8) = 0.75, p = 0.41; main effect of distance from cell body: F(9,74) = 58.82, p < 0.0001; genotype × distance interaction: F(9,74) = 0.57, p = 0.81). Furthermore, no differences were observed in spine density between WT and Shank3ΔC/ΔC mice (Fig. 8B,C; rmANOVA; main effect of genotype: F(1,8) = 1.01, p = 0.34; main effect of distance from cell body: F(3,24) = 32.91, p < 0.0001; genotype × distance interaction: F(3,24) = 0.70, p = 0.56). These findings were repeated in a separate cohort of mice at the younger age of P13–P17 as well to better correspond to the age at which the electrophysiological alteration in mEPSC frequency was observed. In P13–P17 mice, we again found no significant difference in spine density between WT and Shank3ΔC/ΔC mice (data not shown). Therefore, changes in structural synaptic spine number do not appear to account for alterations in synaptic transmission and plasticity in Shank3ΔC/ΔC mice. However, it is possible that a more subtle difference in spine density exists that we may be underpowered to detect.
Figure 8.
No morphological deficits were observed in the CA1 hippocampal neurons in Shank3ΔC/ΔC mice. A, No differences between genotypes were observed in quantitative assessment of branching via Sholl analysis. Legend in A also applies to C. B, Representative examples of WT and Shank3ΔC/ΔC spine density at 90 μm from the soma at 100× magnification. Scale bar, 5 μm in WT (also applies to Shank3ΔC/ΔC). C, No differences between genotypes were observed in spine density in the apical dendrites of CA1 hippocampus pyramidal neurons. n = 20 neurons from 5 mice for each genotype. Error bars represent SEM, *p < 0.05.
Discussion
Role of Shank3 in hippocampus-dependent learning and memory and synaptic transmission
We have identified multiple abnormalities in hippocampus function in a mouse model lacking major naturally occurring isoforms of Shank3. These mice exhibit decreased NMDAR-mediated synaptic transmission, decreased frequency of spontaneous glutamate release, and decreased evoked excitatory synaptic transmission with no change in short-term plasticity or mEPSC amplitude in area CA1 of the hippocampus. This decrease in NMDAR-mediated synaptic transmission is a likely cause of the observed decrease in LTP in the Shank3ΔC/ΔC mice. In turn, the LTP deficits are one potential explanation for the decreased hippocampus-dependent spatial learning abnormalities.
These abnormalities are most likely attributable to the loss of multiple naturally occurring isoforms of Shank3 in the Shank3ΔC/ΔC mutants. However, we cannot rule out the possibility that increases in smaller-molecular-weight forms of Shank3 that appear in the Shank3ΔC/ΔC mice on Western blot with N-terminal and SH3 antibodies could be contributing to these abnormalities. The additional presence of these smaller-molecular-weight isoforms in the Shank3ΔC/ΔC mutants is likely to occur in patients with autism caused by either translocation breakpoints or insertion mutations in exon 21 of Shank3, making them of potential relevance to a subset of autism associated with Shank3 mutations. Furthermore, the presence of additional novel isoforms of Shank3 has not yet been thoroughly evaluated in other published Shank3 mutant models (Bozdagi et al., 2010; Peça et al., 2011; Wang et al., 2011; Yang et al., 2012) and may also account for differences observed in these mutants. Indeed, it is not clear in these other Shank3 mutant models whether the phenotypes are attributable to loss of particular isoforms of Shank3 or to altered ratios of one isoform to another, much less whether novel isoforms appear as a result of these mutations. Mice lacking the entire Shank3 gene coding region will be of interest to compare with existing Shank3 mutant models to resolve these issues and as a model of the 22q13 deletion syndrome or Phelan–McDermid syndrome.
Both spatial learning deficits and decreased LTP in area CA1 of the hippocampus have also been reported in the Shank3e4–9 homozygous mutant mice. This model lacks exons 4–9 coding for the ankryin repeat domain, resulting in loss of only the largest-molecular-weight isoform of Shank3 (SHANK3α; Wang et al., 2011). This finding suggests that loss of this largest isoform alone may be sufficient to produce altered spatial learning and decreased LTP in Shank3 mutants. The NMDA/AMPA ratio using whole-cell recordings was not reported in the Shank3e4–9 homozygous mutant mice. Similar to the present findings in Shank3ΔC/ΔC homozygotes, AMPAR-mediated synaptic transmission was intact in the Shank3e4–9 homozygous mutants as supported by no change in whole-cell mEPSC amplitude. It will be of interest to examine the NMDA/AMPA ratio directly in the Shank3e4–9 homozygous mutants to determine whether loss of only the largest-molecular-weight isoform of Shank3 is sufficient to decrease NMDAR-mediated synaptic transmission.
In addition, LTP deficits in area CA1 of the hippocampus have been reported in the Shank3e4–9 heterozygous mouse model (Bozdagi et al., 2010). However, in this case, extracellular field recording suggests no change in NMDAR-mediated synaptic transmission but rather a decrease in AMPAR-mediated transmission. Furthermore, whole-cell recording in the Shank3e4–9 heterozygotes demonstrated decreased mEPSC amplitude, consistent with a decrease in AMPA-mediated transmission as well as a dramatic increase in mEPSC frequency (Yang et al., 2012). The finding of extracellularly recorded decreases in both LTP and I/O curves were reproduced in a follow-up study by the same group, demonstrating the I/O curve differences in Shank3e4–9 homozygotes as well as heterozygotes (Yang et al., 2012) in conflict with a previous report (Wang et al., 2011). Again, the NMDA/AMPA ratio was not measured in whole-cell recordings. Also in the study by Yang et al., behavioral studies of Shank3e4–9 homozygous and heterozygous mice did not show a statistically significant difference in water maze learning in two, small, male-only cohorts, a finding that also contrasts with previously published work on Shank3e4–9 homozygotes (Wang et al., 2011) and the present study. The different findings in Shank3e4–9 homozygous mice I/O curves using extracellular field recordings (Wang et al., 2011; Yang et al., 2012) and in Shank3e4–9 homozygous water maze (Wang et al., 2011; Yang et al., 2012) remain to be resolved.
In yet another Shank3 homozygous mutant targeting exons 13–16 that code for the PDZ domain of Shank3 (Shank3e13–16) leading to loss of the two higher-molecular-weight isoforms of Shank3 (Shank3α and Shank3β), the minimal hippocampal electrophysiology performed highlights differences with the present findings in Shank3ΔC/ΔC homozygotes. This includes normal extracellularly recorded I/O curves measuring population spike amplitude rather than direct measurement of fEPSPs and no change in the amplitude or frequency of mEPSCs in area CA1 of hippocampus (Peça et al., 2011). Neither the NMDA/AMPA ratio nor LTP were measured in the hippocampus of Shank3e13–16 homozygous mutants (Peça et al., 2011). Curiously, the Shank3e13–16 homozygotes also did not show altered spatial learning in the water maze task with a small cohort of 4- to 5-week-old male mutants (Peça et al., 2011). This difference could be attributable to the different Shank3 mutation, different ages of mice tested, differences in protocols across laboratories, or differences in genetic background. Comparison of the multiple published Shank3 models within the same laboratory under the same conditions will be necessary to resolve these issues.
Interestingly, despite a dramatic increase in mGluR5 in synaptic fractions, we find that mGluR–LTD is not significantly enhanced in Shank3ΔC/ΔC mice. Because group I mGluRs are currently being targeted as novel treatments for other mouse models of autism (Bear et al., 2004; Won et al., 2012; Yang et al., 2012) and Shank3 has been shown to regulate mGluR5 expression and signaling in cultured neurons (Verpelli et al., 2011), additional functional studies of mGluR function in Shank3 mutants will be of critical importance.
Shank3 and social, repetitive, and communication behavior
Social deficits are one of the key diagnostic features of autism (Schreibman, 1988; Mahjouri and Lord, 2012), yet of the two social interaction tests that we administered, we only observed deficits in one trial of the three-chambered social interaction test in which Shank3ΔC/ΔC mice failed to show a preference for social novelty. This phenotype has been reported previously in Shank3e13–16 homozygotes (Peça et al., 2011) and Shank3e4–9 homozygotes (Wang et al., 2011). In the second test using social interaction with a juvenile, Shank3ΔC/ΔC mice did not show any deficits in initial interaction with juveniles or in a subsequent test of social memory. Together, these data suggest primarily intact social interaction with only minimal dysfunction in one task relevant to the social domain.
Surprisingly little phenotype was observed in the other two major behavioral symptoms domains relevant to autism, communication, and repetitive behaviors. An examination of USV during exposure of an adult male to an estrous female showed no changes in number of USVs or in latency to first USV in the Shank3ΔC/ΔC mice. Similarly, in younger mice, we observed no alteration in repetitive grooming behaviors. However, in older Shank3ΔC/ΔC mice, a significant increase in grooming behavior was observed. A thorough temporal/developmental assessment of the development of increased repetitive grooming and similar behaviors may be of interest in future studies.
Shank3 and associated features
Aside from the three core diagnostic features, autism is also associated with motor-coordination impairments (Clarke, 1996; Abu-Dahab et al., 2012), and many patients with 22q13 deletion syndrome exhibit hypotonia and incoordination (Phelan and McDermid, 2012). Shank3ΔC/ΔC mice also show impairment in motor coordination but not in motor learning.
We also tested Shank3ΔC/ΔC mice in tests of anxiety because autism can be associated with anxiety disorders (Gillott et al., 2001). Of the three anxiety-related tasks, elevated plus maze, open field, and dark/light, we only found abnormalities in the dark/light task. Shank3ΔC/ΔC mice showed an increased latency to enter the light side and significantly preferred the dark chamber. Generally, this result alone might lead to a conclusion that Shank3ΔC/ΔC mice have increased anxiety. However, we did not see any differences in the other two anxiety tests. Thus, we interpret the avoidance of light in the dark/light task not as an increase in anxiety but as hypersensitivity to light or perhaps to novelty avoidance. We also tested Shank3ΔC/ΔC mice for acoustic startle response but found no differences. This suggests that Shank3ΔC/ΔC mice have hypersensitivity to only select sensory stimuli.
Some studies report that children with autism respond to novelty with avoidance behaviors (Kootz et al., 1982; Anckarsäter et al., 2006). Our Shank3ΔC/ΔC mice show a marked increase in avoidance of novel inanimate objects as evident in the results from nest-building and marble-burying behavior. Another aberrant response to novelty in Shank3ΔC/ΔC mice is decreased initial locomotor response to a novel environment. This phenotype was only observed in the initial 5 min of exposure with rapid habituation to the locomotor apparatus thereafter. Interestingly, Shank3ΔC/ΔC mice also show a decreased latency to lick their hindpaw on a hotplate, indicating an increase in sensitivity to heat.
Conclusions
Recent studies strongly implicate SHANK3 in autism and Phelan–McDermid syndrome, making a thorough understanding of SHANK3 function in the CNS of critical importance. Our results indicate that Shank3 is critically important for normal synaptic transmission in the hippocampus and for normal spatial learning and memory. Loss of Shank3 leads to minimal social deficits but unveils multiple additional behavioral abnormalities, including motor coordination impairment, novelty avoidance, hypersensitivity to select sensory stimuli, and aberrant locomotor responses to novelty. Additional studies are needed to elucidate the underlying mechanisms of how Shank3 is involved in the above deficits, which brain regions are responsible for such deficits, and whether loss of Shank3 leads to irreversible neurodevelopmental abnormalities or to functional synaptic deficits that can be reversed later in life. Our findings of a decreased NMDA/AMPA ratio at hippocampal synapses and a dramatic increase in mGluR5 localization to synaptosome and PSD fractions suggests these as potential therapeutic targets for future preclinical studies.
Footnotes
This work was funded by National Institute of Child Health and Human Development Grants R01HD069560 (C.M.P.) and R01HD069560-0251 Diversity Supplement (T.C.J.), National Institute of Mental Health Grants R01MH093697 (C.M.P.) and P50MH084020 (P.F.W.), National Institute of Neurological Disorders and Stroke Grant R01NS070301 (P.F.W.), National Institute on Drug Abuse Grant T32DA007290 (J.M.R.), Autism Speaks (C.M.P.), Autism Science Foundation Postdoctoral Fellowship (H.E.S.), Autism Speaks Translational Postdoctoral Fellowship (H.E.S.), Autism Speaks Dennis Weatherstone Predoctoral Fellowship (M.K., M.A.B.), BRAINS for Autism (C.M.P.), The Hartwell Foundation (C.M.P.), and National 973 Basic Research Program of China Grant 20009CB941400 (B.X.).
References
- Abu-Dahab SM, Skidmore ER, Holm MB, Rogers JC, Minshew NJ. Motor and tactile-perceptual skill differences between individuals with high-functioning autism and typically developing individuals ages 5–21. J Autism Dev Disord. 2013;43:2241–2248. doi: 10.1007/s10803-011-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckarsäter H, Stahlberg O, Larson T, Hakansson C, Jutblad SB, Niklasson L, Nydén A, Wentz E, Westergren S, Cloninger CR, Gillberg C, Rastam M. The impact of ADHD and autism spectrum disorders on temperament, character, and personality development. Am J Psychiatry. 2006;163:1239–1244. doi: 10.1176/appi.ajp.163.7.1239. [DOI] [PubMed] [Google Scholar]
- Arons MH, Thynne CJ, Grabrucker AM, Li D, Schoen M, Cheyne JE, Boeckers TM, Montgomery JM, Garner CC. Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. J Neurosci. 2012;32:14966–14978. doi: 10.1523/JNEUROSCI.2215-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Südhof TC, Powell CM. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Kaeser PS, Sudhof TC, Powell CM. RIM1alpha and interacting proteins involved in presynaptic plasticity mediate prepulse inhibition and additional behaviors linked to schizophrenia. J Neurosci. 2010a;30:5326–5333. doi: 10.1523/JNEUROSCI.0328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Südhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010b;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, Dupont B, Tiziano D, Schroer RJ, Neri G, Stevenson RE, Gurrieri F, Schwartz CE. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böckers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F, Richter D, Gundelfinger ED, Kreienkamp HJ. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alpha-fodrin. J Biol Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach JP, van der Zwaag B. Contact in the genetics of autism and schizophrenia. Trends Neurosci. 2009;32:69–72. doi: 10.1016/j.tins.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Clarke A. Rett syndrome. J Med Genet. 1996;33:693–699. doi: 10.1136/jmg.33.8.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RS, Blomberg F, Berzins K, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J Cell Biol. 1977;74:181–203. doi: 10.1083/jcb.74.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SU, del Gaudio D, German JR, Peters SU, Ou Z, Bader PI, Berg JS, Blazo M, Brown CW, Graham BH, Grebe TA, Lalani S, Irons M, Sparagana S, Williams M, Phillips JA, 3rd, Beaudet AL, Stankiewicz P, Patel A, Cheung SW, Sahoo T. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am J Med Genet A. 2010;152A:573–581. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Südhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci U S A. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/S0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafrenière RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, Fombonne E, Joober R, Marineau C, Drapeau P, Rouleau GA. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafrenière RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Côté M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Néri C, Dubé MP, Samuels ME, Marineau C, Stone EA, Awadalla P, Barker PA, Carbonetto S, Drapeau P, Rouleau GA S2D Team. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillott A, Furniss F, Walter A. Anxiety in high-functioning children with autism. Autism. 2001;5:277–286. doi: 10.1177/1362361301005003005. [DOI] [PubMed] [Google Scholar]
- Gong X, Jiang YW, Zhang X, An Y, Zhang J, Wu Y, Wang J, Sun Y, Liu Y, Gao X, Shen Y, Wu X, Qiu Z, Jin L, Wu BL, Wang H. High proportion of 22q13 deletions and SHANK3 mutations in Chinese patients with intellectual disability. PloS One. 2012;7:e34739. doi: 10.1371/journal.pone.0034739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves N, Prince E, Evans DW, Charman T. Repetitive and ritualistic behaviour in children with Prader-Willi syndrome and children with autism. J Intellect Disabil Res. 2006;50:92–100. doi: 10.1111/j.1365-2788.2005.00726.x. [DOI] [PubMed] [Google Scholar]
- Kootz JP, Marinelli B, Cohen DJ. Modulation of response to environmental stimulation in autistic children. J Autism Dev Disord. 1982;12:185–193. doi: 10.1007/BF01531308. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri SE, Palmer M, Segerstrale M, Vesikansa A, Taira T, Collingridge GL. Presynaptic mechanisms involved in the expression of STP and LTP at CA1 synapses in the hippocampus. Neuropharmacology. 2007;52:1–11. doi: 10.1016/j.neuropharm.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Mahjouri S, Lord CE. What the DSM-5 portends for research, diagnosis, and treatment of autism spectrum disorders. Curr Psychiatry Rep. 2012;14:739–747. doi: 10.1007/s11920-012-0327-2. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, Young NB, Barbaro RP, Piven J, Magnuson TR, Crawley JN. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/S0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Ozonoff S, Jensen J. Brief report: specific executive function profiles in three neurodevelopmental disorders. J Autism Dev Disord. 1999;29:171–177. doi: 10.1023/A:1023052913110. [DOI] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE. The 22q13.3 deletion syndrome (Phelan-McDermid Syndrome) Mol Syndromol. 2012;2:186–201. doi: 10.1159/000334260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Südhof TC, Nestler EJ. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/S0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr WG. Short-term presynaptic plasticity. Cold Spring Harbor Perspect Biol. 2012;4:a005702. doi: 10.1101/cshperspect.a005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler J, Bishop SL, Kleinke JR, Lord C. Restricted and repetitive behaviors in young children with autism spectrum disorders. J Autism Dev Disord. 2007;37:73–85. doi: 10.1007/s10803-006-0332-6. [DOI] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10:44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreibman L. Diagnostic features of autism. J Child Neurol [Suppl] 1988;3:S57–S64. doi: 10.1177/088307388800300111. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/S0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Uchino S, Wada H, Honda S, Nakamura Y, Ondo Y, Uchiyama T, Tsutsumi M, Suzuki E, Hirasawa T, Kohsaka S. Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J Neurochem. 2006;97:1203–1214. doi: 10.1111/j.1471-4159.2006.03831.x. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Böckers TM, Dityatev A, Sala C. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011;286:34839–34850. doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Schmeisser MJ, Sala C, Boeckers TM. Scaffold proteins at the postsynaptic density. Adv Exp Med Biol. 2012;970:29–61. doi: 10.1007/978-3-7091-0932-8_2. [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingate MMB, Kirby RS, Pettygrove S, Cunniff C, Meaney F, Schulz E, Miller L, Robinson C, Quintana G, Kaiser MY, Lee LC, Landa R, Newschaffer C, Constantino J, Fitzgerald R, Zahorodny W, Daniels J, Giarelli E, Pinto-Martin J, Levy SE, Nicholas J, Charles J, Zimmerman J, Maenner MJ, Durkin M, Rice C, Baio J, Van Naarden Braun K, Phillips K, Doernberg N, Yeargin-Allsopp M. Prevalence of autism spectrum disorders—Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wöhr M, Roullet FI, Katz AM, Abrams DN, Kalikhman D, Simon H, Woldeyohannes L, Zhang JY, Harris MJ, Saxena R, Silverman JL, Buxbaum JD, Crawley JN. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]