Abstract
Fatty acid metabolism is an attractive route to produce liquid transportation fuels and commodity oleochemicals from renewable feedstocks. Recently, genes and enzymes, which comprise metabolic pathways for producing fatty acid-derived compounds (e.g. esters, alkanes, olefins, ketones, alcohols, polyesters) have been elucidated and used in engineered microbial hosts. The resulting strains often generate products at low percentages of maximum theoretical yields, leaving significant room for metabolic engineering. Economically viable processes will require strains to approach theoretical yields, particularly for replacement of petroleum-derived fuels. This review will describe recent progress toward this goal, highlighting the scientific discoveries of each pathway, ongoing biochemical studies to understand each enzyme, and metabolic engineering strategies that are being used to improve strain performance.
Introduction
Production of fuels and petrochemicals from renewable resources (e.g. photosynthetically produced biomass in plants and algae) is an attractive strategy to meet growing demand while simultaneously curbing the net release of greenhouse gases. A common approach to implementing this strategy is to engineer the metabolism of a microorganism to convert biomass-derived sugars to desired products. Engineered strains can then be cultivated in industrial-scale bioreactors and the resulting products purified using traditional chemical and biochemical engineering techniques. Of the many metabolic pathways found in nature, fatty acid metabolism has received significant attention as a biological route to convert sugars to liquid transportation fuels and higher value oleochemicals (Figure 1). This review will focus on recent progress in developing microbes for converting renewable feed-stocks to fuels and chemicals derived from fatty acid metabolism. It should be noted that other groups are pursuing the same goals using photosynthetic organisms and/or traditional catalytic technologies to produce fuels and chemicals [1–3].
Figure 1.
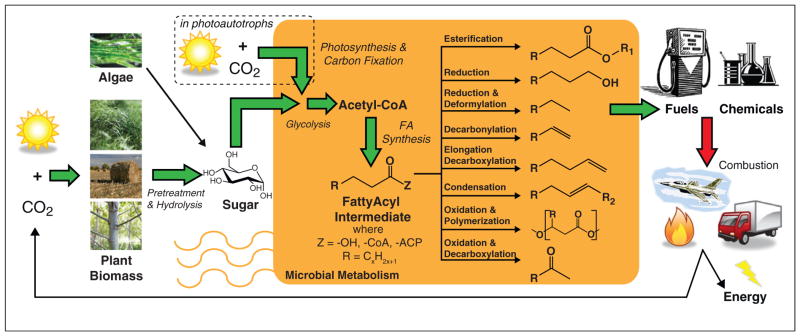
Sustainable production of fatty acid-derived fuels and chemicals. The process begins with the conversion of photosynthetic biomass (e.g. plants or algae) to fermentable intermediates that can be catabolized by cells to generate energy and the building blocks (e.g. acetyl-CoA) required to synthesize anabolic products, including fatty acids. Alternatively, these same chemicals can be derived directly from photosynthesis and carbon fixation. Acetyl-CoA is the direct precursor to fatty acid biosynthetic intermediates that are the starting point for a wide range of chemical products. These products can be used as fuels or chemicals. Upon combustion, the fatty acid-derived products release energy and carbon dioxide that can be recycled to close the carbon cycle.
Fatty acid metabolism is one of a handful of anabolic pathways capable of producing large (>6 carbon) hydrophobic metabolites that are similar to molecules found in high energy petroleum distillates. Other pathways, including isoprenoid and polyketide biosynthesis, have been described elsewhere [4–7]. Higher energy density distillates such as diesel and jet fuel are needed by the heavy transportation sector due to weight and range constraints. Unlike the case for personal vehicles, these constraints also functionally prohibit electricity as a power source, due to the energy storage limitations of modern batteries. Instead, microbially produced fatty acid derivatives have been viewed as a supplement to and potentially a replacement for petrodiesel and jet fuel. This potential has motivated research efforts to metabolically engineer microbes to approach theoretical yields from lignocellulosic feedstocks. Unlike short chain alcohols, the low water solubility of longer carbon chain-length hydrocarbons and esters should result in reduced recovery costs and reduced toxicity in the fermentation broth due to phase separation. In addition, fatty acid derivatives are more likely to be compatible with existing engines as well as transport and storage infrastructure.
In addition to fuels, fatty acid metabolism is a route to many oleochemicals that are currently made from fossil fuels or plant oils. These compounds include oils, waxes, free fatty acids (FFA), fatty alcohols, and bioplastics, which are used as surfactants, lubricants, and materials in many consumer products. Conversion of biomass-derived sugars to these compounds is economically attractive because the ability of microbial catalysts to control product chain length, add precise modifications, and use non-plant oils as feedstocks. Oleochemicals are sold at higher prices than fuels and therefore commercial processes that leverage microbial catalysts will likely target non-fuel applications first.
While the ability of microbes to produce hydrocarbons was discovered decades ago, many of the biochemical pathways for converting intermediates in fatty acid metabolism to downstream products have only been recently elucidated. Pathways for producing fatty acid-derived chemicals start from fatty acyl–acyl carrier protein (ACP) thioesters (in fatty acid biosynthesis), fatty acylcoenzyme A (CoA) thioesters (in fatty acid catabolism), or free fatty acids (from thioesterase cleavage of acylthioesters). The remainder of the review will focus on each product group, recent discoveries, and current challenges to commercial deployment. In general, current research is focusing on identifying homologous pathways in various organisms, characterizing the enzymes that comprise each pathway, and applying metabolic engineering strategies to maximize yields of each product. The major challenges to commercialization lie in increasing yields of products such that they approach the theoretical limits (~0.25–0.40 g/g glucose), engineering strategies for controlling product chain length, and developing strains that can process lignocellulosic hydrolysates.
Biodiesel
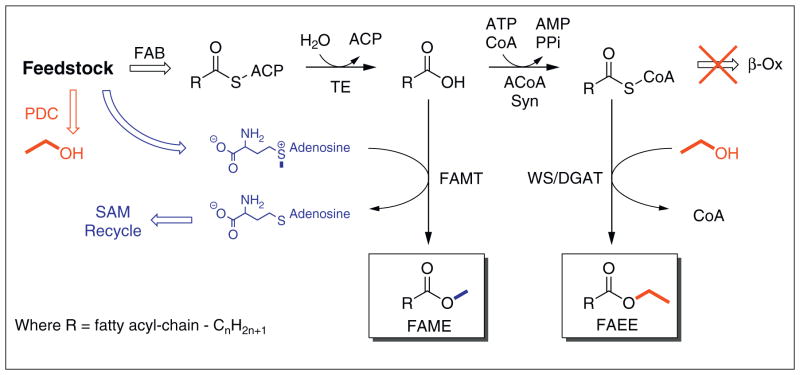
Biodiesel is a mixture of fatty acid methyl esters (FAMEs) that is currently being produced from plant oils, animal fats, and/or FFA using traditional catalysis [8]. Escherichia coli has also been engineered to produce FAME in situ by heterologous expression of a fatty acid O-methyltransfer-ase (FAMT) from Mycobacterium marinum or similar homologues (Figure 2) [9•]. The tested enzymes were most active on C8–C12 saturated and 3-hydroxylated FFA and used S-adenosylmethionine (SAM) as the methyl donor. In this study, the supply of SAM was increased by deletion of metJ, a global regulator that feedback inhibits SAM biosynthesis in response to elevated concentrations of SAM. Yields and titers were very low, with the bottleneck hypothesized to be the lack of thioesterase activity for producing 3-hydroxydecanoic acid, the preferred substrate of the FAMT from M. marinum. Achieving higher yields of FAME will likely require a combination of protein engineering (improving thioesterase and FAMT activities against various fatty acids) and metabolic engineering (optimizing the recycling of SAM).
Figure 2.
Metabolic pathways for synthesis of biodiesel. Microbes use sugars and other renewable feedstocks to produce a wide range of metabolites including ethanol, S-adenosylmethionine (SAM), and fatty acids. Fatty acid metabolism is used to produce long chain acyl-ACP thioesters where typically R = C15H31–C17H35. These acyl-chains which are natively incorporated into membrane lipids can be hydrolyzed by thioesterases (TE) to produce free fatty acids (FFAs). FFAs are the substrate of fatty acid O-methyltransferase (FAMT) which adds a methyl group from SAM to yield fatty acid methyl esters (FAME). Alternatively, FFA can be activated to acyl-CoA thioesters by acyl-CoA synthetases (ACoA Syn). These thioesters can be broken down for energy by the β-oxidation (β-ox) pathway that is often disrupted in engineered strains. Acyl-CoAs can be transesterified by wax ester synthase/acyl-CoA:diacylglycerol acyltransferases (WS/DGAT) using ethanol to yield fatty acid ethyl-esters FAEE. The by-products (e.g. CoA and S-adenosyl-homocysteine) are recycled by additional pathways in the cell.
Fatty acid ethyl esters (FAEEs), which are chemically similar to current biodiesel, can be produced from acyl-CoAs and ethanol using a promiscuous wax ester synthase/acyl-CoA:diacylglycerol acyltransferase (WS/DGAT) first described by the AtfA from Acinetobacter sp. ADP1 (Figure 2) [10••]. Homologues of AtfA have been identified in other γ-proteobacteria and actinomy-cetes and exhibit different substrate specificities [11–13], including an isoprenoid wax ester synthase in Marinobacter hydrocarbonoclasticus that could be used to make highly branched esters with reduced cloud points [14]. To produce FAEEs, fatty acids and ethanol must be fed to a strain harboring AtfA or the atfA expressing strain must be engineered to produce both metabolites. In E. coli, ethanol production can be increased by heterologous expression of pyruvate decarboxylase (Pdc) and alcohol dehydrogenase (AdhB) from the ethanologen Zymomonas mobilis [15]. When E. coli expressing atfA, pdc, and adhB on a plasmid was fed oleate in the fermentation broth, up to 1.28 g/L of FAEE was produced [16]. Similarly, when atfA was expressed in engineered ethanol producing Saccharomyces cerevisiae, 0.24–0.52 g/L of FAEE was produced when cells were fed fatty acids [17]. Expression of atfA in a FFA producing E. coli with exogenous feeding of ethanol produced a titer of 0.4 g/L of FAEE [18•]. The first demonstration of FAEE synthesis without feeding used three plasmids to overexpress atfA (on two separate plasmids), pdc, adhB, tesA′ and fadD (to increase acyl-CoA synthesis from FFA). When E. coli (DH1 ΔfadE) harboring these plasmids was cultivated in broth overlaid with dodecane, the culture produced 30.7 g/L of FAEE, representing approximately 7% of the maximum theoretical yield [18•]. Fed batch cultivation of a similar strain increased production to 0.922 g/L at the expense of yield (reported as 0.025 g FAEE g−1 glucose) [19]. A similar strain was used to produce biodiesel from a minimal media formulated with ionic liquid pretreated switch-grass, albeit at low rates (100 mg/L in 3200 hours) [20]. It should be noted that this strain coexpressed genes for producing ethyl esters and cellulases to release sugars from the pretreated biomass, a first step toward consolidated bioprocessing to produce advanced biofuels.
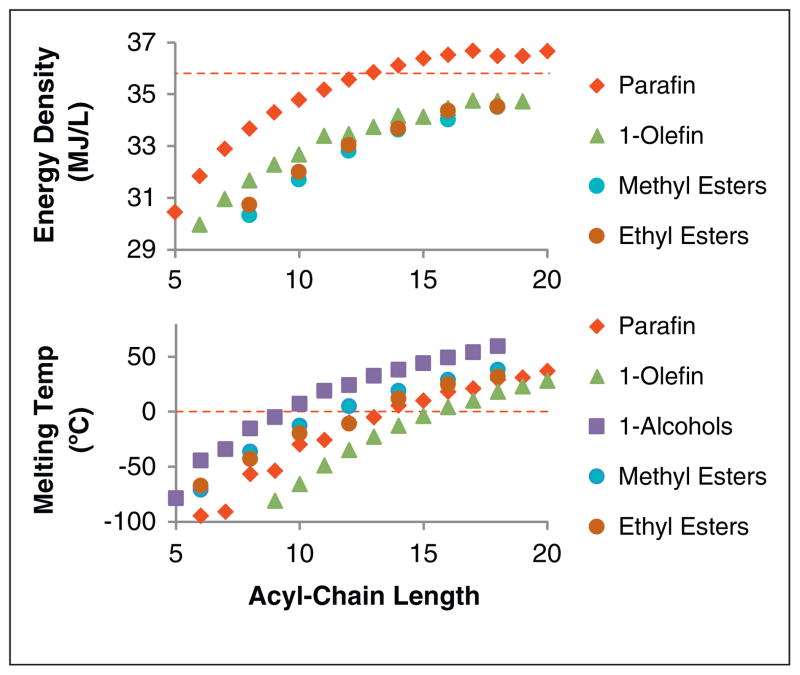
These initial demonstrations confirmed the ability of E. coli to produce biodiesel de novo from renewable carbon sources. While promising, three challenges remain: firstly increasing yields to approach theoretical limits; secondly attaining these yields from biomass-derived feedstocks; and finally tailoring FAME/FAEE composition to meet desired fuel properties (e.g. energy density and melting point, illustrated in Figure 3, and cetane number [ASTM > 40]). In the latter challenge, the preferred chain lengths, modifications, and degree of homogeneity (i.e. single component or blends) will depend on the demands of the final application. Product yields are a critical issue that will determine if microbial hydrocarbons will be competitive economically. At current prices, sugar (~$0.16–0.20 lb−1) will contribute $3.00–4.50 per gallon of FAEE, if produced at 95% of theoretical yield (Figure 4). The theoretical yield (TY) is defined here as the maximum stoichiometric yield of product given the known metabolic network of the cell, in addition to any heterologously introduced pathways. Significant efforts to decrease the cost of sugars are underway, but feedstock costs are likely to remain a major percentage of final fuel costs, as it is the case with current petroleum derived fuels. The challenges to achieving theoretical yields of FAEE include deregulating control and maximizing flux through fatty acid biosynthesis (FAB), balancing the rates of fatty acid synthesis, acyl-CoA synthesis, ethanol production, and FAEE synthesis, and developing cultivation strategies to minimize the amount of feedstock that is consumed for cell growth and maintenance. A recent synthetic biology study aimed at balancing the supply of ethanol and acyl-CoA succeeded in improving FAEE titer (1.5 g/L) and yield (reported as 28% of TY) [21••]. Substrate supply was controlled using a dynamic sensor-regulator system that comprised two FadR-regulated promoters. DNA binding by FadR is modulated by the presence of acyl-CoA. Therefore, the presence of excess acyl-CoA-led to derepression of promoters controlling transcription of genes involved in ethanol synthesis and conversion of acyl-CoA to FAEE enabling ethanol synthesis and conversion of acyl-CoA to FAEE (consumption of acyl-CoA). The final strain incorporated engineered FadR-responsive promoters that optimized FAEE yields, albeit with significant room for additional improvement.
Figure 3.
The properties of fatty acid derivatives depend on the length of the acyl chain. For example, the molar energy content increases with each additional carbon, but the volumetric energy density plateaus after 12–14 carbons near the density of petrodiesel (36 MJ/L). Similarly, the melting temperatures of alkanes, olefins, and esters corresponding to the common membrane fatty acids (C16, C18) are above 0 °C, which complicates their use as neat (i.e. single component) fuels in cold climates. These examples illustrate the need to control chain length in order to obtain useful chemical products. The properties of fatty acids are also influenced by incorporation of branches and other chemical modifications. Data were collected from CRC Handbook of Chemistry and Physics, NIST WebBook, and Materials Safety Data Sheets.
Figure 4.
Impact of raw material costs as a function of glucose price given product yields at 95% of the theoretical limit. Error bars represent yields ±5% of theoretical. Values were calculated from the following equation.
Alkanes, fatty aldehydes, and fatty alcohols
Bacteria have been known to synthesize hydrocarbons for many decades [22] and the search for enzymes responsible has been fraught with difficulties [23]. However, recent work has uncovered several pathways for producing long-chain (C10 < x < C20) and very-long chain (>C20) alkanes. For example, a bioinformatic comparison of cyanobacteria capable of producing and not producing alkanes identified two genes that were later shown to encode enzymes that produce fatty aldehydes and convert them to alkanes by removing the carbonyl carbon [24•]. The first enzyme was proven to be an acyl-ACP reductase that cleaves the thioester bond and uses NADPH to reduce the fatty acid to an aldehyde (Figure 5). Subsequent work re-classified the second enzyme as an aldehyde deformylase that uses oxygen and a protein reductive system (NADPH, ferridoxin, ferridoxin reductase) to generate an alkane, formate, and water [25,26]. When the acyl-ACP reductase and aldehyde deformylase were heterologously expressed in E. coli, up to 300 mg/L of pentadecane, pentadecene, and heptadecene was produced. The alkane composition matched the predominant fatty acids (16:0, 16:1, and 18:1) found in E. coli. The successful demonstration of alkane production motivates further metabolic engineering to improve yields. Microbially derived hydrocarbons possess higher energy densities and higher cloud points than current biodiesel blends, enabling year-round usage in all climates [27,28]. To leverage these advantages, challenges including a large NADPH requirement, balancing the protein reductive system for the aldehyde deformylase, and potential toxicity caused by aldehyde intermediates will need to be solved. Inhibition of aldehyde deformylase by hydrogen peroxide (with respect to O2) was also recently reported [29]. Performance was improved in vitro by expression of a fusion protein of aldehyde deformylase and catalase.
Figure 5.
Synthesis of alkanes and fatty alcohols via fatty aldehydes. Fatty aldehydes are made by reductive cleavage of acyl-thioesters. Acyl-ACPs, intermediates in fatty acid biosynthesis (FAB), are substrates of acyl-ACP reductases that use NADPH as a reducing equivalent. Acyl-ACPs are also substrates for thioesterases (TE) that produce free fatty acids. Free fatty acids can be activated by acyl-CoA synthetases to yield acyl-CoAs that are reductively cleaved by acyl-CoA reductases. Fatty aldehydes are reduced to fatty alcohols by aldehyde reductases or deformylated to alkanes.
Fatty aldehydes (and theoretically alkanes) can also be produced by acyl-CoA reductases, which have been identified in many species [30–32,33•]. In this pathway (Figure 5), substrate specificity is provided by acyl-ACP thioesterases (individual thioesterases target a wide range of chain lengths [34]) which produce FFA that can be converted to acyl-CoAs by acyl-CoA synthetases (e.g. E. coli FadD). When acyl-CoA reductases are expressed in E. coli in the presence of acyl-CoAs, a mixture of fatty aldehydes and fatty alcohols is detected, suggesting either spontaneous oxidation of aldehydes to fatty alcohols or the existence of other reductive enzymes capable of acting on long chain substrates. Fatty alcohols have higher selling prices than diesel, are widely used in industry, and are currently derived from plant oils or petrochemical sources. These reasons motivate metabolic engineering efforts to construct fatty alcohol producing strains. Of the efforts to characterize acyl-CoA reductases, only a handful reported titers higher than 10 mg/L of fatty alcohol. Overexpression of TesA′, overexpression of FadD, and heterologous expression of acr1, an acyl-CoA reductase from Acinetobacter calcoaceticus, in an E. coli ΔfadE strain resulted in production of approximately 100 mg/L of C16 and C18 fatty alcohols [18•]. Swapping the acyl-ACP thioesterase from TesA′ to those from Umbellularia californica or Cuphea hookeriana shifted the predominant chain length to C12 and C14, respectively, but with unspecified titers. In order to further increase yields, it is likely that all of the conversion steps after FAB will need to be carefully balanced to avoid the accumulation of intermediates, analogous to FAEE synthesis. Optimization of expression levels of the U. californica thioesterase, fadD, and an acyl-CoA reductase more than doubled fed-batch titers in a recent study from 210 to 449 mg/L of C12–C14 alcohols [35]. Recently, an NADPH-dependent fatty aldehyde reductase has been isolated from Marinobacter aquaeolei VT8 which possesses both acyl-CoA reductase and aldehyde reductase activity [33•,36]. This enzyme could be used to prevent accumulation of toxic aldehydes in a fatty alcohol producer. During the preparation of this review, a carboxylic acid reductase from M. marinum was found to exhibit substantial activity toward reduction of medium-chain and long-chain length FFAs to the corresponding fatty aldehydes [37]. When coupled with a native aldehyde reductase from E. coli (YjgB) or aldehyde deformylase, titers of over 350 mg/L of fatty alcohols and 2 mg/L of alkanes were achieved.
α-Olefins
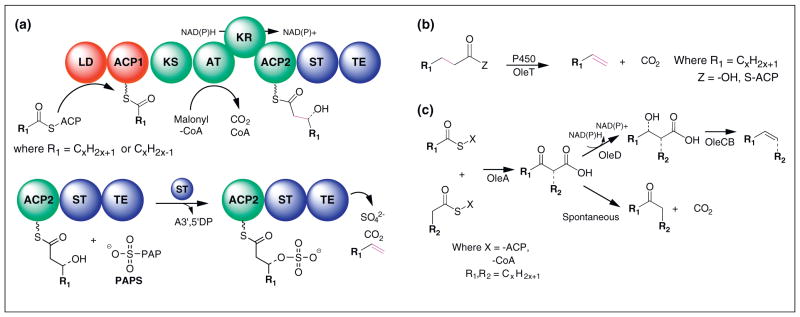
In addition to alkanes, several bacterial species produce α-olefins, hydrocarbons with terminal double bonds, (Figure 6). This class of compound could be blended with diesel fuels but would have higher value as chemical building blocks. Native olefins are typically either one carbon shorter or one carbon longer than the dominant fatty acids found in the cell membrane. Synechococcus sp. PCC 7002, a cyanobacteria, produces two α-olefins, 1-nonadecene and 1,14-nonadecadiene, that are one carbon larger than the largest fatty acids [22,38]. A gene, called ols (for olefin synthase), encoding a large protein sharing homology with type I polyketide synthases was recently shown to be responsible for producing these two olefins [38]. Ols has a domain architecture that suggests that an elongated β-hydroxyacyl chain is generated by standard polyketide enzymology. A predicted sulfotransferase domain is hypothesized to activate the β-hydroxyl group by addition of sulfate from an activated intermediate (e.g. 3′-phosphoadenosine 5′-phosphosulfate). Sulfation would provide the activation necessary to allow subsequent dehydration and decarboxylation to yield 1-nonadecene. While titers from PCC 7002 were increased 2-fold to 5-fold for 1-nonadecene and 1,14-nonadecadiene, respectively, by replacement of the native ols promoter with a stronger promoter, titers remain in the sub μg/mL range. Heterologous expression of ols was recently demonstrated after significant optimization, but alkene products were not detected in E. coli [39]. Engineering of the ols gene (e.g. replacing the loading domain) could lead to a set of pathways for producing a wide range of α-olefins. Unfortunately, expression of polyketide synthases in E. coli is not facile and efforts may need to shift to other hosts (e.g. Actinobacteria).
Figure 6.
Biosynthesis of olefins. (a) In cyanobacteria, olefins are formed via an elongation–decarboxylation mechanism catalyzed by a type I megasynthase. Synthesis begins when a fatty acyl chain is transferred from a discrete acyl–acyl-carrier protein (ACP) to the ACP1 domain. A typical polyketide extension module consisting of ketosynthase, acyltransferase, and ketoreductase domains extends the acyl-chain by two carbons donated from malonyl-CoA and reduces the beta-ketogroup to a hydroxyl. This position is activated by transfer of sulfate group from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) via the sulfotransferase domain. The terminal thioesterase (TE) domain catalyzes release of carbon dioxide, a sulfate ion and the olefin product. (b) Alternatively, terminal double bonds are introduced by decarboxylation catalyzed by cytochrome P450 (OleT) leading to olefins one carbon shorter than their substrates. (c) Internal olefins and ketones are produced by a set of enzymes that operate on acyl-thioesters. The pathway begins by OleA catalyzed condensation of two acyl-chains. The remaining enzymes are responsible for reducing and dehydrating the OleA product. Alternatively, spontaneous decarboxylation can produce ketones.
A second route for terminal alkene production, ident-ified in a Jeotgalicoccus species, is catalyzed by a cyto-chrome P450 enzyme named OleT (Figure 6) [40]. When OleT was heterologously expressed in E. coli, 1-pentadecene and 1,10-heptadecadiene (one carbon less than the dominant fatty acids in E. coli) were produced at unreported titers. Similarly, 1-heptadecene was produced when stearic acid (18:0) was fed to E. coli expressing OleT and in in vitro reactions using purified OleT. These data suggest that OleT can utilize both acyl-ACPs and FFA as substrates. Combining expression of thioesterases with OleT could lead to efficient production of medium chain length α-olefins as long as high levels of functional OleT expression (P450 expression is notoriously difficult) can be achieved in a facile host such as E. coli.
Oils, waxes, long chain olefins, and ketones
While not likely to be used directly as fuels, high molecular weight oils and waxes could be synthesized by microorganisms and used as feedstocks for producing fuels and chemicals via traditional refining methods. These compounds also have a variety of industrial applications as lubricants, food additives, and plasticizers. Many oleaginous bacteria and fungi accumulate triacyl-glyceride (TAG) oils at high yields [41] and metabolic engineering has succeeded increasing production from these strains (e.g. Yarrowia lipolytica [42]). The physical properties of oils can be modulated by introducing acyl chains of various lengths. For example, a diacylgycerol acetyltransferase from Euonymus alatus (burning bush) incorporates an acetyl group in place of the third acyl chain to produce a low viscosity oil [43]. Heterologous expression of oil synthesis pathways in oleaginous microbes is therefore a promising strategy to produce oils from biomass feedstocks in high yield.
Waxes have also been produced in E. coli using a wax ester/diacylglycerol acetyltransferase (WS/DGAT, e.g. AtfA) [44]. Similar long-chain (>C20) alkenes (with internal double bonds, Figure 6) and ketones are produced in several bacteria by a set of enzymes named OleABCD [45]. While the details of the OleABCD pathway are not completely elucidated, it has been shown that OleA catalyzes the head-to-head condensation of two acyl-thioe-sters (between the carbonyl of one acyl chain and the alpha carbon of the second) and OleD catalyzes the reduction of an internal ketone that is essential for the subsequent dehydration (hypothetically catalyzed by OleC and/or OleB) that yields the final alkene [45–47]. Heterologous expression of OleA from Micrococcus luteus in a FFA producing E. coli (DH1 ΔfadE, plasmid based TesA′), generated small quantities of long-chain length (C27–C29) monoke-tones [48] that are hypothesized to be spontaneous dec-arboxylation products [46]. Similarly, when the three gene operon (homologs of OleB and OleC are fused in M. luteus) was expressed in the same FFA producing E. coli, four internal alkene species (C27–C29 in size with 2 or 3 desaturations) were observed. As in the case of α-olefins, identification and characterization of the OleABCD pathway enable metabolic engineering efforts to produce ketones and olefins. Unlike the case for α-olefins, the enzymes involved are reasonably sized, express well in E. coli, and have minimal cofactor requirements.
Free fatty acids
While not useful as fuels, free fatty acids (FFAs) are commodity chemicals that are commonly derived from plant oils such as coconut (for C12–C14) and palm (for C16–C18). FFAs are feedstocks for chemicals used in cleaning products, agrochemicals, biocidal agents, textile processing agents, and polymer additives and can also be used as a feedstock for ex vivo production of biodiesel. FFAs are the most direct product that can be derived from FAB and yields of over 70% of theoretical have been achieved [49]. Introduction of an efficiently expressed acyl-ACP thioes-terase deregulates FAB and results in greatly increased carbon flux through the pathway. Several recent reviews have highlighted thioesterase expression and additional efforts that increase flux through FAB to FFA [50,51].
Methyl ketones and polyesters
Methyl ketones are another class of fatty acid-derived products, which could have value as biofuels or chemical feedstocks. Recent work has demonstrated production of C11–C15 methyl ketones in a strain of E. coli engineered to produce FFA and guide them through β-oxidation to β-ketoacyl-CoA thioesters (Figure 7) [52•]. This was accomplished by replacing FadE activity with a soluble acyl-CoA oxidase from M. luteus, overexpressing FadB (generates β-ketoacyl-CoA from enoyl-CoA), and deleting fadA (used to produce acetyl-CoA and a shorter acyl-CoA from β-ketoacyl-CoA). Additional overexpression of a poorly characterized acyl-CoA thioesterase, FadM, was found to significantly increase titer to approximately 50 mg/L, suggesting its role is as a β-ketoacyl-CoA thioes-terase. Released β-ketoacids were hypothesized to spontaneously decarboxylate to yield methyl ketones. Use of a decane overlay further improved titers to 380 mg/L [52•].
Figure 7.
Synthesis of chemicals via intermediates in β-oxidation. Fatty acids generated by thioesterase (TE) cleavage or scavenged from the environment enter β-oxidation as acyl-CoA synthesized by acyl-CoA synthetases. Oxidation of acyl-CoA by FadE yields enoyl-CoA that can be hydrated by enoyl-CoA hydratases (FadB or PhaJ) to generate 3-hydroxyacyl-CoAs of the (S)-stereochemistry and (R)-stereochemistry, respectively. 3-(R)-hydroxyacyl-CoA can be polymerized (red highlights) by polyhydroxyalkanoate (PHA) synthases (PhaC) to yield medium chain length polyester (mcl-PHA). 3-(S)-hydroxyacyl-CoA is further oxidized by FadB to a 3-ketoacyl-CoA that is ordinarily cleaved by FadA to generate an acyl-CoA and acetyl-CoA. Alternatively, ketoacyl-CoA can be hydrolyzed by FabM to yield a keto-acid that spontaneously decarboxylates to yield a methyl ketone (blue highlights). To facilitate the synthesis of mcl-PHA and methyl ketones, it is necessary to knockout competing pathways at branch points (FadB for mcl-PHA, FadA for methyl ketone).
A final category of fatty acid-derived products are bio-plastics generated from polymerizing hydroxylated acyl-CoAs to form polyhydroxyalkanoates with pendant medium-chain length alkyl groups (Figure 7). This class of bioplastics has been generated by feeding vegetable oils to bacterial cultures, which enter β-oxidation to generate polymerizable 3-hydroxyacyl-CoA monomers [53]. However, the iterative nature of β-oxidation generates a spectrum of monomer chain lengths, yielding undefined heteropolymers. Alternative strategies disrupt normal β-oxidation while taking advantage of an acyl-ACP thioes-terase to generate more uniform acyl chain lengths (mostly C12) [54–56].
Concluding remarks
Fatty acid metabolism offers the means to produce a wide range of high value chemicals and energy dense transportation fuels from renewable feedstocks. With the recent discovery of the genes and enzymes involved, attention can turn to engineering strains to produce target compounds in high yield. Balancing the supply of substrates and cofactors has proven to be important in improving yields, but identifying and circumventing the remaining barriers (regulation of fatty acid biosynthesis, product toxicity, and maximizing carbon flux through the engineered pathway) will be critical to achieve industrially viable yields. Furthermore, understanding the mechanisms by which these enzymes operate will lead to opportunities to engineer or evolve variants that produce products of non-native chain length at higher rates. Lastly, identifying and engineering hosts that can simultaneously produce fatty acid derived products and provide advantages when using lignocellulosic feedstocks (lignotoxin tolerance, consolidated bioprocessing, thermal tolerance, solvent tolerance, acid tolerance, generally regarded-as-safe) will be essential for biotechnology to play a leading role in a biomass-based chemical economy.
Acknowledgments
This manuscript was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Sciences DE-FC02-07ER64494) and the National Science Foundation (CBET-1149678). R.M.L. was supported as a trainee in the Chemistry-Biology Interface Training Program (NIH) and by the Department of Chemical and Biological Engineering Dahlke-Hougen Fellowship.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Scott SA, Davey MP, Dennis JS, Horst I, Howe CJ, Lea-Smith DJ, Smith AG. Biodiesel from algae: challenges and prospects. Curr Opin Biotechnol. 2010;21:277–286. doi: 10.1016/j.copbio.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Jones CS, Mayfield SP. Algae biofuels: versatility for the future of bioenergy. Curr Opin Biotechnol. 2012;23:346–351. doi: 10.1016/j.copbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Alonso DM, Bond JQ, Dumesic JA. Catalytic conversion of biomass to biofuels. Green Chem. 2010;12:1493–1513. [Google Scholar]
- 4.Huffer S, Roche CM, Blanch HW, Clark DS. Escherichia coli for biofuel production: bridging the gap from promise to practice. Trends Biotechnol. 2012;30:538–545. doi: 10.1016/j.tibtech.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Rodriguez S, Keasling JD. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opin Biotechnol. 2011;22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 7.Wackett LP. Engineering microbes to produce biofuels. Curr Opin Biotechnol. 2011;22:388–393. doi: 10.1016/j.copbio.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Vyas AP, Verma JL, Subrahmanyam N. A review on FAME production processes. Fuel. 2010;89:1–9. [Google Scholar]
- 9•.Nawabi P, Bauer S, Kyrpides N, Lykidis A. Engineering Escherichia coli for biodiesel production utilizing a bacterial fatty acid methyltransferase. Appl Environ Microbiol. 2011;77:8052–8061. doi: 10.1128/AEM.05046-11. This article describes the discovery and initial engineering of enzymes capable of conferring FAME production to E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Kalscheuer R, Steinbuchel A. A novel bifunctional wax ester synthase/acyl-CoA:diacylglycerol acyltransferase mediates wax ester and triacylglycerol biosynthesis in Acinetobacter calcoaceticus ADP1. J Biol Chem. 2003;278:8075–8082. doi: 10.1074/jbc.M210533200. The manuscript describes the discovery of a permiscous wax ester/diacylgycerol acyltransferase that has enabled the in vivo synthesis of fatty acid ethyl esters. [DOI] [PubMed] [Google Scholar]
- 11.Waltermann M, Stoveken T, Steinbuchel A. Key enzymes for biosynthesis of neutral lipid storage compounds in prokaryotes: properties, function and occurrence of wax ester synthases/acyl-CoA:diacylglycerol acyltransferases. Biochimie. 2007;89:230–242. doi: 10.1016/j.biochi.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Barney BM, Wahlen BD, Garner E, Wei J, Seefeldt LC. Differences in substrate specificities of five bacterial wax ester synthases. Appl Environ Microbiol. 2012;78:5734–5745. doi: 10.1128/AEM.00534-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi SB, Valle-Rodriguez JO, Khoomrung S, Siewers V, Nielsen J. Functional expression and characterization of five wax ester synthases in Saccharomyces cerevisiae and their utility for biodiesel production. Biotechnol Biofuels. 2012:5. doi: 10.1186/1754-6834-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holtzapple E, Schmidt-Dannert C. Biosynthesis of isoprenoid wax ester in Marinobacter hydrocarbonoclasticus DSM 8798: identification and characterization of isoprenoid coenzyme A synthetase and wax ester synthases. J Bacteriol. 2007;189:3804–3812. doi: 10.1128/JB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987;53:2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalscheuer R, Stolting T, Steinbuchel A. Microdiesel. Escherichia coli engineered for fuel production. Microbiology. 2006;152:2529–2536. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 17.Yu KO, Jung J, Kim SW, Park CH, Han SO. Synthesis of FAEEs from glycerol in engineered Saccharomyces cerevisiae using endogenously produced ethanol by heterologous expression of an unspecific bacterial acyltransferase. Biotechnol Bioeng. 2012;109:110–115. doi: 10.1002/bit.23311. [DOI] [PubMed] [Google Scholar]
- 18•.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. This manuscript demonstrates successful production of FFA, FAEE, and fatty alcohols in engineered E. coli using laboratory media and a renewable feedstock (xylan) [DOI] [PubMed] [Google Scholar]
- 19.Duan YK, Zhu Z, Cai K, Tan XM, Lu XF. De novo biosynthesis of biodiesel by Escherichia coli in optimized fed-batch cultivation. PLoS One. 2011:6. doi: 10.1371/journal.pone.0020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. This article demonstrates how an acyl-CoA sensor can be used to balance FAEE synthesis activity with the supply of ethanol and acyl-CoA. A synthetic biology strategy was used to convert FadR-responsive promoters into a sensor for optimizing FAEE synthesis in E. coli. The resulting strain generated the highest reported titers of FAEE to date. [DOI] [PubMed] [Google Scholar]
- 22.Winters K, Parker PL, Vanbaale C. Hydrocarbons of blue-green algae: geochemical significance. Science. 1969;163:467–468. doi: 10.1126/science.163.3866.467. [DOI] [PubMed] [Google Scholar]
- 23.Wackett LP, Frias JA, Seffernick JL, Sukovich DJ, Cameron SM. Genomic and biochemical studies demonstrating the absence of an alkane-producing phenotype in Vibrio furnissii M1. Appl Environ Microbiol. 2007;73:7192–7198. doi: 10.1128/AEM.01785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Schirmer A, Rude MA, Li XZ, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329:559–562. doi: 10.1126/science.1187936. This manuscript details a bioinformatic study that led to the identification of enzymes that catalyze the production of alkanes in cyanobacteria. [DOI] [PubMed] [Google Scholar]
- 25.Li N, Chang W-c, Warui DM, Booker SJ, Krebs C, Bollinger JM. Evidence for only oxygenative cleavage of aldehydes to alk(a/e)nes and formate by cyanobacterial aldehyde decarbonylases. Biochemistry. 2012;51:7908–7916. doi: 10.1021/bi300912n. [DOI] [PubMed] [Google Scholar]
- 26.Li N, Norgaard H, Warui DM, Booker SJ, Krebs C, Bollinger JM. Conversion of fatty aldehydes to alka(e)nes and formate by a cyanobacterial aldehyde decarbonylase: cryptic redox by an unusual dimetal oxygenase. J Am Chem Soc. 2011;133:6158–6161. doi: 10.1021/ja2013517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knothe G. Biodiesel and renewable diesel: a comparison. Prog Energy Combust Sci. 2010;36:364–373. [Google Scholar]
- 28.Rottig A, Wenning L, Broker D, Steinbuchel A. Fatty acid alkyl esters: perspectives for production of alternative biofuels. Appl Microbiol Biotechnol. 2010;85:1713–1733. doi: 10.1007/s00253-009-2383-z. [DOI] [PubMed] [Google Scholar]
- 29.Andre C, Kim SW, Yu XH, Shanklin J. Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc Natl Acad Sci U S A. 2013;110:3191–3196. doi: 10.1073/pnas.1218769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doan TT, Carlsson AS, Hamberg M, Bulow L, Stymne S, Olsson P. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J Plant Physiol. 2009;166:787–796. doi: 10.1016/j.jplph.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Teerawanichpan P, Qiu X. Fatty acyl-CoA reductase and wax synthase from Euglena gracilis in the biosynthesis of medium-chain wax esters. Lipids. 2010;45:263–273. doi: 10.1007/s11745-010-3395-2. [DOI] [PubMed] [Google Scholar]
- 32.Reiser S, Somerville C. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J Bacteriol. 1997;179:2969–2975. doi: 10.1128/jb.179.9.2969-2975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Willis RM, Wahlen BD, Seefeldt LC, Barney BM. Characterization of a fatty acyl-CoA reductase from Marinobacter aquaeolei VT8: a bacterial enzyme catalyzing the reduction of fatty acyl-CoA to fatty alcohol. Biochemistry. 2011;50:10550–10558. doi: 10.1021/bi2008646. This article describes the characterization of an acyl-CoA reductase that catalyzes both the reductive cleavage of the acyl-CoA thioester and the reduction of the fatty aldehyde to yield a fatty alcohol. This enzyme was discovered in a marine, oil-degrading bacterium and offers the ability to channel flux from acyl-CoA to fatty alcohol without releasing large amounts of fatty aldehyde. [DOI] [PubMed] [Google Scholar]
- 34.Cantu DC, Chen YF, Reilly PJ. Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 2010;19:1281–1295. doi: 10.1002/pro.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng YN, Li LL, Liu Q, Yang JM, Wang XW, Liu W, Xu X, Liu H, Zhao G, Xian M. Optimization of fatty alcohol biosynthesis pathway for selectively enhanced production of C12/14 and C16/18 fatty alcohols in engineered Escherichia coli. Microb Cell Fact. 2012;11:65. doi: 10.1186/1475-2859-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofvander P, Doan TT, Hamberg M. A prokaryotic acyl-CoA reductase performing reduction of fatty acyl-CoA to fatty alcohol. FEBS Lett. 2011;585:3538–3543. doi: 10.1016/j.febslet.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Akhtar MK, Turner NJ, Jones PR. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc Natl Acad Sci U S A. 2013;110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in alpha-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Environ Microbiol. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendez-Perez D, Gunasekaran S, Orler VJ, Pfleger BF. A translation-coupling DNA cassette for monitoring protein translation in Escherichia coli. Metab Eng. 2012;14:298–305. doi: 10.1016/j.ymben.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 40.Rude MA, Baron TS, Brubaker S, Alibhai M, Del Cardayre SB, Schirmer A. Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol. 2011;77:1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walther TC, Farese RV. Lipid droplets and cellular lipid metabolism. In: Kornberg RD, editor. Annual Review of Biochemistry. Vol. 81. 2012. pp. 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi: 10.1016/j.ymben.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Durrett TP, McClosky DD, Tumaney AW, Elzinga DA, Ohlrogge J, Pollard M. A distinct DGAT with sn-3 acetyltransferase activity that synthesizes unusual, reduced-viscosity oils in Euonymus and transgenic seeds. Proc Natl Acad Sci U S A. 2010;107:9464–9469. doi: 10.1073/pnas.1001707107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalscheuer R, Stoveken T, Luftmann H, Malkus U, Reichelt R, Steinbuchel A. Neutral lipid biosynthesis in engineered Escherichia coli: jojoba oil-like wax esters and fatty acid butyl esters. Appl Environ Microbiol. 2006;72:1373–1379. doi: 10.1128/AEM.72.2.1373-1379.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sukovich DJ, Seffernick JL, Richman JE, Gralnick JA, Wackett LP. Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA. Appl Environ Microbiol. 2010;76:3850–3862. doi: 10.1128/AEM.00436-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonnett SA, Papireddy K, Higgins S, del Cardayre S, Reynolds KA. Functional characterization of an NADPH dependent 2-alkyl-3-ketoalkanoic acid reductase involved in olefin biosynthesis in Stenotrophomonas maltophilia. Biochemistry. 2011;50:9633–9640. doi: 10.1021/bi201096w. [DOI] [PubMed] [Google Scholar]
- 47.Frias JA, Richman JE, Erickson JS, Wackett LP. Purification and characterization of OleA from Xanthomonas campestris and demonstration of a non-decarboxylative Claisen condensation reaction. J Biol Chem. 2011;286:10930–10938. doi: 10.1074/jbc.M110.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beller HR, Goh E-B, Keasling JD. Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl Environ Microbiol. 2010;76:1212–1223. doi: 10.1128/AEM.02312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F, Ouellet M, Batth TS, Adams PD, Petzold CJ, Mukhopadhyay A, Keasling J. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab Eng. 2012;14:653–660. doi: 10.1016/j.ymben.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Handke P, Lynch SA, Gill RT. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng. 2011;13:28–37. doi: 10.1016/j.ymben.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Lennen RM, Pfleger BF. Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 2012;30:659–667. doi: 10.1016/j.tibtech.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Goh E-B, Baidoo EEK, Keasling JD, Beller HR. Engineering of bacterial methyl ketone synthesis for biofuels. Appl Environ Microbiol. 2012;78:70–80. doi: 10.1128/AEM.06785-11. This article describes metabolic engineering of E. coli for production of methyl ketones. The strategy involves genetic manipulations of beta-oxidation and overexpression of FadM, which is hypothesized to be a beta-ketoacyl-CoA thioesterase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao X, Chen JC, Wu Q, Chen GQ. Polyhydroxyalkanoates as a source of chemicals, polymers, and biofuels. Curr Opin Biotechnol. 2011;22:768–774. doi: 10.1016/j.copbio.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Agnew D, Stevermer A, Youngquist J, Pfleger B. Engineering Escherichia coli for production of mcl-PHA homopolymer from glucose. Metab Eng. 2012;14:705–713. doi: 10.1016/j.ymben.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rehm BH, Steinbuchel A. Heterologous expression of the acyl–acyl carrier protein thioesterase gene from the plant Umbellularia californica mediates polyhydroxyalkanoate biosynthesis in recombinant Escherichia coli. Appl Microbiol Biotechnol. 2001;55:205–209. doi: 10.1007/s002530000541. [DOI] [PubMed] [Google Scholar]
- 56.Wang HH, Zhou XR, Liu QA, Chen GQ. Biosynthesis of polyhydroxyalkanoate homopolymers by Pseudomonas putida. Appl Microbiol Biotechnol. 2011;89:1497–1507. doi: 10.1007/s00253-010-2964-x. [DOI] [PubMed] [Google Scholar]