Abstract
Salvia miltiorrhiza Bunge, a perennial plant of Lamiaceae, accumulates abietane-type diterpenoids of tanshinones in root, which have been used as traditional Chinese medicine to treat neuroasthenic insomnia and cardiovascular diseases. However, to date the biosynthetic pathway of tanshinones is only partially elucidated and the mechanism for their root-specific accumulation remains unknown. To identify enzymes and transcriptional regulators involved in the biosynthesis of tanshinones, we conducted transcriptome profiling of S. miltiorrhiza root and leaf tissues using the 454 GS-FLX pyrosequencing platform, which generated 550,546 and 525,292 reads, respectively. RNA sequencing reads were assembled and clustered into 64,139 unigenes (29,883 isotigs and 34,256 singletons). NCBI non-redundant protein databases (NR) and Swiss-Prot database searches anchored 32,096 unigenes (50%) with functional annotations based on sequence similarities. Further assignments with Gene Ontology (GO) terms and KEGG biochemical pathways identified 168 unigenes referring to the terpenoid backbone biosynthesis (including 144 MEP and MVA pathway genes and 24 terpene synthases). Comparative analysis of the transcriptomes identified 2,863 unigenes that were highly expressed in roots, including those encoding enzymes of early steps of tanshinone biosynthetic pathway, such as copalyl diphosphate synthase (SmCPS), kaurene synthase-like (SmKSL) and CYP76AH1. Other differentially expressed unigenes predicted to be related to tanshinone biosynthesis fall into cytochrome P450 monooxygenases, dehydrogenases and reductases, as well as regulatory factors. In addition, 21 P450 genes were selectively confirmed by real-time PCR. Thus we have generated a large unigene dataset which provides a valuable resource for further investigation of the radix development and biosynthesis of tanshinones.
Introduction
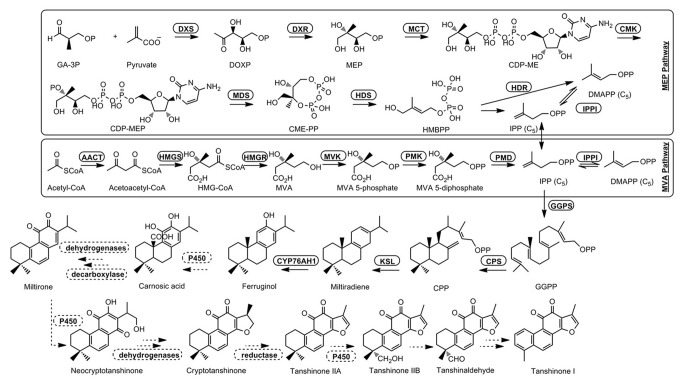
Salvia miltiorrhiza Bunge is a perennial plant in the genus Salvia of the Lamiaceae family. Its dried root or rhizome is called Danshen in traditional Chinese medicine, and was recorded in first pharmaceutical monograph Shennong’s Classic of Materia Medica (A.D. 102-200). S. miltiorrhiza has been cultivated throughout Eastern Asia and used to treat and prevent cardiovascular, cerebrovascular, hyperlipidemia and acute ischemic stroke diseases [1]. The active ingredients in S. miltiorrhiza are considered to contain both hydrophilic and lipophilic components. The hydrophilic phenolic acids include rosmarinic acid, salvianolic acid B, lithospermic acid and dihydroxyphenyllactic acid or Danshensu, and they may function as antibacterial, anti-oxidative and antiviral reagents [2,3]. And the lipophilic diterpenoid components are generally known as tanshinones, including structurally related tanshinone I, tanshinone IIA, cryptotanshinone, and dihydrotanshinone I. All these diterpenoids share the abietane-type skeletons, and tanshinone IIA is considered to be the most important bioactive component [4,5] (Figure 1).
Figure 1. Biosynthetic pathway of tanshinone compounds.
The solid arrows denote known steps and the dashed arrows denote hypothetical steps. Modified from Ma et al. and Gao et al. [7,13]. Enzymes of 2-C-methyl-D-erythritol 4- phosphate (MEP) pathway are follows: 1-deoxy-D-xylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (MCT), 4-diphosphocytidyl-2-Cmethyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MDS), 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR); and enzymes of mevalonate (MVA) pathway are acetyl-CoA acetyltransferase (AACT), 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), mevalonate kinase (MVK), 5-phosphomevalonate kinase (PMK) and 5-diphosphomevalonate decarboxylase (PMD). Isopentenyl diphosphate isomerase (IPPI) catalyzes the isomerisation of dimethylallyl dihosphate (DMAPP) to isopentenyl diphosphate (IPP) whereas conversion of IPP to geranylgeranyl diphosphate (GGPP) is catalysed by geranylgeranyl diphosphate synthase (GGPPS). Hypothetical tanshinones biosynthetic pathway was deduced by the consideration of identified diterpenoid natural products from S. miltiorrhiza [56,57]. These steps involve series hydroxylation, dehydrogenation and reduction reactions catalysed by cytochrome P450s, dehydrogenases and reductases.
In plant cells, terpenoids are synthesized through either the cytoplasmic mevalonate (MVA) or the plastidic 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway, with possible cross-talk of the common precursors of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) [6]. As diterpenoid compounds, the abietane-type tanshinones are proposed to be derived mainly from the MEP pathway, starting from the conversion of geranylgeranyl diphosphate (GGPP) to ent-copalyl diphosphate (CPP) and to miltiradiene. After hydroxylation, decarboxylation, oxidation and reduction steps, the miltiradiene is converted into cryptotanshinone, tanshinone I, tanshinone IIA or tanshinone IIB (Figure 1) [7].
Based on sequence homology, a number of MVA and MEP pathway enzymes, as well as two diterpenoid pathway enzymes of geranylgeranyl diphosphate (SmGGPPS) synthase and ent-copalyl diphosphate synthase (SmCPS), have been reported [7,8]. However, up to now only two enzymes specific to the tanshinones biosynthetic pathway have been identified: one is the kaurene synthase-like (SmKSL), a diterpene synthase that utilizes copalyl diphosphate (CPP) as substrate to produce miltiradiene [7]; the other is the P450 monooxygenase CYP76AH1, which catalyses the conversion of miltiradiene to ferruginol [9]. It is of great interest to identify enzymes catalysing the remaining steps of the pathway.
Medicinal plants, which produce different classes of natural products, cover a wide range of plant taxa and most of them have only limited EST, transcriptome and genome data. Next generation sequencing (NGS) platforms provide highly efficient tools to discover novel enzymes and transcription factors from these non-model species. In recent years, genomic approaches have been widely used for discovering and characterizing secondary metabolism pathways and the related genes. For S. miltiorrhiza, a cDNA library of whole plantlet was constructed and 10,228 ESTs were generated [10]; a cDNA microarray was used to identify 114 differentially expressed cDNAs in S. miltiorrhiza hairy root [11]; 56,774 unigenes in transcriptome of S. miltiorrhiza over entire growing cycle were obtained by Illumina deep sequencing [12]; and by searching S. miltiorrhiza draft genome (unavailable to public), 40 terpenoid biosynthesis-related genes encoding all enzymes involved in the biosynthesis of universal isoprene precursors of IPP and DMAPP, were proposed [13].
Biosynthesis and accumulation of secondary metabolites are often tissue-specific, and related genes of enzymes and regulators also show organ- or tissue-specific expression patterns [14-16]. Tanshinones are highly enriched in roots, with only a low or trace amount in aerial organs of S. miltiorrhiza, such as leaves [17]. Moreover, both the accumulation of diterpenoid tanshinones and expressions of related pathway genes in hairy root cultures of S. miltiorrhiza can be induced by biotic (such as the carbohydrate fraction of yeast extract), abiotic (Ag+ and Cd2+) elicitors, or phytohormones (salicylic acid and methyl jasmonate) [18-23].
Such spatial distribution of tanshinones suggests a root-preferential expression pattern of biosynthetic pathway genes. In contrast to previous approaches that were focused solely on single or mixed tissues, we profiled transcriptomes of root and leaf tissues separately and performed a comparative analysis, which provides a comprehensive insight into tanshinones biosynthesis and regulation, as well as the biology of root (rhizome) development and the secondary metabolism in roots of perennial plants.
Results and Discussion
Transcriptome sequencing, de novo assembly and sequence clustering
Tanshinones accumulate mainly in S. miltiorrhiza root, with only a trace amount in aerial tissues [17]. To make sure that the root and leaf samples under investigation had a striking difference in tanshinones accumulation, we performed High Performance Liquid Chromatography (HPLC) analyses of their methanol extracts. We found that the levels of tanshinone I, cryptotanshinone, dihydrotanshinone I and tanshinone IIA were high in root, but barely detected in leaf (Figure S1). These results confirmed previous observations and ensured that our root and leaf samples were suitable for subsequent comparative analyses of transcriptomes.
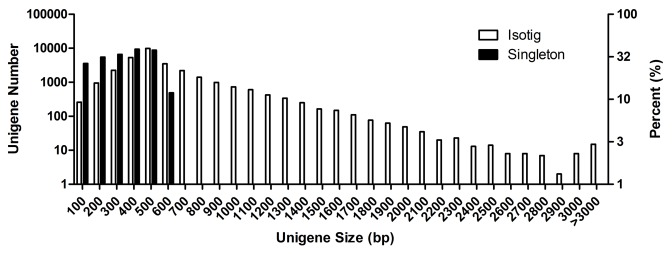
The root and leaf RNA samples were profiled by pyrosequencing with Roche’s 454 GS-FLX system, which generated 550,546 and 525,292 reads, with the average lengths of 325 bp and 375 bp, respectively. After trimming the adapter and low quality reads and removing those shorter than 50 bp, 455,552 and 470,863 high quality reads were obtained from root and leaf libraries, respectively. These reads were combined and assembled into 16,806 isotigs (N50 = 750 bp) and 84,116 singletons. Using a sequence similarity cutoff of 95%, the assembled sequences were clustered into 64,139 unigenes (including 29,883 isotigs and 34,256 singletons), with a mean length of 413 bp and total size of 26.4 Mb (Table 1; Figure 2).
Table 1. Summary of sequencing data of S. miltiorrhiza root and leaf transcriptomes.
| Leaf | Root | |
|---|---|---|
| Total number of raw reads | 525,292 | 550,546 |
| Total number of clean reads | 470,863 | 455,552 |
| Total clean nucleotides (nt) | 158,851,743 | 141,393,767 |
| Total number of isotigs | 16,806 | |
| N50 of the isotigs | 750 bp | |
| Total number of singletons | 84,116 | |
| Total number of unigenes | 64,139 | |
| Average unigene length | 413 bp | |
Figure 2. Sequence length distribution of unigenes in the S. miltiorrhiza transcriptomes of root and leaf.
Functional annotation and pathway analysis
Unigenes were searched against the NCBI non-redundant protein database (NR) and the Swiss-Prot database using blastx program with E-value cut-off of 1e-5. Totally 32,010 unigenes had at least one match to known protein sequences in the NCBI NR database and 18,201 unigenes found hits in the Swiss-Prot database. After consolidation, 32,096 unigenes (50%) were functional annotated.
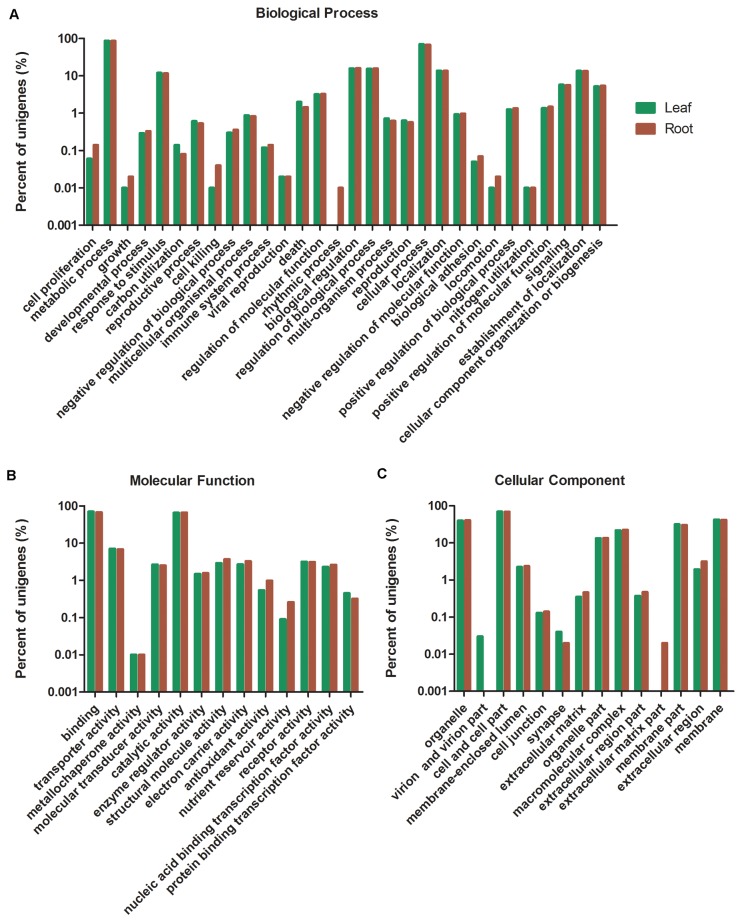
Gene Ontology (GO) assignment was performed to functional categorize these annotated unigenes, which resulted in 24,394 unigenes mapped to at least one GO term, of which 21,364 were assigned to the “biological process”, 23,136 to the “molecular function”, and 10,662 to the “cellular component”. Based on origins of the assembled reads, from root or leaf pools, the enrichment of unigenes to GO terms was analysed. The counts for the most categories were similar in root and leaf libraries, such as in the category of “biological process”, ‘metabolic process’ was dominant (86.56% and 85.99% in leaf and root, respectively, the same below), and ‘cellular process’ also had a high-percentage (70.53% and 67.61%); regarding to the “molecular function” category, ‘binding’ (70.29% and 67.48%) and ‘catalytic activity’ (65.48% and 65.88%) were the most enriched; within the category of “cellular component”, ‘cell and cell part’ (69.66% and 68.81%) was the most highly enriched (Figure 3). On the other hand, unigenes classified to the terms of ‘nutrient reservoir activity’, ‘rhythmic process’, ‘locomotion’ and ‘extracellular matrix part’ were more abundant in root (percentage > 2 fold), whereas those related to ‘virion and virion part’ were more in leaf (Figure 3).
Figure 3. Functional classification of unigenes expressed in S. miltiorrhiza leaf and root.
Gene Ontology (GO) terms are summarized in three main categories of biological process (A), molecular function (B) and cellular component (C).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway assignments were then performed to provide alternative functional annotations to enzymes of biochemical pathways and their Enzyme Commission (EC) numbers. This analysis resulted in 802 unigenes referring to the biosynthesis of secondary metabolites, of which 168 to terpenoids and 389 to other metabolites, including flavonoids and alkaloids (Table S1).
Differential expression analysis
We calculated the normalized expression values (RPKM, Reads Per Kilobase per Million mapped reads) of each unigene in root and leaf libraries, and those with > 2-fold change with a false discovery rate (FDR) < 0.01 were considered as differentially expressed genes. Totally 2,863 unigenes were found to have more abundant transcripts in root and 2,561 in leaf, of which 831 and 852 were annotated, respectively.
Among the differentially expressed genes, those function in chloroplast development and photosynthesis exhibited the most significant differential expression levels between the root and leaf tissues (FDR < 0.01). Genes that encode enzymes of the chlorophyll biosynthesis pathway (such as chlorophyll synthase), chlorophyll A/B binding proteins, and components of photosystem I/II and light-harvesting complex were highly expressed in leaf, but not expressed in root. Similarly, the genes involved in carbohydrate metabolism, such as glucose-1-phosphate adenylyltransferase and glucose-6-phosphate 1-dehydrogenase, and fatty acid metabolism, such as beta-ketoacyl-ACP synthase II, acyl-[acyl-carrier-protein] desaturase, acyl-CoA thioesterase), were also expressed at much higher levels in leaf than in root, in agreement with the fact that plant leaf is the centre for biomass and energy production (Table S2).
On the contrary, transcripts for transport proteins, including those specific or non-specific for sugars, irons and lipids, were highly abundant in root, which is consistent with their functions in nutrient absorption. Moreover, transcripts of putative pathogen resistance genes were also detected in root, suggesting an active interaction between root and soil organisms (Table S3).
Isoprenoid biosynthesis
Full length cDNAs of isoprenoid biosynthetic enzymes, including those of MVA and MEP pathways, as well as isoprenyl diphosphate synthases of geranyl diphosphate synthase (GPPS), farnesyl diphosphate synthase (FPPS) and geranylgeranyl diphosphate synthase (GGPPS) [13], were searched against both leaf and root transcriptome libraries to estimate their expression levels. Transcripts of all these genes were detected in both libraries (Table 2). In general, transcripts of MEP pathway genes were more abundant in leaf, as revealed by much higher numbers of reads of 1-deoxy-D-xylulose 5-phosphate synthase 1 (SmDXS1), 1-deoxy-D-xylulose 5-phosphate reductoisomerase (SmDXR) and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 2 (SmHDR2) genes in leaf than in root. In contrast, the genes in the MVA pathway were more actively expressed in root according to numbers of reads mapped to genes for acetyl-CoA C-acetyltransferase 1 (SmAACT1), 3-hydroxy-3-methylglutaryl-CoA reductase (SmHMGRs) and isopentenyl diphosphate isomerase 1 (SmIPPI1) (Table 2). Moreover, some of the genes belonging to the same family showed distinctive expression patterns. For example, of the four SmDXS genes analysed, SmDXS1 was specifically expressed in leaf, whereas SmDXS2 was up-regulated in root (Table 2), suggesting their functional specialization during evolution.
Table 2. Expression of unigenes in isoprenoid biosynthesis pathways.
| Pathway | Gene name | Unigene | Reads in leaf | Reads in root |
|---|---|---|---|---|
| MVA | AACT 1 | isotig00545 | 4 | 54 |
| AACT 2 | isotig22400 | 0 | 2 | |
| isotig27922 | 3 | 0 | ||
| HMGS | isotig01307 | 23 | 49 | |
| HMGR 1 | isotig04635 | 7 | 18 | |
| HMGR 2 | isotig01508 | 2 | 43 | |
| HMGR 3 | isotig03994 | 22 | 147 | |
| isotig07175 | 14 | 126 | ||
| gen_HITXW8201CK83K | 0 | 1 | ||
| gen_HITXW8201BFCSU | 0 | 1 | ||
| HMGR 4 | isotig07145 | 31 | 484 | |
| MVK | isotig10296 | 11 | 4 | |
| PMK | isotig21613 | 0 | 2 | |
| gen_HITXW8201A60UW | 0 | 1 | ||
| PMD | isotig02363 | 0 | 27 | |
| IPPI 1 | isotig01015 | 21 | 146 | |
| IPPI 2 | isotig34210 | 2 | 0 | |
| MEP | DXS 1 | isotig00083 | 1171 | 1 |
| DXS 2 | isotig11491 | 0 | 26 | |
| isotig10422 | 0 | 11 | ||
| DXS 3 | isotig28922 | 2 | 1 | |
| isotig11378 | 4 | 14 | ||
| DXS 4 | isotig00662 | 7 | 5 | |
| isotig29585 | 4 | 0 | ||
| DXS 5 | — | |||
| DXR | isotig06138 | 31 | 3 | |
| isotig09197 | 13 | 1 | ||
| MCT | isotig01252 | 11 | 16 | |
| ye_HN6LTMS01DEI5U | 1 | 0 | ||
| CMK | isotig00089 | 36 | 50 | |
| MDS | isotig11395 | 7 | 4 | |
| HDS | isotig00067 | 98 | 128 | |
| HDR 1 | isotig03934 | 5 | 13 | |
| gen_HITXW8201BTHGM | 0 | 1 | ||
| gen_HITXW8201EN94F | 0 | 1 | ||
| HDR 2 | isotig04251 | 89 | 0 | |
| isotig11365 | 17 | 0 | ||
| isotig01210 | 106 | 0 | ||
| Monoterpene | GPPS LSU | isotig05039 | 2 | 69 |
| isotig20070 | 0 | 3 | ||
| gen_HITXW8201D6C8K | 0 | 1 | ||
| GPPS SSUII 1 | gen_HITXW8201DT9BE | 0 | 1 | |
| GPPS SSUII 2 | isotig10575 | 44 | 44 | |
| isotig10082 | 232 | 72 | ||
| isotig27077 | 6 | 0 | ||
| Sesquiterpene | FPPS | isotig00761 | 26 | 65 |
| Diterpene | GGPPS 1 | isotig04334 | 95 | 324 |
| GGPPS 2 | isotig22076 | 0 | 2 | |
| GGPPS 3 | isotig10232 | 19 | 468 |
MVA, mevalonate pathway; AACT, acetyl-CoA acetyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MVK, mevalonate kinase; PMK, 5-phosphomevalonate kinase; PMD, 5-diphosphomevalonate decarboxylase; IPPI, Isopentenyl diphosphate isomerase; MEP, Enzymes of 2-C-methyl-D-erythritol 4-phosphate pathway; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2-Cmethyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; GPPS, geranyl diphosphate synthase; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase.
In plant cell, monoterpenes are synthesized in plastid using geranyl diphosphate (GPP) as precursor, whereas sesquiterpenes and triterpenes are produced in cytoplasm using farnesyl diphosphate (FPP) as precursor. The GPPS (both large and small subunits) and FPPS genes exhibited similar expression levels in leaf and root. However, transcripts of geranylgeranyl diphosphate synthase (GGPPS), which produces the precursor for diterpenoids, including tanshinones, were highly abundant and enriched in root (Table 2), with the RPKM values of SmGGPPS1 being 910.88 in root and 258.41 in leaf; 9.51 in root and 0 in leaf for SmGGPPS2; and 1988.85 in root and 78.33 in leaf for SmGGPPS3, respectively.
Terpene synthases
Plants produce a rich array of terpenoids with diverse structures, which play important roles in both basic biological processes and interactions with environmental factors. Terpenoids are synthesized by terpene synthases (TPSs), which comprises of mono-, sesqui- or di-terpene synthases. According to phylogenetic relationships, TPSs are also classified into TPS-a, b, c, d, e/f, g and h subfamilies [24].
From the dataset 24 TPS unigenes were identified. Based on sequence homology to functionally characterized TPSs in the NCBI NR database, 13 unigenes were annotated as (3S)-linalool synthase, myrcene/ocimene synthase or 1.8-cineole synthase, which are all monoterpene biosynthesis [25-30]; and 7 as α-humulene/β-caryophyllene synthase or (+)-α-barbatene synthase, which are sesquiterpene biosynthesis [31-34]. In addition, one copy of ent-kaurene synthase (a diterpene synthase) and 3 fragments of squalene synthases (triterpene synthase) were also identified.
Most of these TPS genes had low levels of transcripts in both root and leaf libraries. Comparatively, 4 monoterpene synthases (isotig 01344, 05322, 05141, 12563) and 1 sesquiterpene synthase (isotig 08178) showed higher expression levels in root, whereas 1 monoterpene synthase (isotig 07874) and 2 sesquiterpene synthases (isotig 34119, 14598) in leaf (Table 3).
Table 3. Terpene synthase genes of S. miltiorrhiza.
| Terpene synthase | Unigene ID | Length(bp) |
No. in Leaf
|
No. in Root | Annotation | Gene bank accession ID | |
|---|---|---|---|---|---|---|---|
| Mono- | isotig21756 | 465 | 0 | 2 | (3S)-linalool synthase | CAD10147 | |
| ye_HN6LTMS01EHVGF | 462 | 1 | 0 | (3S)-linalool synthase | CAD57081 | ||
| isotig05322 | 711 | 0 | 9 | myrcene/ocimene synthase | NP_189209 | ||
| isotig05141 | 725 | 1 | 176 | myrcene/ocimene synthase | ABD77416 | ||
| isotig12563 | 450 | 2 | 13 | myrcene/ocimene synthase | EEE79235 | ||
| ye_HN6LTMS01EP8XE | 391 | 1 | 0 | myrcene/ocimene synthase | AES84848 | ||
| ye_HN6LTMS01BJR9J | 442 | 1 | 0 | myrcene/ocimene synthase | ACM89961 | ||
| ye_HN6LTMS01AIOKP | 459 | 1 | 0 | myrcene/ocimene synthase | ABU87404 | ||
| isotig07874 | 596 | 15 | 0 | myrcene/ocimene synthase | AFI47927 | ||
| isotig01344 | 1206 | 0 | 38 | 1.8-cineole synthase | EEF01161 | ||
| isotig20942 | 396 | 0 | 2 | 1.8-cineole synthase | ABD77416 | ||
| ye_HN6LTMS01DSMZC | 425 | 1 | 0 | 1.8-cineole synthase | ACN42009 | ||
| ye_HN6LTMS01BY84P | 453 | 1 | 0 | 1.8-cineole synthase | ABP01684 | ||
| Sesqui- | isotig08178 | 576 | 0 | 13 | α-humulene/β-caryophyllene synthase | AAX16076 | |
| ye_HN6LTMS01E1WK7 | 389 | 1 | 0 | α-humulene/β-caryophyllene synthase | CBI18625 | ||
| ye_HN6LTMS01CQ9FP | 451 | 1 | 0 | α-humulene/β-caryophyllene synthase | ADK73618 | ||
| isotig30406 | 378 | 2 | 0 | α-humulene/β-caryophyllene synthase | ADK73618 | ||
| ye_HN6LTMS01BB18N | 440 | 1 | 0 | α-humulene/β-caryophyllene synthase | ADV24747 | ||
| isotig14598 | 342 | 7 | 0 | α-humulene/β-caryophyllene synthase | ADK73619 | ||
| isotig34119 | 454 | 7 | 0 | (+)-α-barbatene synthase | CAH10288 | ||
| Di- | isotig23951 | 419 | 1 | 1 | ent-kaurene synthase | CBI32839 | |
| Tri- | gen_HITXW8201A8A9N | 425 | 0 | 1 | squalene synthases | AFK29284 | |
| gen_HITXW8201DAZL3 | 361 | 0 | 1 | squalene synthases | AAV58897 | ||
| gen_HITXW8201A3HRU | 406 | 0 | 1 | squalene synthases | AER23670 | ||
Prediction of candidate enzymes in tanshinone biosynthetic pathway
Because tanshinones are highly accumulated in root, the genes encoding enzymes of tanshinones biosynthesis are expected to show preferential, if not exclusive, expression in root. We first examined the expression pattern of known enzymes in the pathway. Early steps of tanshinones biosynthesis has been partially characterized: the precursor GGPP is converted to CPP by SmCPS [7], then to miltiradiene by the diterpene synthase kaurene synthase-like (SmKSL) enzyme [7], followed by hydroxylation to ferruginol by a P450 monooxygenase, CYP76AH1 [9]. As expected, the transcripts of SmCPS, SmKSL and CYP76AH1 were apparently more abundant in root than in leaf. While SmCPS and SmKSL genes were specifically expressed in root, the CYP76AH1 expression in root was 3.7-fold higher in root than in leaf (Table 4).
Table 4. Known and candidate genes involved in tanshinones biosynthesis.
| Gene | Unigene ID | RPKM in leaf | RPKM in root | Product |
|---|---|---|---|---|
| Cpalyl diphosphate synthase (SmCPS) | isotig01513 | 0 | 241.03 | Cpalyl diphosphate (CPP) |
| Kurene synthase-like (SmKSL) | isotig26056 | 0 | 16.34 | Miltiradiene |
| SmCYP76AH1 | isotig05166 | 5.88 | 27.37 | Ferruginol |
| Cytochrome P450s | isotig06040 | 0.00 | 173.28 | Unknown |
| isotig11627 | 4.45 | 275.21 | ||
| isotig06191 | 3.20 | 701.62 | ||
| isotig06719 | 0.00 | 688.94 | ||
| isotig00280 | 59.77 | 352.42 | ||
| isotig04519 | 69.75 | 463.00 | ||
| isotig04821 | 89.05 | 491.53 | ||
| isotig13353 | 15.26 | 330.35 | ||
| isotig00944 | 11.25 | 170.66 | ||
| isotig12561 | 47.05 | 451.06 | ||
| isotig04473 | 41.63 | 334.71 | ||
| isotig00674 | 21.93 | 135.62 | ||
| isotig12886 | 4.87 | 245.79 | ||
| isotig01966 | 0.00 | 88.54 | ||
| isotig14439 | 5.95 | 276.17 | ||
| isotig04633 | 0.00 | 113.34 | ||
| isotig04603 | 0.00 | 109.85 | ||
| isotig03664 | 22.73 | 161.40 | ||
| isotig05120 | 26.50 | 151.73 | ||
| isotig13720 | 10.53 | 173.60 | ||
| isotig15194 | 14.96 | 238.98 | ||
| isotig01982 | 0.00 | 47.49 | ||
| isotig10408 | 0.00 | 90.47 | ||
| isotig09263 | 27.23 | 156.36 | ||
| isotig09616 | 0.00 | 81.78 | ||
| isotig05760 | 12.52 | 93.56 | ||
| isotig03639 | 5.04 | 59.80 | ||
| isotig05574 | 15.22 | 94.16 | ||
| isotig13458 | 0.00 | 85.12 | ||
| isotig12292 | 4.64 | 86.16 | ||
| isotig09252 | 7.76 | 80.04 | ||
| isotig14674 | 6.29 | 103.70 | ||
| isotig01290 | 17.46 | 61.18 | ||
| isotig12891 | 63.55 | 196.53 | ||
| isotig09542 | 0.00 | 49.01 | ||
| isotig13691 | 0.00 | 59.97 | ||
| isotig12983 | 0.00 | 55.56 | ||
| isotig12207 | 0.00 | 52.54 | ||
| isotig13148 | 0.00 | 56.47 | ||
| isotig10175 | 4.11 | 59.27 | ||
| isotig01300 | 10.51 | 43.33 | ||
| isotig03088 | 9.42 | 48.55 | ||
| isotig09140 | 0.00 | 39.80 | ||
| isotig06948 | 0.00 | 34.65 | ||
| isotig25188 | 0.00 | 48.18 | ||
| isotig13449 | 0.00 | 53.07 | ||
| isotig12432 | 0.00 | 48.39 | ||
| isotig11783 | 4.50 | 55.62 | ||
| isotig11894 | 9.07 | 65.45 | ||
| isotig09674 | 19.96 | 82.29 | ||
| isotig02125 | 38.90 | 86.53 | ||
| isotig06295 | 6.45 | 43.21 | ||
| isotig13770 | 0.00 | 44.05 | ||
| isotig11385 | 22.16 | 82.22 | ||
| isotig12590 | 4.75 | 48.93 | ||
| isotig05973 | 12.63 | 9559.75 | ||
| isotig00853 | 37.86 | 2210.09 | ||
| isotig09109 | 354.60 | 969.42 | ||
| isotig12661 | 0.00 | 34.87 | ||
| isotig08939 | 0.00 | 561.04 | ||
| isotig00890 | 9.52 | 21.27 | ||
| isotig07498 | 6.98 | 57.53 | ||
| Double band reductase (SmDBR) | Isotig01665 | 136.05 | 313.59 | Unknown |
| Short-chain dehydrogenase (SmSD) | Isotig06117 | 6.34 | 98.08 | Unknown |
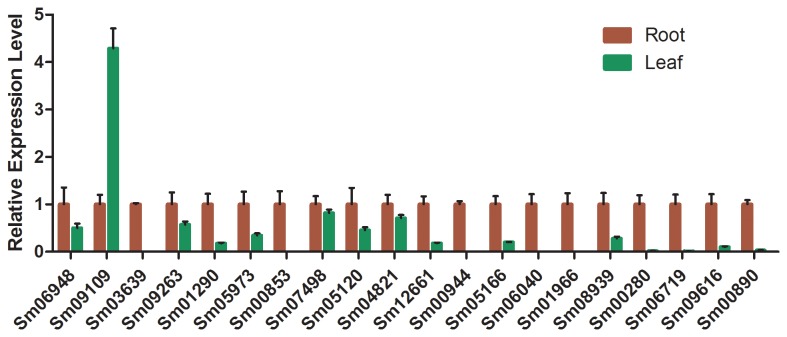
Cytochrome P450s are widely involved in the biosynthesis of secondary metabolites , including phenolic compounds, flavonoids, isoprenoids and alkaloids [35]. In our hypothetical tanshinones biosynthetic pathway, several steps are deduced to be catalysed by P450s (Figure 1). To identify additional candidate genes, RPKM values of P450 family unigenes in the two libraries were compared, which resulted in 63 P450 unigenes with > 2-fold (FDR < 0.05) expression levels in root than in leaf (Table 4). To validate this, 21 P450 unigenes were selected and their expression patterns were analysed by quantitative real-time RT-PCR (qRT-PCR), of which 20 showed higher expression levels in root, and only one showed a reverse pattern (Figure 4). These data suggested that the expression patterns deduced from the RPKM values in our transcriptome analyses are reliable.
Figure 4. Relative expression levels of P450 genes in S. miltiorrhiza root and leaf.
Total RNAs were extracted from root and leaf of 1-year-old S. miltiorrhiza and expression levels of P450s were analysed by quantitative real-time PCR. ACTIN was used as the internal reference gene and the relative abundance of each P450 genes in roots were compared to those in leaf, which were all set to be 1. Error bars indicate standard deviations of three biological replicates.
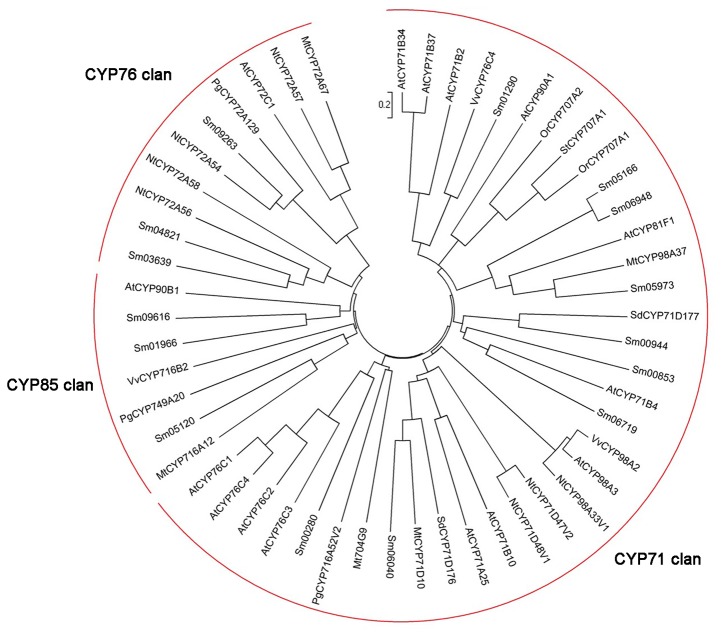
We then used 5’ and 3’ RACE (rapid amplification of cDNA end) to isolate full length cDNAs (with likely complete open reading frames) of 15 P450 genes. Phylogenetic analysis of the deduced protein sequences with P450s from other plants revealed that 9 of them (Sm01290, Sm05166, Sm06948, Sm05973, Sm00944, Sm00853, Sm06719, Sm06040 and Sm00280) belong to the CYP71 clan, 3 (Sm05120, Sm01966 and Sm09616) to the CYP85 clan, and the rest 3 (Sm03639, Sm04821 and Sm09263) to the CYP72 clan (Figure 5).
Figure 5. Phylogenetic analysis of CYP450s from S. miltiorrhiza and other plants.
Amino acid sequences were aligned using CLUSTALW program, and evolutionary distances were calculated using MEGA4 software with the Poisson correction method. Nt, Nicotiana tabacum; Pg, Panax ginseng; Mt, Medicago truncatula; Vv, Vitis vinifera; Sd, Scoparia dulcis; Or, Orobanche ramose; St, Solanum tuberosum; Sm, Salvia miltiorrhiza. The GenBank/EMBL/DDBJ accession numbers of the sequences are ABC69417.1 (NtCYP72A54), AEY75218.1 (PgCYP72A129), ABC69414.1 (NtCYP72A57), ABC69422.1 (NtCYP72A58), ABC69419.1 (NtCYP72A56), ABC59075.1 (MtCYP72A67), ABC59086.1 (MtCYP98A37), XP_002283338 (VvCYP98A2), ABC69384.1 (NtCYP98A33V1), NP_189259.1 (AtCYP71B4), NP_195459.1 (AtCYP81F1), NP_182079.1 (AtCYP76C4), NP_182081.1 (AtCYP76C2), ADA70805.1 (SdCYP71D176), XP_003617706.1 (MtCYP71D10), NP_200536.3 (AtCYP71B10), NP_680107.1 (AtCYP71A25), XP_002266024.1 (VvCYP716B2), NP_850337.1 (AtCYP98A3), NP_850439.1 (AtCYP76C1), NP_172767.1 (AtCYP71B2), XP_002276576.1 (VvCYP76C4), NP_173149.1 (AtCYP72C1), XP_003592376.1 (Mt704G9), AEY75214.1 (PgCYP749A20), NP_196188.1 (AtCYP90A1), AFO63032.1 (PgCYP716A52V2), ABC59076.1 (MtCYP716A12), NP_190635.1 (AtCYP90B1), NP_182082.2 (AtCYP76C3), AFP74115.1 (OrCYP707A2), ABA55732.1 (StCYP707A1), AFP74114.1 (OrCYP707A1), ADA70806.1 (SdCYP71D177), ABC69395.1 (NtCYP71D47V2), ABC69397.1 (NtCYP71D48V1), NP_189261.1 (AtCYP71B34), NP_189264.3 (AtCYP71B37).
The CYP71 clan P450s are in the A-type P450 clade, and contains the most of P450 families involved in plant secondary metabolism, such as CYP73, CYP98, CYP76 and CYP706 families [35]. For example, the rice CYP701A8 gene encodes an ent-kaurene oxidase that catalyses C3α-hydroxylation of ent-sandaracopimaradiene and ent-cassadiene, both are biosynthetic intermediates of the oryzalexin and the phytocassane families of phytoalexins [36]. In addition, a diterpenoid biosynthetic gene cluster was identified on rice chromosome 2, in which two genes encoding CYP71 clan members of CYP76M7 and CYP71Z7 catalyse the early and later steps in phytocassane biosynthesis, respectively [37,38]. Thus, the S. miltiorrhiza CYP71 clan members can be of particular interests in further elucidation of biosynthetic pathway of tanshinones. Due to distinct catalytic activities of P450 monooxygenases, differentially expressed P450 genes of the CYP85 and CYP72 clans also hold the potential to be enzymes of the pathway.
Biosynthesis of tanshinones involves downstream steps of decarboxylation, oxidation and reductions, which can be catalysed by decarboxylase, dehydrogenase and reductase, respectively [7,9]. The dehydrogenase-catalysed dehydration steps are likely involved in the conversion of ferruginol to cryptotanshinone, whereas the conversion of cryptotanshinone to tanshinone IIA requires a double band reduction at the position between C15-C16, which may be catalysed by a reductase (Figure 1). Unigene dataset was searched with Zingiber zerumbet short-chain dehydrogenase (ZSD1, AB480831) [39] and Artemisia annua aldehyde Δ11(13) reductase 2 (DBR2, EU704257) [40] to identify candidate dehydrogenases and reductases. One gene showed 66% nucleotide sequence identity to ZSD1 (named SmSD) and another showed 69% identity to DBR2 (named SmDBR). Both genes were expressed at significantly higher levels in root than in leaf, with 15.5-fold of RPKM value (FDR < 0.01) for SmSD and 2.3-fold (FDR < 0.01) for SmDBR (Table 4), suggesting a high possibility of SmSD and SmDBR being involved in the biosynthesis of tanshinones.
Prediction of transcription factors in tanshinones biosynthetic pathway
In plants, transcription factors of different families have been found to regulate secondary metabolism pathways, including those of WRKY, AP2/ERF, MYB and bHLH families [41,42]. In our unigene dataset, 735 genes were annotated as transcription factors, including zinc finger (148), AP2/ERF (82), MYB (82) and WRKY (61) family members. Among these, 209 were expressed in both root and leaf tissues, 67 showed a higher expression level in root and 45 in leaf (Table S4 and S5). Of the root up-regulated transcription factors, members of the AP2/ERF and GRAS families were markedly enriched, with 8 AP2/ERF genes up-regulated in root versus 2 in leaf, and 5 GRAS genes enriched in root versus only 1 in leaf (Table 5). Considering the root-specific accumulation of tanshinones, such higher expressions of these transcription factors in root suggest that they are worthy of further investigation.
Table 5. Summary of transcription factor unigenes of S. miltiorrhiza.
| TFs family | Number of genes detected | Up-regulated in root | Up-regulated in leaf |
|---|---|---|---|
| WRKY | 61 | 8 | 6 |
| MYB | 82 | 8 | 6 |
| AP2/ERF | 82 | 8 | 2 |
| MYC | 9 | 0 | 2 |
| GRAS | 30 | 5 | 1 |
| zinc finger | 148 | 11 | 7 |
| bHLH | 42 | 5 | 4 |
| bZIP | 34 | 3 | 2 |
| Others | 247 | 19 | 15 |
| Total | 735 | 67 | 45 |
The AP2/ERF members have been shown regulate secondary metabolism pathways in several medicinal plants. In A. annua, two JA-responsive AP2/ERF proteins AaERF1 and AaERF2 positively regulate genes encoding amorpha-4,11-diene synthase (ADS, a sesquiterpene synthase) and CYP71AV1, two key enzymes of artemisinin biosynthesis [43]. In Catharanthus roseus, the AP2/ERF proteins of ORCA2 and ORCA3 regulate terpenoid indole alkaloid metabolism through binding to the promoter of the strictosidine synthase (STR) gene and activating its expression [44,45]. In present analysis, 82 unigenes were annotated as AP2/ERF family transcription factors, of which 8 were drastically abundant in root and thus may be related to root-specific production of tanshinones.
Root plays a vital role in plant growth and development. The transcription factors of GRAS family, such as SHORT-ROOT (SHR) and SCARECROW (SCR), have been reported to play a critical role in root cortex development [46-48]. SHR protein acts as both a signal from the stele and an activator of endodermal cell fate whereas SCR mediats cortex cell division [49]. Moreover, it has been proposed that SHR and SCR have a conserved function in determining media cell fates and multiplication of cell layers in rice and Arabidopsis [50]. From the S. miltiorrhiza transcriptome, two genes were identified as SHR and SCR homologs, respectively. Tanshinones accumulate mainly in cortex of S. miltiorrhiza root, thus the identification of GRAS family factor genes expressed in root may provide an insight into the relation between cortex development and biosynthesis of tanshinones.
Conclusions
This study generates the largest dataset of unigenes for S. miltiorrhiza and provides candidate enzymes involved in tanshinones biosynthesis based on their root-preferential expression pattern, including CYP450s, dehydrogenases and reductases. Candidate transcription factors were also identified, which are of great interests in further investigation of the relationship between biosynthesis of tanshinones and root development. Our data are also of great value to understanding the biosynthesis and regulation of secondary metabolites in perennial plants.
Materials and Methods
Plant tissue collection
Seeds of Salvia miltiorrhiza Bunge were collected from Anhui Province, China, and grown in field in Shanghai Chenshan Botanical Garden with approval of Shanghai Chenshan Botanical Garden. No protected plant species was sampled for this research. Leaf and root materials from 1-year-old plants were collected, immediately frozen in liquid nitrogen and stored at -80 °C prior to RNA extraction.
RNA extraction
Total RNAs from roots and leaves of three replicates were extracted using the TRIzol Reagent (Invitrogen) and treated with DNase I (Takara) according to manufacturer’s instructions. RNA quality was examined using 1% agarose gel and the concentration was determined using a Nanodrap spectrophotometer (Thermo). Root and leaf RNA pools were prepared by mixing equal amounts of three RNA replicates.
cDNA library construction and 454 sequencing
Reverse transcription was performed with the SMART IITM cDNA Synthesis Kit (Clontech, USA) according to manufacturer’s instructions. Double stranded cDNAs were separated on 2% agarose gel, and those >100 bp were recovered. Concentrations of cDNAs were determined by Bioanalyzer 2100 (Agilent, Germany), and subjected to pyrosequencing with the GS-FLX Titanium Instrument (Roche). Image and signal processing were performed using 454 Life Science software (Roche). All sequence data have been deposited to the National Center for Biotechnology Information’s Sequence Read Archive (SRA) with the accession number of SRR1005880.
Assembly
To obtain high quality clean sequences, SeqClean was used to trim adapter sequences and Lucy (version 1.20p) was used to remove low quality sequences and those < 50 bp. Clean reads from root and leaf libraries were assembled into 16,806 isotigs and 84,116 singletons using Newbler (Roche, version 2.6) software. To reduce the redundancy, clustering was performed with CD-HIT (version 4.0), and isotig and singleton sequences with minimum 95% identity were merged into a single representative unigene.
Annotation of unigenes
Unigenes were used as query sequences to search against the non-redundant protein (NR) database at NCBI (http://www.ncbi.nlm.nih.gov) and the Swiss-Prot protein database (http://www.ebi.ac.uk/uniprot) with E-value cutoff of 1e-5. The annotations of the best hits were recorded. Gene Ontology (GO) (http://www.geneontology.org/) were further used to category the function of the unigenes by Blast2GO [51], and the unigenes were assigned to biological functions on the macro levels of “biological process”, “cellular component” and “molecular function”. The Kyoto Encyclopedia of Genes and Genome (KEGG) pathways database (http://www.genome.jp/kegg/) were assigned to unigenes by KEGG Automatic Annotation Server (KAAS) [52].
Differential expression analysis
Gene expression levels of unigenes in roots or leaves were normalized and calculated as reads per kb per million reads (RPKM) values during the assembly and clustering process. Significance of differential gene expression between the root and leaf tissues was determined by a p-value < 0.05 by random test and corrected using the false discovery rate (FDR).
Determination of tanshinones by HPLC
Determination of tanshinones contents was carried out with an Agilent 1260 Infinity HPLC system as described [53], and equipped with a diode array detector (DAD, G4212B) using a SB-C18 Analytical HPLC Column (4.6 x 250 mm, 5 um, Agilent). The mobile phase consisted of 0.1% (vol/vol) formic acid methanol solution (A) and 0.1% (vol/vol) formic acid aqueous solution (B), with a gradient elution from 37% to 80% at a flow rate of 1.0 mL∙min-1. The HPLC chromatogram was monitored at 270 nm and the column temperature was set at 30 °C. The dihydrotanshinone I, tanshinone I, cryptotanshinone and tanshinone IIA were determined by comparing to authorized standards (WuXi App. Tec., China).
Quantitative real-time RT-PCR
Total RNA were reverse transcribed with the ReverTra Ace -α- kit (Toyobo) according to manufacturer’s protocol. Real-time PCR was performed using the SYBR® Premix Ex Taq™ II (Perfect Real Time) kit (Takara) on a Mastercycler system (Eppendorf, Germany) with gene-specific primer pairs (Table S6). ACTIN was used as the internal reference gene [54]. The relative expression value was calculated via the 2-ΔΔCt method [55].
Full-length cDNA verification and cloning of CYP450s
The 5’ and 3’ RACE (rapid amplification of cDNA end) were performed by the 5’-Full RACE Kit and the 3’-Full RACE Core Set Ver.2.0 (Takara) according to manufacturer’s instructions. PCR products were cloned into the pMD18-T Vector (Takara) for Sanger sequencing.
Full-length cDNAs were obtained by assembling fragments obtained by 5’ and 3’ RACE in combination with annotated unigene sequences. P450 sequences from other plant species were obtained by searching the GenBank database at NCBI. The amino acid sequence alignment of P450 proteins was performed with CLUSTALW program of MEGA 4 software with default parameters. Phylogenetic tree was built with the neighbour-joining method with MEGA 4 program.
Supporting Information
HPLC analyses of tanshinones in S. miltiorrhiza root and leaf. S. miltiorrhiza root (A) and leaf (B).
(TIF)
Functional classification of metabolic pathways related to secondary metabolism by KEGG.
(XLSX)
List of unigenes expressed more abundantly in S. miltiorrhiza leaf.
(XLSX)
List of unigenes that are more abundant in S. miltiorrhiza root.
(XLSX)
Transcription factors that showed higher expression levels in S. miltiorrhiza root.
(XLSX)
Transcription factors that showed higher expression in S. miltiorrhiza leaf.
(XLSX)
List of oligonucleotide primer sequences. All primers used for full-length cDNA cloning and qRT-PCR analyses of P450s in present study are listed.
(XLSX)
Acknowledgments
The authors are grateful to Shanghai Oebiotech CO. LTD. for sequencing and data analysis, Jiawei Wang, Zhonghua Tang and Juan Guo for helpful discussions.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant No. 31200222), and the Special Fund for Shanghai Landscaping Administration Bureau Program (grant No. F132424 and F112418). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhou L, Zuo Z, Chow MS (2005) Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol 45: 1345-1359. doi: 10.1177/0091270005282630. PubMed: 16291709. [DOI] [PubMed] [Google Scholar]
- 2. Huang B, Yi B, Duan Y, Sun L, Yu X et al. (2008) Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol Biol Rep 35: 601-612. doi: 10.1007/s11033-007-9130-2. PubMed: 17805988. [DOI] [PubMed] [Google Scholar]
- 3. Xiao Y, Zhang L, Gao S, Saechao S, Di P et al. (2011) The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PLOS ONE 6: e29713. doi: 10.1371/journal.pone.0029713. PubMed: 22242141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dong Y, Morris-Natschke SL, Lee KH (2011) Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat Prod Rep 28: 529-542. doi: 10.1039/c0np00035c. PubMed: 21225077. [DOI] [PubMed] [Google Scholar]
- 5. Li YI, Elmer G, Leboeuf RC (2008) Tanshinone IIA reduces macrophage death induced by hydrogen peroxide by upregulating glutathione peroxidase. Life Sci 83: 557-562. doi: 10.1016/j.lfs.2008.08.003. PubMed: 18762198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laule O, Fürholz A, Chang HS, Zhu T, Wang X et al. (2003) Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A 100: 6866-6871. doi: 10.1073/pnas.1031755100. PubMed: 12748386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao W, Hillwig ML, Huang L, Cui G, Wang X et al. (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11: 5170-5173. doi: 10.1021/ol902051v. PubMed: 19905026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kai G, Liao P, Zhang T, Zhou W, Wang J et al. (2010) Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnology and Bioprocess Engineering 15: 236-245. doi: 10.1007/s12257-009-0123-y. [DOI] [Google Scholar]
- 9. Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L et al. (2013) CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proceedings of the National Academy of Sciences. PubMed: 23812755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan Y, Wang Z, Tian W, Dong Z, Spencer DF (2010) Generation and analysis of expressed sequence tags from the medicinal plant Salvia miltiorrhiza. Sci China Life Sci 53: 273-285. doi: 10.1007/s11427-010-0005-8. PubMed: 20596838. [DOI] [PubMed] [Google Scholar]
- 11. Cui G, Huang L, Tang X, Zhao J (2011) Candidate genes involved in tanshinone biosynthesis in hairy roots of Salvia miltiorrhiza revealed by cDNA microarray. Mol Biol Rep 38: 2471-2478. doi: 10.1007/s11033-010-0383-9. PubMed: 21082262. [DOI] [PubMed] [Google Scholar]
- 12. Wenping H, Yuan Z, Jie S, Lijun Z, Zhezhi W (2011) De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 98: 272-279. doi: 10.1016/j.ygeno.2011.03.012. PubMed: 21473906. [DOI] [PubMed] [Google Scholar]
- 13. Ma Y, Yuan L, Wu B, Li X, Chen S et al. (2012) Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot 63: 2809-2823. doi: 10.1093/jxb/err466. PubMed: 22291132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murata J, Roepke J, Gordon H, De Luca V (2008) The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell 20: 524-542. doi: 10.1105/tpc.107.056630. PubMed: 18326827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H, Penmetsa RV, Yuan M, Gong L, Zhao Y, et al. (2012) Development and characterization of BAC-end sequence derived SSRs, and their incorporation into a new higher density genetic map for cultivated peanut (Arachis hypogaea L.). BMC plant biology 12: 10 [DOI] [PMC free article] [PubMed]
- 16. Xu YH, Wang JW, Wang S, Wang JY, Chen XY (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol 135: 507-515. doi: 10.1104/pp.104.038612. PubMed: 15133151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li JT, Dong JE, Liang ZS, Shu ZM, Wan GW (2008) [Distributional difference of fat-soluble compounds in the roots, stems and leaves of four Salvia plants]. Fen zi xi bao sheng wu xue bao = Journal of molecular cell biology / Zhongguo xi bao sheng wu xue xue hui zhu ban 41: 44-52. [PubMed] [Google Scholar]
- 18. Ge X, Wu J (2005) Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sciences 168: 487-491. doi: 10.1016/j.plantsci.2004.09.012. [DOI] [Google Scholar]
- 19. Ge X, Wu J (2005) Induction and potentiation of diterpenoid tanshinone accumulation in Salvia miltiorrhiza hairy roots by beta-aminobutyric acid. Appl Microbiol Biotechnol 68: 183-188. doi: 10.1007/s00253-004-1873-2. PubMed: 15672269. [DOI] [PubMed] [Google Scholar]
- 20. Zhang C, Yan Q, Cheuk WK, Wu J (2004) Enhancement of tanshinone production in Salvia miltiorrhiza hairy root culture by Ag+ elicitation and nutrient feeding. Planta Med 70: 147-151. doi: 10.1055/s-2004-815492. PubMed: 14994193. [DOI] [PubMed] [Google Scholar]
- 21. Shi M, Kwok KW, Wu JY (2007) Enhancement of tanshinone production in Salvia miltiorrhiza Bunge (red or Chinese sage) hairy-root culture by hyperosmotic stress and yeast elicitor. Biotechnol Appl Biochem 46: 191-196. doi: 10.1042/BA20060147. PubMed: 17014425. [DOI] [PubMed] [Google Scholar]
- 22. Yan Q, Wu J, Liu R (2011) Modeling of tanshinone synthesis and phase distribution under the combined effect of elicitation and in situ adsorption in Salvia miltiorrhiza hairy root cultures. Biotechnol Lett 33: 813-819. doi: 10.1007/s10529-010-0484-5. PubMed: 21120586. [DOI] [PubMed] [Google Scholar]
- 23. Kai G, Liao P, Xu H, Wang J, Zhou C et al. (2012) Molecular mechanism of elicitor-induced tanshinone accumulation in Salvia miltiorrhiza hairy root cultures. Acta Physiologiae Plantarum 34: 1421-1433. doi: 10.1007/s11738-012-0940-z. [DOI] [Google Scholar]
- 24. Lamien-Meda A, Nell M, Lohwasser U, Börner A, Franz C et al. (2010) Investigation of antioxidant and rosmarinic acid variation in the sage collection of the genebank in Gatersleben. J Agric Food Chem 58: 3813-3819. doi: 10.1021/jf903993f. PubMed: 20187608. [DOI] [PubMed] [Google Scholar]
- 25. Ben Farhat M, Jordán MJ, Chaouech-Hamada R, Landoulsi A, Sotomayor JA (2009) Variations in essential oil, phenolic compounds, and antioxidant activity of tunisian cultivated Salvia officinalis L. J Agric Food Chem 57: 10349-10356. doi: 10.1021/jf901877x. PubMed: 19886685. [DOI] [PubMed] [Google Scholar]
- 26. Kamatou GP, Viljoen AM, Gono-Bwalya AB, van Zyl RL, van Vuuren SF et al. (2005) The in vitro pharmacological activities and a chemical investigation of three South African Salvia species. J Ethnopharmacol 102: 382-390. doi: 10.1016/j.jep.2005.06.034. PubMed: 16099614. [DOI] [PubMed] [Google Scholar]
- 27. Santos-Gomes PC, Seabra RM, Andrade PB, Fernandes-Ferreira M (2003) Determination of phenolic antioxidant compounds produced by calli and cell suspensions of sage (Salvia officinalis L.). Journal of plant physiology 160: 1025-1032. [DOI] [PubMed] [Google Scholar]
- 28. Basappa Maheswarappa N, Subbaiah V, Muthupalani M, Yamagani PK, Mohan K et al. (2013) Antioxidant activity of carnosic acid and rosmarinic acid in raw and cooked ground chicken patties. J Sci Food Agric: ([MedlinePgn:]) PubMed: 23740828. [DOI] [PubMed] [Google Scholar]
- 29. Zu G, Zhang R, Yang L, Ma C, Zu Y et al. (2012) Ultrasound-Assisted Extraction of Carnosic Acid and Rosmarinic Acid Using Ionic Liquid Solution from Rosmarinus officinalis. Int J Mol Sci 13: 11027-11043. doi: 10.3390/ijms130911027. PubMed: 23109836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fuhrman B, Volkova N, Rosenblat M, Aviram M (2000) Lycopene synergistically inhibits LDL oxidation in combination with vitamin E, glabridin, rosmarinic acid, carnosic acid, or garlic. Antioxid Redox Signal 2: 491-506. doi: 10.1089/15230860050192279. PubMed: 11229363. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y, Smuts JP, Dodbiba E, Rangarajan R, Lang JC et al. (2012) Degradation study of carnosic acid, carnosol, rosmarinic acid, and rosemary extract (Rosmarinus officinalis L.) assessed using HPLC. J Agric Food Chem 60: 9305-9314. doi: 10.1021/jf302179c. PubMed: 22881034. [DOI] [PubMed] [Google Scholar]
- 32. Liu T, Sui X, Zhang R, Yang L, Zu Y et al. (2011) Application of ionic liquids based microwave-assisted simultaneous extraction of carnosic acid, rosmarinic acid and essential oil from Rosmarinus officinalis. J Chromatogr A 1218: 8480-8489. doi: 10.1016/j.chroma.2011.09.073. PubMed: 21999917. [DOI] [PubMed] [Google Scholar]
- 33. Yesil-Celiktas O, Sevimli C, Bedir E, Vardar-Sukan F (2010) Inhibitory effects of rosemary extracts, carnosic acid and rosmarinic acid on the growth of various human cancer cell lines. Plant Foods Hum Nutr 65: 158-163. doi: 10.1007/s11130-010-0166-4. PubMed: 20449663. [DOI] [PubMed] [Google Scholar]
- 34. Jordan MJ, Lax V, Rota MC, Loran S, Sotomayor JA (2012) Relevance of carnosic acid, carnosol, and rosmarinic acid concentrations in the in vitro antioxidant and antimicrobial activities of Rosmarinus officinalis (L.) methanolic extracts. J Agric Food Chem 60: 9603-9608. [DOI] [PubMed] [Google Scholar]
- 35. Bak S, Beisson F, Bishop G, Hamberger B, Höfer R et al. (2011) Cytochromes P450. Arabidopsis Book, 9: e0144 PubMed: 22303269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Q, Hillwig ML, Wu Y, Peters RJ (2012) CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol 158: 1418-1425. doi: 10.1104/pp.111.187518. PubMed: 22247270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11alpha-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315-3325. doi: 10.1105/tpc.108.063677. PubMed: 19825834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu Y, Hillwig ML, Wang Q, Peters RJ (2011) Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett 585: 3446-3451. doi: 10.1016/j.febslet.2011.09.038. PubMed: 21985968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Okamoto S, Yu F, Harada H, Okajima T, Hattan J et al. (2011) A short-chain dehydrogenase involved in terpene metabolism from Zingiber Zerumbet. FEBS J 278: 2892-2900. doi: 10.1111/j.1742-4658.2011.08211.x. PubMed: 21668645. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A et al. (2008) The molecular cloning of artemisinic aldehyde Delta11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem 283: 21501-21508. PubMed: 18495659. [DOI] [PubMed] [Google Scholar]
- 41. De Geyter N, Gholami A, Goormachtig S, Goossens A (2012) Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci 17: 349-359. doi: 10.1016/j.tplants.2012.03.001. PubMed: 22459758. [DOI] [PubMed] [Google Scholar]
- 42. Yang CQ, Fang X, Wu XM, Mao YB, Wang LJ et al. (2012) Transcriptional regulation of plant secondary metabolism. J Integr Plant Biol 54: 703-712. doi: 10.1111/j.1744-7909.2012.01161.x. PubMed: 22947222. [DOI] [PubMed] [Google Scholar]
- 43. Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ et al. (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5: 353-365. doi: 10.1093/mp/ssr087. PubMed: 22104293. [DOI] [PubMed] [Google Scholar]
- 44. Menke FL, Champion A, Kijne JW, Memelink J (1999) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J 18: 4455-4463. doi: 10.1093/emboj/18.16.4455. PubMed: 10449411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. The Plant journal : for cell and molecular biology 25: 43-53. [DOI] [PubMed]
- 46. Sánchez C, Vielba JM, Ferro E, Covelo G, Solé A et al. (2007) Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol 27: 1459-1470. doi: 10.1093/treephys/27.10.1459. PubMed: 17669736. [DOI] [PubMed] [Google Scholar]
- 47. Dhondt S, Coppens F, De Winter F, Swarup K, Merks RM et al. (2010) SHORT-ROOT and SCARECROW regulate leaf growth in Arabidopsis by stimulating S-phase progression of the cell cycle. Plant Physiol 154: 1183-1195. doi: 10.1104/pp.110.158857. PubMed: 20739610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Torres-Galea P, Huang L-F, Chua N-H, Bolle C (2006) The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Molecular Genetics and Genomics 276: 13-30. doi: 10.1007/s00438-006-0123-y. PubMed: 16680434. [DOI] [PubMed] [Google Scholar]
- 49. Nakajima K, Benfey PN (2002) Signaling in and out: control of cell division and differentiation in the shoot and root. Plant Cell 14 Suppl: S265-S276. PubMed: 12045282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ et al. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421-425. doi: 10.1126/science.1139531. PubMed: 17446396. [DOI] [PubMed] [Google Scholar]
- 51. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674-3676. doi: 10.1093/bioinformatics/bti610. PubMed: 16081474. [DOI] [PubMed] [Google Scholar]
- 52. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35: W182-W185. doi: 10.1093/nar/gkm321. PubMed: 17526522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li MH, Chen JM, Peng Y, Wu Q, Xiao PG (2008) Investigation of Danshen and related medicinal plants in China. J Ethnopharmacol 120: 419-426. doi: 10.1016/j.jep.2008.09.013. PubMed: 18930799. [DOI] [PubMed] [Google Scholar]
- 54. Yang Y, Hou S, Cui G, Chen S, Wei J et al. (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 37: 507-513. doi: 10.1007/s11033-009-9703-3. PubMed: 19680786. [DOI] [PubMed] [Google Scholar]
- 55. Guterman I, Shalit M, Menda N, Piestun D, Dafny-Yelin M et al. (2002) Rose scent: genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14: 2325-2338. doi: 10.1105/tpc.005207. PubMed: 12368489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X, Morris-Natschke SL, Lee KH (2007) New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 27: 133-148. doi: 10.1002/med.20077. PubMed: 16888751. [DOI] [PubMed] [Google Scholar]
- 57. Baskan S, Oztekin N, Erim FB (2007) Determination of carnosic acid and rosmarinic acid in sage by capillary electrophoresis. Food Chemistry 101: 1748-1752. doi: 10.1016/j.foodchem.2006.01.033. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC analyses of tanshinones in S. miltiorrhiza root and leaf. S. miltiorrhiza root (A) and leaf (B).
(TIF)
Functional classification of metabolic pathways related to secondary metabolism by KEGG.
(XLSX)
List of unigenes expressed more abundantly in S. miltiorrhiza leaf.
(XLSX)
List of unigenes that are more abundant in S. miltiorrhiza root.
(XLSX)
Transcription factors that showed higher expression levels in S. miltiorrhiza root.
(XLSX)
Transcription factors that showed higher expression in S. miltiorrhiza leaf.
(XLSX)
List of oligonucleotide primer sequences. All primers used for full-length cDNA cloning and qRT-PCR analyses of P450s in present study are listed.
(XLSX)