Abstract
Circulating tumor cells (CTCs) are one of the most crucial topics in rare cell biology and have become the focus of a significant and emerging area of cancer research. While CTC enumeration is a valid biomarker in prostate cancer, the current FDA-approved CTC technology is unable to detect CTCs in a large portion of late stage prostate cancer patients. Here we introduce the NanoVelcro CTC Chip, a device composed of a patterned silicon nanowire substrate (SiNW) and an overlaid polydimethylsiloxane (PDMS) chaotic mixer. Validated by two institutions participating in the study, the NanoVelcro Chip assay exhibits very consistent efficiency in CTC-capture from patient samples. The utilized protocol can be easily replicated at different facilities. We demonstrate the clinical utility of the NanoVelcro Chip by performing serial enumerations of CTCs in prostate cancer patients after undergoing systemic therapy. Changes in CTC numbers after 4–10 weeks of therapy were compared with their clinical responses. We observed a statistically significant reduction in CTCs counts in the clinical responders. We performed long-term follow up with serial CTC collection and enumeration in one patient observing variations in counts correlating with treatment response. This study demonstrates the consistency of the NanoVelcro Chip assay over time for CTC enumeration and also shows that continuous monitoring of CTC numbers can be employed to follow responses to different treatments and monitor disease progression.
Keywords: Nanomaterials, microfluidics, circulating tumor cell, prostate cancer, diagnostics
1. Introduction
Rare cell biology is an emerging field of science that studies minor populations of cells that are known to play crucial roles in biological systems. In addition to stem cells, circulating tumor cells (CTCs) have been recognized as one of the most important topics in this field. In the solid tumor setting, CTCs are regarded as a population of rare cancer cells that have detached from a primary tumor and/or metastatic lesions that have entered the circulation. These cells can travel and potentially seed new metastatic sites [1, 2]. Metastasis is the hallmark of progression to an incurable state for most solid tumors. This process leads to pain and suffering for most cancer patients and eventually ends in death. While the molecular mechanisms of metastatic progression are still largely unknown, there is evidence that CTCs are shed well before the onset of macroscopic metastasis [3–5]. A better understanding of CTC biology may provide new insights into the progression from localized to metastatic disease. Many groups have proposed that CTCs may be used as a “liquid biopsy” for tumors in the clinical setting, providing convenient access to surrogate tissue when metastatic lesions are difficult to biopsy. However, detection and characterization of CTCs are technically challenging due to the extremely low abundance (a few to hundreds of cells per mL) of CTCs among a high number (109 cells/mL) of background cells in blood, even in patients with advanced diseases [6–8]. Over the past decade, significant technological innovation and scientific thought has been devoted to creating a reliable method for the isolation and detection of CTCs. In response to this effort, a wide array of technologies has been developed based on different operation mechanisms.
The most commonly used CTC enrichment techniques fall into one of the three categories: immunomagnetic separation, flow cytometry, and microfluidic chips. Immunomagnetic separation methodologies use magnetic beads conjugated with positive capture agents (e.g. anti-EpCAM antibody) [9] and negative depletion agents (e.g. anti-CD45 antibody to remove white blood cells) [10, 11]. This separation method is the approach used by the CellSearch™ assay, the only FDA-approved CTC diagnostic technology for clinical use. Flow cytometry is one of the most powerful technologies for detection and isolation of cell subpopulations [12]. This technique uses a combination of object size and protein expression characteristics to sort cells. Nonetheless, due to the extremely low abundance of CTCs and the inability to confirm them by morphology, it is practically challenging to apply flow cytometry to accurately enumerate CTCs from whole blood. Recently, several microfluidic-based technologies have recently been developed by the Toner [13–15] and Soper groups [16, 17]. These microfluidic devices improved CTC-capture efficiencies by enhancing contact frequency between the CTCs and the capture substrates. However, the majority of these technologies suffer from the large vertical depth of their 3D device features (micropillars or herringbones). They also require multiple cross-sectional imaging scans (that are time consuming and generate huge image files) in order to avoid out-of-focus or superimposed images of device immobilized CTCs. The time consuming and labor intensive nature of these methods severely limits their applicability to routine clinical practice.
CTC enumeration has been validated as a biomarker for response to therapy in prostate, colon, and breast cancer and is being explored in other settings [9, 18, 19]. Despite the recognized value of serum prostate specific antigen (PSA) measurement in many settings, it has become clear to clinicians that better biomarkers for the monitoring of prostate cancer (PC) treatment response are still needed to guide clinical decision-making given the discordance of serum PSA and clinical behavior in many settings. The introduction and validation of the CellSearch™ assay as a biomarker of response to therapy for advanced PC and has created new opportunities for innovation in cancer research. Despite the validation of this approach, the CellSearch™ assay is considered a relatively crude measurement tool. This assay is unable to detect CTCs in more than 50% of metastatic castration-resistant prostate cancer (mCRPC) patients. Moreover, the majority of patients are found to have less than 2 CTCs in 7.5ml of blood in those CTC-positive cases [20]. This number has a very low dynamic range making statistical analysis difficult. Furthermore, this approach is subject to errors in the identification of CTCs.
Our nanostructured CTC chips present a novel mechanism for CTC capture. In our previous publications, we pioneered the concept of NanoVelcro substrates, by which anti-EpCAM antibody coated silicon nanowires (SiNW) were constructed to immobilize CTCs [21, 22]. The uniqueness of our approach lies in the use of a nanostructured substrate where the enhanced local topographic interactions between the anti-EpCAM-coated nanosubstrates and nano-scaled cellular surface components (e.g., microvilli) are analogous to the working principle of Velcro™ [23–25]. To further improve the CTC-capture efficiency, we integrated an overlaid polydimethylsiloxane (PDMS) chaotic mixer generating vertical flows that enhance contacts between CTCs and the capture substrate. Compared to the aforementioned stationary capturing device, the resultant NanoVelcro device has a dynamic capturing mechanism allowing up to 85% CTC capture efficiency [22]. Side-by-side analytical validation studies using both artificial and patient CTC samples suggested that the sensitivity of our NanoVelcro device outperformed [22] that of CellSearch™.
With our previous publications [21, 22] demonstrating the proof-of-concept, here we introduce the “NanoVelcro Chip,” optimized from our previous devices for CTC capture and enumeration in clinical use. The CTC-capture efficiency of the chip has been validated jointly by the Uro-Oncology teams at the University of California, Los Angeles (UCLA) and the Samuel Oschin Comprehensive Cancer Institute at Cedars-Sinai Medical Center (CSMC). We also demonstrate the ability to reliably identify CTCs from a patient over a period of treatment to monitor CTC changes in response to these various therapies.
2. Materials and methods
2.1. NanoVelcro Chip fabrication
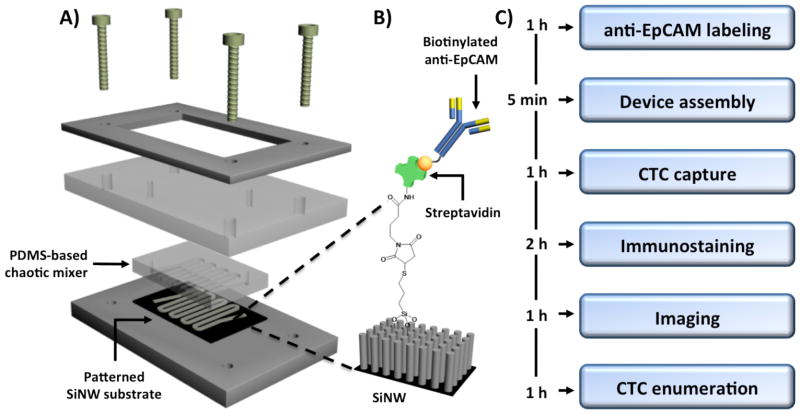
The NanoVelcro CTC chip is composed of three parts: (I) a serpentine chaotic mixer chip made of PDMS, (ii) a patterned SiNW substrate with high-affinity anti-EpCAM coating, and (iii) a home-machined holder set to sandwich a well-aligned PDMS mixer chip with the SiNW substrate (Fig 1). The PDMS mixer chips were fabricated using a standard soft-lithography method. In order to minimize system error caused by inconsistency of PDMS mixer chips, a silicon mold with a desired configuration was made by dry etching, and then framed in a metal container. Well-mixed PDMS was poured onto this mold and cured in an oven at 80°C. After 48 h of baking, the cured PDMS layer was peeled off and punched with two through holes at both ends of the channel for tubing connection. Consistent channel structure and PDMS thickness were achieved using this method.
Fig. 1.
Device configuration and experimental workflow: (A) A NanoVelcro CTC Chip is composed of an overlaid PDMS-based chaotic mixer, a patterned silicon nanowire (SiNW) substrate, and a multilayer chip holder to assemble both functional components together. (B) Silanation reaction and NHS chemistry were employed to covalently link streptavidin onto the SiNW substrate, allowing conjugation of biotinylated anti-EpCAM prior to CTC detection studies. (C) The workflow enables CTC capture and enumeration in 6 hours.
The fabrication method for the SiNW substrate was reported earlier [22]. Briefly, a patterned layer of photoresist (AZ 5214, AZ Electronic Materials USA Corp.) was deposited on the silicon wafers using standard photolithography technique. Then a chemical etching solution composed of deionized water, HF (4.6M), and silver nitrate (0.02M) was applied to the wafer to construct SiNWs with an average length of 15 μm at the uncovered surface area defined by the photoresist pattern. After that, boiling aqua regia (3:1 (v/v) HCl/HNO3) was added to the nanostructured substrate for 15 minutes to remove the silver film. The remaining photoresist was then removed by repeatedly rinsing with acetone and ethanol. After DI water rinsing and nitrogen blow-dry, the substrate was ready for subsequent streptavidin coating.
Streptavidin coating was also conducted following a previously established protocol. Basically, the substrate was first treated with 4 % (v/v) 3-mercaptopropyl trimethoxysilane in ethanol for 45 minutes at room temperature, and then with the coupling agent N-maleimidobutyryloxy succinimide ester (GMBS, 0.25 mM in DMSO) for another 30 minutes. After incubation with streptavidin (SA, 10 μg/ml in 1 x PBS) for 1 hour, the streptavidin-coated substrate was stored at 4° C to avoid activity decay before usage.
Prior to each test, the PDMS chaotic mixer chip and streptavidin-coated substrate were sandwiched together using a home-machined holder set consisting of (i) one stainless steel bottom plate, (ii) one PMMA (poly(methyl methacrylate)) clapboard, (iii) one stainless steel top frame, and (iv) a screw at each corner (Fig 1A). To form a complete device, the PDMS chip and SiNW substrate in accurate alignment were placed on the center of the bottom plate. Then PMMA clapboard was carefully laid on top to ensure the two through holes on PDMS chip were located right in the middle of their corresponding bores pre-machined on the clapboard. Finally, the top frame was anchored to the PMMA clapboard and bottom plate by screws to form a constant pressure seal to prevent leakage. The anti-EpCAM solution was then loaded into the channel by a syringe pump (KDS 200, KD scientific) for conjugation. Before sample injection, the channel required multiple PBS washings to remove free anti-EpCAM.
Compared to our previous design (Table 1), this new version of the NanoVelcro CTC chip has a smaller surface area and equivalent CTC capture performance. This may dramatically shorten the time spent on whole chip scanning while reducing total reagent consumption for each test. Also, removing the SiNW at the turning avoids the non-specific trapping of cells and other blood components caused by local flow resistance.
Table 1.
Comparison of the fabrication specifications between the previous reported device and the NanoVelcro CTC Chip
| Previous device | NanoVelcro CTC Chip | |
|---|---|---|
| PDMS mold | Su-8 structures on silicon wafer | Dry etched silicon wafer |
| Channel length | 88 cm | 22 cm |
| SNP configuration | Serpentine line | Disconnected straight lines |
2.3. Cells
PC3 and LNCaP cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). C4-2 cells, an androgen-independent derivative of LNCaP cells were provided by Dr. Leland Chung who developed this subline [26]. Cells were cultured in RPMI1640 supplemented with 2 mM L-glutamine (Invitrogen), 5% fetal bovine serum (Omega Scientific), penicillin (100 unit/mL) and streptomycin (100 μg/mL) at 37°C in humidified atmosphere with 5% CO2. Before use, cells were detached with sodium citrate (0.015M sodium citrate with 0.135M KCl), washed and then resuspended in Ca2+ and Mg2+ free PBS. Suspended cells were stained with a lipophilic tracer DiO (Invitrogen), serial diluted and then enumerated in a 48 well plate in triplicate. The wells with around 30 to 300 cells were counted for accurate cell concentration determination. Then cell suspensions with specific number of cells were spiked into PBS or blood from healthy donors.
2.4. Blood samples
Blood samples were obtained from prostate cancer patients and healthy volunteers at UCLA and CSMC under Institutional Review Board approved protocols at each site. Blood used for cell line spiking studies was obtained from healthy volunteer donors aged 20 to 40. All blood specimens were collected into EDTA-containing vacutainer tubes (BD bioscience) and processed within 24 h.
2.5. NanoVelcro CTC capture workflow (Fig. 1C)
A CellSearch™ CTC control kit containing 200/mL of pre-stained EpCAM-positive SK-BR-3 breast cancer cells was spiked into PBS as a model system for parameter optimization. The chemical conjugation of anti-EpCAM was achieved by loading a solution of biotinylated anti-EpCAM (25 μL, 10 μg/mL in PBS with 1.0 % (w/v) BSA and 0.1 % (w/v) sodium azide) into the assembled chip using a syringe pump (KDS200, KD scientific). After incubation for 30 minutes, the microchannels were washed with 200 μL of PBS. Then 1.0 mL of the sample solution was loaded and flowed through the CTC capture platform at different flow rates. After the capture, 100 μL of PBS was loaded into the chip followed by 300 μL of 4 % paraformaldehyde (PFA, Sigma-Aldrich) for cell fixation. After 15 minutes of PFA fixation, the NanoVelcro chip was dissembled and the SiNW substrate with captured cells was examined under a fluorescence microscope. The distribution of captured cells was recorded for optimization of parameters.
2.6. Immunocytochemistry staining and imaging
The patient blood sample study was performed by a similar procedure, but needed additional steps for CTC identification using immunohistochemistry. After fixation, the SiNW substrate was detached from the holder set and rinsed with PBS. Three hundred microliters of 0.5% Triton X-100 in PBS was loaded onto the substrate for cell permeabilization for 10 minutes. Then PE-conjugated anti-cytokeratin antibody (BD bioscience) and FITC-conjugated anti-CD45 (BD bioscience) diluted with 2% BSA in PBS were added onto the chip and incubated for 2 hours at room temperature. After thorough washing, the substrate was put onto a glass slide and DAPI mounting solution (Invitrogen) was added along with a cover slide.
The imaging system was composed of an upright fluorescence microscope (Eclipse 90i, Nikon) with the NIS-Element imaging software (Nikon), a precision motorized stage (ProScan II system, Prior Scientific) and fluorescent light source (SPECTRAX, Lumencor). We first scanned through the substrate under 4X objective, creating a mosaic micrograph for the determination of fluorescence intensity of all automatically counted events. X-Y scatter plots summarizing the CK and CD45 expressions of individual cells (including CTCs and WBCs) on a NanoVelcro substrate were exported and plotted in Microsoft Excel. After identifying potential cancer cell populations in the Excel plot, we set the limitations in the NIS-Element review and previewed the 4X micrographs. Micrographs at 10X were taken for each of the candidate cells identified by the 4X objective. The final enumeration was based on the criteria of CK+/CD45−/DAPI+, 40 μm>diameter >10 μm – parameters similar to those utilized by the CellSearch™ assay [27].
2.7. Statistical analysis
We retrospectively calculated the CTC changes before the treatment and after 4–10 weeks of treatment initiation. The CTC responses in the responder group and non-responder group were statistically analyzed by the Wilcoxon rank sum test.
3. Results
3.1. Capture condition optimization
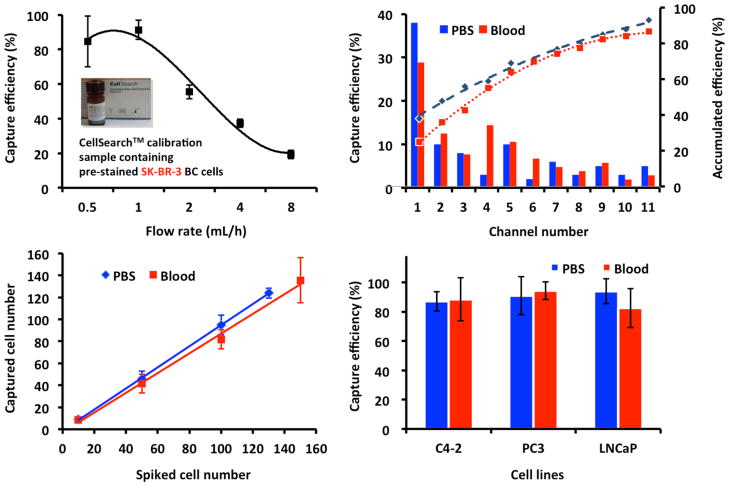
We spiked 0.3 mL of the CellSearch™ calibration sample, containing 100 pre-stained EpCAM-positive SK-BR-3 breast cancer cells, into 0.7 mL of PBS as a model system. After labeling with biotinylated anti-EpCAM, samples were pumped through the NanoVelcro CTC chips at different flow rates (Fig. 2A). The results showed that the capture efficiency was optimized at the rate of 0.5 to 1 mL/h. We chose a flow rate of 0.5mL/h for experiments to decrease shearing force in order to preserve potentially fragile CTCs.
Fig. 2. Optimization of the capture efficiency.
(A) Cell-capture efficiency of NanoVelcro CTC Chip at flow rates of 0.5, 1, 2, 4, and 8 mL/h. Error bars show standard deviations (n=3–4). CellSearch™ calibration samples containing 100 pre-stained EpCAM-positive SK-BR-3 breast cancer cells were spiked into PBS as a model system. (B) The cell distribution and accumulative cell-capture efficiency in a NanoVelcro CTC Chip were assessed in PBS and normal blood. (C) Cell-capture efficiency at different cell numbers ranging from 10–150 cells mL-1 in two different types of samples: whole blood and PBS buffer. (D) The capture efficiency observed for different prostate cancer cell lines in PBS and blood.
The cell distribution and cumulative cell-capture efficiency were assessed along the length of the total channel. Samples utilized included cancer cells spiked into PBS and cells spiked into healthy donor blood. These tests showed that over 60% of the captured cells were located in the first 4 channels, with the majority captured in the first 1 to 2 channels. This finding was consistent across the PBS and donor blood model systems (Fig. 2B). The capture efficiencies were also constant across different numbers of cells in PBS and blood (Fig. 2C). Finally, we tested different prostate cancer cell lines on the NanoVelcro Chips. Three commonly utilized prostate cancer lines, LNCaP, C4-2, and PC3, showed a similar capture efficiency of 80–95% in both PBS and blood (Fig. 2D).
3.2. Immunostaining and enumeration in the cell line model system
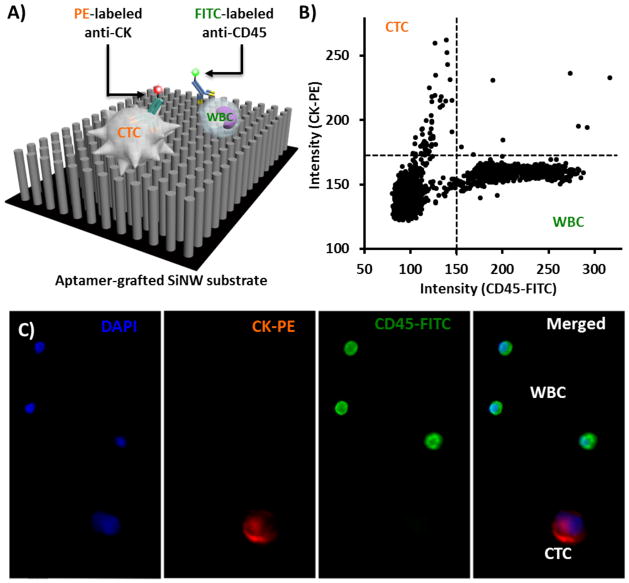
Using methods analogous to the CellSearch™ assay, immunofluorescence staining for CK was used as positive selection (epithelial cell marker) and for CD45 as negative selection (leukocytes) (Fig. 3A). PC3 cells spiked into the healthy donor blood were isolated using the NanoVelcro capture protocol. After initial processing of the blood through the chip, the substrate was stained for CK and CD45. A low power mosaic micrograph was created using a scanning microscope (Fig. S1). DAPI staining and cell size were used to exclude cell debris and any uncertain cellular objects. Candidate cell events were defined as DAPI+/size>10 μm. These events are represented on a scatter plot (Fig. 3B). CK+/CD45− cells were selected and imaged under a 10X objective. Experienced operators then read all the micrographs and the CTCs were manually enumerated. CTCs were defined as CK+/CD45−/DAPI+ events with a diameter of 10–40 μm with morphological features consistent with epithelial cells (Fig. 3C).
Fig. 3. Immunofluorescence (IF) staining of CTCs immobilized on NanoVelcro substrates.
(A) Schematic representation of an IF protocol developed for identification of CTCs (CK+/CD45−/DAPI+, 40 μm>diameter >10 μm) from non-specifically captured WBCs (CK−/CD45+/DAPI+, 40 μm>diameter >10 μm) and cell debris. (B) An XX-Y scatter plot that summarizes the CK and CD45 expressions of individual cells (including CTCs and WBCs) on a NanoVelcro substrate helps to identify candidate cancer cells. (C) Typical micrographs of a CTC and WBCs immobilized on a NanoVelcro substrate.
3.3. CTC capture and enumeration in patient samples from 2 institutions
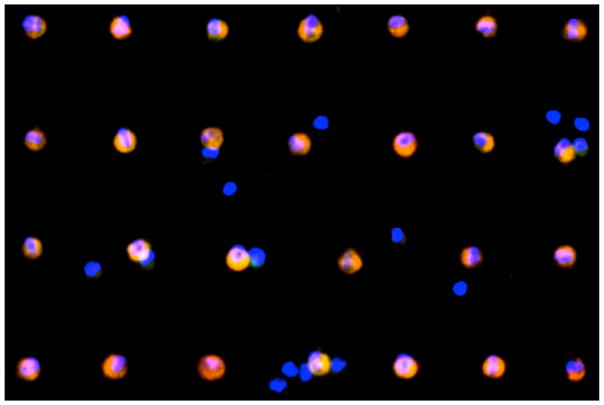
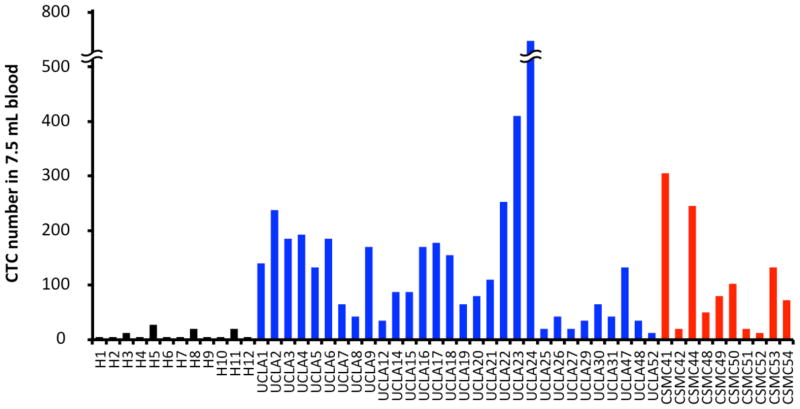
Using optimized protocols, patient samples from both UCLA and CSMC were processed on NanoVelcro Chips for CTC capture and enumeration. The blood samples from different healthy donors were also processed for circulating epithelial cell (CEC) enumeration. A total of 40 patients plus 12 healthy donors participated in the enumeration study (Table 1). The NanoVelcro Chip successfully captured CTCs with preservation of morphology (Fig. 4). CEC events in healthy patients were consistently lower (0–2 CECs/1mL of blood) than CTCs from prostate cancer patients that were captured with very high efficiency (1–99 CTCs/1mL of blood) at both institutions (Fig. 5). The CEC capture established the background level of non-cancerous epithelial cells detected by our assay allowing for proper enumeration of CTCs.
Fig. 4.
Mosaic image of 27 CTCs from patient UCLA19
Fig. 5.
The enumerated CTCs (CK+/CD45−/DAPI+, 40 μm>diameter >10 μm) from 1mL blood of healthy donors and patients at their first visit (Normalized to 7.5mL scale for comparison with CellSearch method). Blue columns represent the enumerated CK+CD45− circulating epithelial cells in healthy donor’s blood; blue columns represent UCLA patient samples; red represents CSMC patient samples.
3.4. CTC number before and after therapeutic interventions
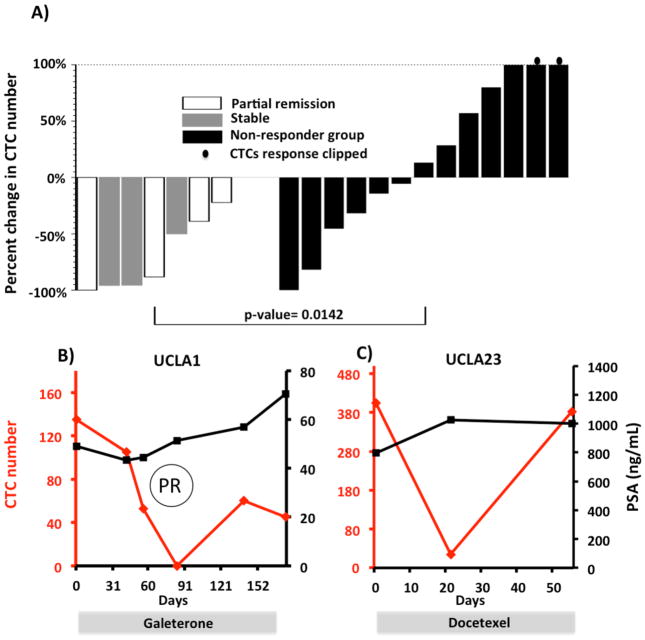
After validating our ability to consistently capture and enumerate CTCs in prostate cancer patients, we compared CTC numbers before and after the initiation of different treatments. Only 14 patients of the original 40 received new treatments during our follow-up study. A total of 19 series of measurements were recorded. The on-treatment CTC enumerations were typically done 4–10 weeks after commencing a new therapy (Table S1). Clinical responses were determined by the treating physician independent of CTC enumeration. CTC counts before and after the treatments were measured. Patients were stratified based on clinical responses. As demonstrated in Fig. 6A, patients with disease progression showed variable CTC changes, whereas patients with stable disease or clinical benefit experienced consistent decreases in CTC counts (p=0.014). Serial CTC number and serum PSA measurements of patients UCLA1 and UCLA23 are shown in Fig. 6B and 6C. For patient UCLA1, CTC number significantly decreased temporally corresponding to a radiographic documented tumor regression after starting galeterone (TOK-001). Despite his radiographic partial response, his serum PSA concentration continued to increase. In this case, CTC number appeared to be a more accurate indicator of disease activity. In comparison, patient UCLA23 had no measurable disease by radiograph. After the initiation of docetaxel therapy, his CTC number decreased followed by a quick rebound to his original CTC number. This correlated temporally with clinical deterioration. During this period, his serum PSA concentration did not increase.
Fig. 6.
Patient CTC numbers before and after treatment. (A) Percent change of the patients’ CTC number 4–10 weeks after treatment initiation. Left panel showed the CTC responses in the clinical responder group; right panel showed CTC responses in the clinical non-responder group. (B,C) The serial CTC changes of patient UCLA1 (B) and patient UCLA23 (C) are plotted along with serum PSA concentration and treatment type. PR stands for partial responses assessed in UCLA1 by the RECIST criteria. His reference lymph node (LN) sizes assessed by imaging were 6.2cm on day 2, 3.9 cm on day 86 and 3.1 cm on day 177.
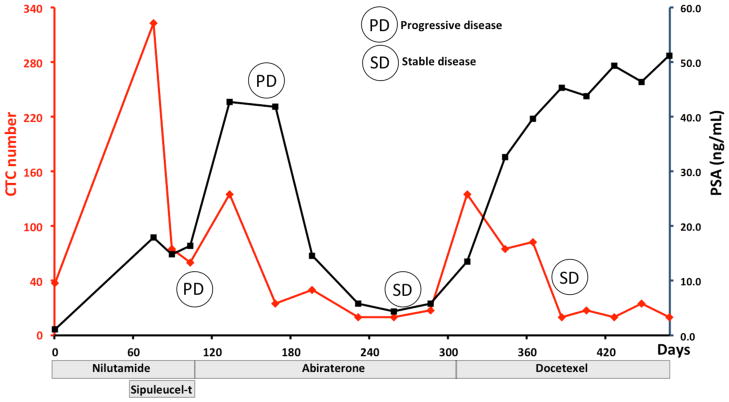
Patient UCLA8 provided blood samples over the course the course of a year during which he received four different therapeutic interventions: nilutamide, sipuleucel-t, abiraterone, and docetaxel. CTC counts initially dropped after the initiation of nilutamide and sipuleucel-t, and then rebounded as signs of treatment failure emerged. This finding was comparable to patient UCLA23. Furthermore, as his disease stabilized by docetaxel therapy, his CTC number remained low despite PSA progression.
4. Discussion
4.1. Novelty and advantages of the NanoVelcro Chip
In comparison with other CTC capturing technologies, the NanoVelcro Chip has several prominent advantages. First, the miniature size of NanoVelcro Chip combined with its SiNW substrate allows for both temporally quick cell capture and rapid imaging. Compared to the microfluidic device developed by Toner [14], our substrate is not only smaller in surface area, but also is able to capture cells onto a 2D-surface. The imaging of CTC chips such as those utilized by the Toner group requires considerable time and processing power given the extremely large image data sets needed to cover the entire 3D-structure of their microposts. The NanoVelcro Chip captures CTCs onto a very narrow focal plane allowing for a simple 2D scan to cover all the captured events. This greatly facilitates the remainder of the processing including gating of the cell size, DAPI, CD45-FITC and CK-PE intensity to identify potential CTCs. The candidate CTCs are manually confirmed, and all debris-like cells are eliminated from our enumeration. Secondly, the isolation and enumeration protocol is fairly simple and user-friendly and can be easily transferred to any laboratory with basic human sample handling techniques. Most importantly, our platform yielded consistent detection of CTCs in both localized patients undergoing prostatectomy and metastatic patients with castration-resistant disease (Fig. 5). Our previous studies with side-by-side CTC enumeration comparing our platform to the current standard showed that the NanoVelcro Chip constantly captured more CTCs than CellSearch [22]. Furthermore, our NanoVelcro device captures CTCs in 1 mL of blood as compared to the 7.5 mL required for the CellSearch assay. This minimizes the required phlebotomy for patients who are typically anemic due to disease while allowing for repeat testing on a single tube of blood to help avoid technical errors.
4.2. Clinical applications of the NanoVelcro Chip
There continues to be a profound need for reliable biomarkers for monitoring PC disease activity. The serum PSA assay has come under great scrutiny given the large number of factors, which can impact its accuracy including flare responses to both new and older treatments. We have demonstrated the serial CTC enumeration can be performed in PC patients before and after starting treatment. Our study also shows that the capture efficiency of the NanoVelcro Chip is reliable and capable of monitoring dynamic changes in CTC number over times. In this early experience, our data showed that declines in CTC number paralleled clinical benefit, In contrast, we observed a variety of CTC number alterations in clinical non-responders. In the group of patients who did not experience clinical benefit from starting a new therapy, the CTCs typically decreased initially then rebounded quickly (Fig. 6C). This observation may reflect a rapid response of the CTCs sensitive to the treatment followed by continued shedding of resistant clones into the circulation.
It has been reported that CTC number determined by CellSearch is a better overall survival predictor than PSA [28]. When we compared serial CTC enumerations with corresponding serum PSA concentrations, UCLA1 had disease amenable to RECIST (Response Evaluation Criteria in Solid Tumors)– the standard for radiographic evaluation. We noted reduction in tumor size that temporally coincided with a decrement in CTC number. This finding was discordant with his PSA that rose in response. This was due to PSA flare, a well-recognized phenomenon that coincides several PC treatments (Fig. 6B). PSA is optimally used when considered over time and in relation to treatment. Our preliminary work suggests that the same holds true for CTC number. We observed that all patients with regressing or stable disease showed a drop in CTC number. This suggests that serial CTC enumeration may predict response to therapy. As shown in Fig. 7, we observed that the overall trends of his CTC number were similar to that of his serum PSA changes. When correlating with his radiographic assessments, his CTC and PSA both showed an initial decrease and a subsequent rise at the time when he progressed through nilutamide and sipuleucel-t. Interestingly, in response to abiraterone, he showed an initial PSA and CTC increase along with radiographic progression. His disease stabilized (by serum PSA and imaging studies) after 3 months of treatment. This corresponded temporally to a stabilization of his CTC number. We also noted that his decline in CTC counts more strongly paralleled his clinical and radiographic response to docetaxel. This again is in contrast to his serum PSA which rose. These findings suggested that serial CTC enumerations with the NanoVelcro Chip may have predictive values in assessing response to therapy.
Fig. 7.
Serial CTC and PSA changes of UCLA8 are plotted during which multiple treatment responses and progressions are documented.
In the current literature, there are reports indicating that CTC numbers change in response to different therapies in different cancers [29, 30]. However, the predictive value of CTC numbers was less clear, likely due to the relatively low sensitivity of the CellSearch™ assay. Its limited dynamic range creates a need for very large cohorts to provide adequate power to demonstrate a correlation between CTC number and clinical outcome. As the NanoVelcro Chip generally demonstrates much higher capture efficiency compared to the CellSearch™ assay [22], CTC enumeration by the NanoVelcro Chip may yield a wider dynamic range and may be a better predictor of drug response and disease progression.
4.3. Limitations and future directions of the NanoVelcro Chip
The NanoVelcro Chip clearly has a robust capture efficiency and reliable consistency. Despite this, we have not yet demonstrated that the NanoVelcro Chip can be used to obtain crucial molecular information from captured CTCs. Our future efforts will be devoted to characterizing the molecular signatures of CTCs, including genomic, transcriptional and proteomic analysis. Since the captured CTCs are alive, these cells could potentially be expanded ex vivo for further molecular interrogation. We recognize the greater challenge of obtaining biologically functional CTCs that can be used for ex vivo culture. This would allow for characterizing their behavioral characteristics. Such a process would create a new means of evaluating drug sensitivity and resistance in individual patients. Aware of these urgent needs, we are currently evaluating alternative substrates to enable the physical isolation of CTCs. Using a quantitative immunocytochemistry approach previously described by our group [34], multiparametric molecular analysis may also provide us with extra information on gene expression, mutation, amplification, and deletion. This may provide insight into how these changes may alter cell signaling in cancer.
5. Conclusions
We have demonstrated the reliable, consistent, and efficient capture of cancer cells using the NanoVelcro Chip with both cancer cells spiked into normal human blood and CTCs from patients both at UCLA and CSMC. Our technique allows serial capture of CTCs from patients who are receiving therapy. Our results suggest a relationship between increase in CTC number and disease progression. In selected patients, we detected reductions in CTC number in response to different therapeutic approaches.
Based on our preliminary success in applying the NanoVelcro Chip to the clinical setting, we are looking forward to further translating this technology to the clinic with the goal of determining prognosis and providing early insight into treatment response. We believe that these data will ultimately help guide clinical decision making to benefit patients.
Supplementary Material
Supplemental Figure S1. A mosaic micrograph (obtained by 4X objective) of a NanoVelcro substrate after CTC capture study. This substrate was automatically scanned by a software-controlled immunofluorescence microscope. The CTC candidates were identified based on their DAPI, CK and CD45 immunofluorescence levels and sizes before the operator confirmed them under a 10X objective.
Supplemental Table S1. Summary of patients’ treatments and their clinical and CTC responses
Table 2.
Clinicopathologic characteristics of 40 patients participating in our CTC enumerations
| Variable | Categories | Evaluable patients at UCLA (n=30) | Evaluable patients at CSMC (n=10) |
|---|---|---|---|
| Age at baseline | Median (Range) | 70.5 (43–88) | 67 (52–79) |
| Gleason Score | 3+2 | 1 (3%) | 0 (0%) |
| 3+3 | 1 (3%) | 4 (40%) | |
| 3+4 | 6 (20%) | 4 (40%) | |
| 4+3 | 5 (17%) | 1 (10%) | |
| 4+4 | 4 (13%) | 1 (10%) | |
| 4+5 | 2 (7%) | 0 (0%) | |
| 5+4 | 2 (7%) | 0 (0%) | |
| 5+5 | 1 (3%) | 0 (0%) | |
| Unknown/Other | 8 (27%) | 0 (0%) | |
| Stage | II | 0 (0%) | 6 (60%) |
| III | 0 (0%) | 2 (20%) | |
| IV | 30 (100%) | 2 (20%) | |
| Baseline PSA (ng/mL) | Median (Range) | 50.25 (0.18– >5000) | 5.6 (4.26–293.7) |
Highlights.
We construct NanoVelcro CTC Chip for highly efficient capture and enumeration of circulating tumor cells in prostate cancer patients
NanoVelcro CTC Chip can be easily replicated to another institute with similar performance
NanoVelcro CTC Chip assay detected CTC decrease when patients had clinical responses to treatments
NanoVelcro CTC Chip has the capability to perform serial CTC enumerations across a timespan of 460 days with clinically relevant results.
Acknowledgments
The research endeavors at UCLA were supported by a Creativity Award from Prostate Cancer Foundation (PCF), UCLA Prostate Cancer SPORE Program, and research grants (R21 CA151159 and R33 CA157396) from NIH/NCI Innovative Molecular Analysis Technologies (IMAT) Program. The research endeavors at Cedars Sinai Medical Center were supported by DoD Idea Award (W81XWH-11-1-0422) and PCF Young Investigator Award.
References
- 1.Pantel K, Brakenhoff RH. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson DR, Dracopoli N, Gumbs Petty B, Compton C, Cristofanilli M, Deisseroth A, Hayes DF, Kapke G, Kumar P, Lee JS, Liu MC, McCormack R, Mikulski S, Nagahara L, Pantel K, Pearson-White S, Punnoose EA, Roadcap LT, Schade AE, Scher HI, Sigman CC, Kelloff GJ. J Transl Med. 2012;10:138. doi: 10.1186/1479-5876-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaiser J. Science. 2010;327:1072–1074. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]
- 4.Bernards R, Weinberg RA. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- 5.Criscitiello C, Sotiriou C, Ignatiadis M. Curr Opin Oncol. 2010;22:552–558. doi: 10.1097/CCO.0b013e32833de186. [DOI] [PubMed] [Google Scholar]
- 6.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LWMM, Uhr JW. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zieglschmid V, Hollmann C, Böcher O. Critical Reviews in Clinical Laboratory Sciences. 2005;42:155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 9.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 10.Jacob K, Sollier C, Jabado N. Expert Rev Proteomics. 2007;4:741–756. doi: 10.1586/14789450.4.6.741. [DOI] [PubMed] [Google Scholar]
- 11.Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Biotechnology and bioengineering. 2009;102:521–534. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan AL, Vantyghem SA, Tuck AB, Chambers AF, Chin-Yee IH, Keeney M. Cytometry A. 2005;65:4–14. doi: 10.1002/cyto.a.20132. [DOI] [PubMed] [Google Scholar]
- 13.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Science translational medicine. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olmos D, Arkenau HT, Ang JE, Ledaki I, Attard G, Carden CP, Reid AH, A’Hern R, Fong PC, Oomen NB, Molife R, Dearnaley D, Parker C, Terstappen LW, de Bono JS. Ann Oncol. 2009;20:27–33. doi: 10.1093/annonc/mdn544. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 20.Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Lancet Oncol. 2009;10:233–239. doi: 10.1016/S1470-2045(08)70340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew Chem Int Ed Engl. 2009;48:8970–8973. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Angew Chem Int Ed Engl. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer KE, Aleman BJ, Tao SL, Daniels RH, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano Lett. 2009;9:716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis ASG, Varde M. J Natl Cancer I. 1964;33:15. [PubMed] [Google Scholar]
- 25.Liu WF, Chen CS. Advanced drug delivery reviews. 2007;59:1319–1328. doi: 10.1016/j.addr.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thalmann GN, Anezinis PE, Chang SM, Zhau HE, Kim EE, Hopwood VL, Pathak S, von Eschenbach AC, Chung LW. Cancer research. 1994;54:2577–2581. [PubMed] [Google Scholar]
- 27.Attard G, de Bono JS. Current opinion in genetics & development. 2011;21:50–58. doi: 10.1016/j.gde.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 28.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 29.Bidard FC, Mathiot C, Degeorges A, Etienne-Grimaldi MC, Delva R, Pivot X, Veyret C, Bergougnoux L, de Cremoux P, Milano G, Pierga JY. Ann Oncol. 2010;21:1765–1771. doi: 10.1093/annonc/mdq052. [DOI] [PubMed] [Google Scholar]
- 30.Lowes LE, Lock M, Rodrigues G, D’Souza D, Bauman G, Ahmad B, Venkatesan V, Allan AL, Sexton T. Clin Transl Oncol. 2012;14:150–156. doi: 10.1007/s12094-012-0775-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. A mosaic micrograph (obtained by 4X objective) of a NanoVelcro substrate after CTC capture study. This substrate was automatically scanned by a software-controlled immunofluorescence microscope. The CTC candidates were identified based on their DAPI, CK and CD45 immunofluorescence levels and sizes before the operator confirmed them under a 10X objective.
Supplemental Table S1. Summary of patients’ treatments and their clinical and CTC responses