Abstract
Phase-I drug metabolizing enzymes catalyze reactions of hydrolysis, reduction, and oxidation of drugs and play a critical role in drug metabolism. However, the functions of most phase-I enzymes are not mature at birth, which markedly affects drug metabolism in newborns. Therefore, characterization of the expression profiles of phase-I enzymes and the underlying regulatory mechanisms during liver maturation is needed for better estimation of using drugs in pediatric patients. The mouse is an animal model widely used for studying the mechanisms in the regulation of developmental expression of phase-I genes. Therefore, we applied RNA sequencing to provide a “true quantification” of the mRNA expression of phase-I genes in the mouse liver during development. Liver samples of male C57BL/6 mice at 12 different ages from prenatal to adulthood were used for defining the ontogenic mRNA profiles of phase-I families, including hydrolysis: carboxylesterase (Ces), paraoxonase (Pon), and epoxide hydrolase (Ephx); reduction: aldo-keto reductase (Akr), quinone oxidoreductase (Nqo), and dihydropyrimidine dehydrogenase (Dpyd); and oxidation: alcohol dehydrogenase (Adh), aldehyde dehydrogenase (Aldh), flavin monooxygenases (Fmo), molybdenum hydroxylase (Aox and Xdh), cytochrome P450 (P450), and cytochrome P450 oxidoreductase (Por). Two rapidly increasing stages of total phase-I gene expression after birth reflect functional transition of the liver during development. Diverse expression patterns were identified, and some large gene families contained the mRNA of genes that are enriched at different stages of development. Our study reveals the mRNA abundance of phase-I genes in the mouse liver during development and provides a valuable foundation for mechanistic studies in the future.
Introduction
Drug-metabolizing enzymes play a central role in the elimination of drugs from the body. The metabolic reactions of hydrolysis, reduction, and oxidation usually introduce a small functional group to the substrate or convert an existing functional group to a new group that may be further conjugated with a large and bulky water-soluble cosubstrate. The reactions associated with hydrolysis, reduction, and oxidation are often called phase-I reactions, and the reactions associated with conjugation are often referred as phase-II reactions. The liver is the major organ for phase-I drug metabolism; however, before birth, the liver functions as a hematopoietic organ. A functional transition occurs in the liver after birth, and most of the drug-metabolizing enzymes mature during this period. Changes in expression of some phase-I enzymes during liver maturation in humans have been reported, including cytochrome P450 (P450) (Stevens et al., 2003, 2008; Koukouritaki et al., 2004; Croom et al., 2009), carboxylesterase (CES) (Yang et al., 2009; Zhu et al., 2009), paraoxonase (PON) (Cole et al., 2003; Huen et al., 2009), alcohol dehydrogenase (ADH) (Smith et al., 1971), and flavin monooxygenase (FMO) (Cherrington et al., 1998; Koukouritaki et al., 2002; Hines, 2006). The dynamic changes in the ontogenic expression of these genes are thought to be responsible for the substantial pharmacokinetic differences between newborns and adults, and this contributes to differences in therapeutic efficacy and adverse drug reactions in pediatric patients (Kearns et al., 2003; Blake et al., 2005; Hines, 2007, 2008, 2013). An in-depth understanding of the regulatory mechanisms of the ontogeny of phase-I enzymes is needed for safer and more effective drug therapy for pediatric patients.
Several limitations exist in studies of the developmental regulation of phase-I enzymes with human samples. The first limitation is ethical and technical issues in recruiting human subjects and obtaining suitable human samples (Rowell and Zlotkin, 1997). Second, variations in human metabolic capacity, which may be caused by genetic or environmental factors, can interfere with studies aimed to reveal the regulatory mechanisms that are only due to age. Furthermore, mechanistic loss-of-function or gain-of-function strategies are not applicable directly in human samples. Animal models are advantageous in overcoming these limitations. In recent years, mice and rats have surpassed other laboratory animals as the experimental models of choice for the study of physiology, metabolism, and disease (Muruganandan and Sinal, 2008; Hrycay and Bandiera, 2009). The advantages of these models include rapid growth, easy maintenance, and the development of genetic manipulation techniques for mechanistic studies with gain-of-function and loss-of-function strategies. Several laboratories, including ours, have examined the ontogenic gene expression profiles in mouse or rat livers for some phase-I genes, including P450s (Hart et al., 2009; Cui et al., 2012b), Ces (Zhu et al., 2009), aldo-keto reductase (Akr) (Pratt-Hyatt et al., 2013), Adh and aldehyde dehydrogenase (Aldh) (Smolen et al., 1990; Alnouti and Klaassen, 2008), Pon (Li et al., 1997), and Fmo (Falls et al., 1995; Cherrington et al., 1998; Janmohamed et al., 2004). The developmental expression patterns of some phase-I genes in mice and rats are similar to those in humans.
Previous studies quantified phase-I gene expression on the mRNA level by either Northern blot, reverse-transcription polymerase chain reaction, microarrays, or multiplex suspension bead arrays, which only provide relative quantification of a given gene and do not allow a quantitative comparison of genes in different families. With the development of next-generation sequencing technologies such as RNA sequencing (RNA-Seq), it is possible to define a whole transcriptome with low background noise, no upper limit for quantification, and a high degree of reproducibility for both technical and biologic replicates (Mortazavi et al., 2008; Nagalakshmi et al., 2008). More importantly, RNA-Seq quantifies the true abundance of mRNA molecules in biologic samples and enables comparison of the expression of all genes (Malone and Oliver, 2011). We have reported that RNA-Seq can reveal ontogenic patterns of P450s (Peng et al., 2012), phase-II enzymes (Lu et al., 2013), transporters (Cui et al., 2012a), and epigenetic modifiers (Lu et al., 2012) in the mouse liver during maturation.
In this report, RNA-Seq was used to systematically quantify the mRNA expression of major non-P450 phase-I genes in the mouse liver during postnatal maturation to define the ontogenic profiles of these mRNAs. The groups included enzymes catalyzing reactions in hydrolysis (carboxylesterase, paraoxonase, and epoxide hydrolase), reduction (aldo-keto reductase, quinone oxidoreductase, and dihydropyrimidine dehydrogenase), and oxidation (alcohol dehydrogenase, aldehyde dehydrogenase, flavin monooxygenases, molybdenum hydroxylase, and cytochrome P450 oxidoreductase). The purpose of this study was to generate comprehensive information on the ontogeny of mRNAs of phase-I genes in the livers of mice, which will form the foundation for determining the regulatory mechanisms controlling the various transcription patterns of phase-I genes during liver maturation.
Materials and Methods
Animals.
Eight-week-old C57BL/6 breeding pairs of mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were housed according to the American Animal Association Laboratory animal care guidelines and were bred under standard conditions in the Laboratory Animal Resources Facility at the University of Kansas Medical Center (KUMC). The use of these mice was approved by the Institute of Laboratory Animal Resources at KUMC. Liver samples (n = 3) were collected at the following 12 ages: day −2 (gestational day 17), day 0 (right after birth and before the start of suckling), day 1 (exactly 24 hours after birth), and days 3, 5, 10, 15, 20, 25, 30, 45, and 60 (collected at approximately 9:00 AM). These ages represent the periods of prenatal (day −2), neonatal (days 0–10), juvenile (days 15–30), and young adult (days 45–60). Due to potential variations caused by the estrous cycle in maturing adult female mice, only male livers were used for this study. The livers were immediately frozen in liquid nitrogen after removal and stored at −80°C.
Total RNA Extraction, Sequencing Library Construction, and RNA-Seq.
RNA extraction, library construction, and RNA-Seq were performed as previously described elsewhere (Peng et al., 2012).
RNA-Seq Data Analysis.
After the sequencing images were generated by the sequencing platform, the pixel-level raw data collection, image analysis, and base calling were performed using Illumina’s Real Time Analysis (RTA) software (Illumina, San Diego, CA). The output bcl files were converted to qseq files by Illumina BCL Converter 1.7 software and subsequently converted to FASTQ files for downstream analysis. The RNA-Seq reads from the FASTQ files were mapped to the mouse reference genome (NCBI37/mm9) by Tophat 1.2.0 (http://tophat.cbcb.umd.edu/). The output files in BAM (binary sequence alignment) format were analyzed by Cufflinks 1.0.3 (http://cufflinks.cbcb.umd.edu/) to estimate the transcript abundance (Trapnell et al., 2010). The mRNA abundance was expressed as the number of fragments per kilobase of exon per million reads mapped (FPKM).
Data Visualization and Statistics.
The significance of the observed expression (measured FPKM) of a gene at a given age relative to null expression (zero FPKM) was determined by the drop-in-deviance F test of the fitted FPKM values to a Poisson log linear regression model with a zero intercept that permits extra Poisson variation. The resulting P values were adjusted for multiple-hypothesis testing by the Benjamini-Hochberg method (FDR-BH; Benjamini and Hochberg, 1995). Phase-I drug metabolizing enzymes that were significantly expressed (FDR-BH ≤ 0.05) in at least one of the 12 time-points were selected for analysis. Genes that were significantly differentially expressed between at least two time points during liver development were determined by the drop-in-deviance F test of the fitted FPKM values to a full
where AGE is an indicator variable representing the 12 ages, j=1...11) and reduced
Poisson-log-linear regression model accounting for extra Poisson variation. The resulting P values were adjusted for false discovery as before, and those genes with a FDR-BH ≤ 0.05 were considered significantly differentially expressed over time. Two-way hierarchical clustering dendrograms were generated by JMP (version 10; SAS Institute, Inc., Cary, NC) to determine the expression patterns of the phase-I genes during liver development.
Results
Total Expression and Proportions of Individual Phase-I Families during Liver Maturation
Transcript abundances of the 186 mouse phase-I genes were calculated by Cufflinks and presented as FPKM values in Supplemental Table 1. If the Benjamini-Hochberg adjusted P value of the drop-in-deviance F test (FDR-BH) for gene expression in at least one of the 12 ages was less than 0.05 for a phase-I gene, then that gene was considered to be expressed in the liver during maturation. Table 1 lists the number of genes in each category that are significantly expressed at various developmental stages of prenatal (day −2), neonatal (day 5), adolescence (day 25), and adult (day 60) as well as the total number of genes expressed during liver development (day −2 to day 60) in each phase-I family. Of the 186 mouse phase-I genes, 136 genes were expressed in the liver during maturation, but only about half of them (64) were expressed in the prenatal liver.
TABLE 1.
Number of expressed genes in each phase I family in mouse livers at specific ages of prenatal (day −2), neonatal (day 5), adolescence (day 25), and adult (day 60), as well as during development (days −2 to 60)
Numbers in brackets represent total number of genes in the family.
| Gene Family | Prenatal Day −2 | Neonatal Day 5 | Adolescence Day 25 | Adult Day 60 | Liver Days −2 to 60 |
|---|---|---|---|---|---|
| Hydrolysis | |||||
| Ces (18) | 4 | 11 | 13 | 11 | 14 |
| Pon (3) | 2 | 3 | 3 | 3 | 3 |
| Ephx (4) | 2 | 2 | 2 | 2 | 2 |
| Reduction | |||||
| Akr (16) | 9 | 14 | 12 | 10 | 14 |
| Nqo (2) | 2 | 2 | 2 | 2 | 2 |
| Dpyd (1) | 0 | 1 | 0 | 1 | 1 |
| Oxidation | |||||
| Adh (6) | 2 | 3 | 3 | 3 | 5 |
| Aldh (20) | 13 | 15 | 14 | 13 | 15 |
| Aox+Xdh (5) | 0 | 2 | 3 | 3 | 3 |
| Fmo (7) | 3 | 3 | 2 | 2 | 5 |
| P450 (103) | 26 | 55 | 59 | 50 | 71 |
| Por (1) | 1 | 1 | 1 | 1 | 1 |
| Total (186) | 64 | 112 | 114 | 101 | 136 |
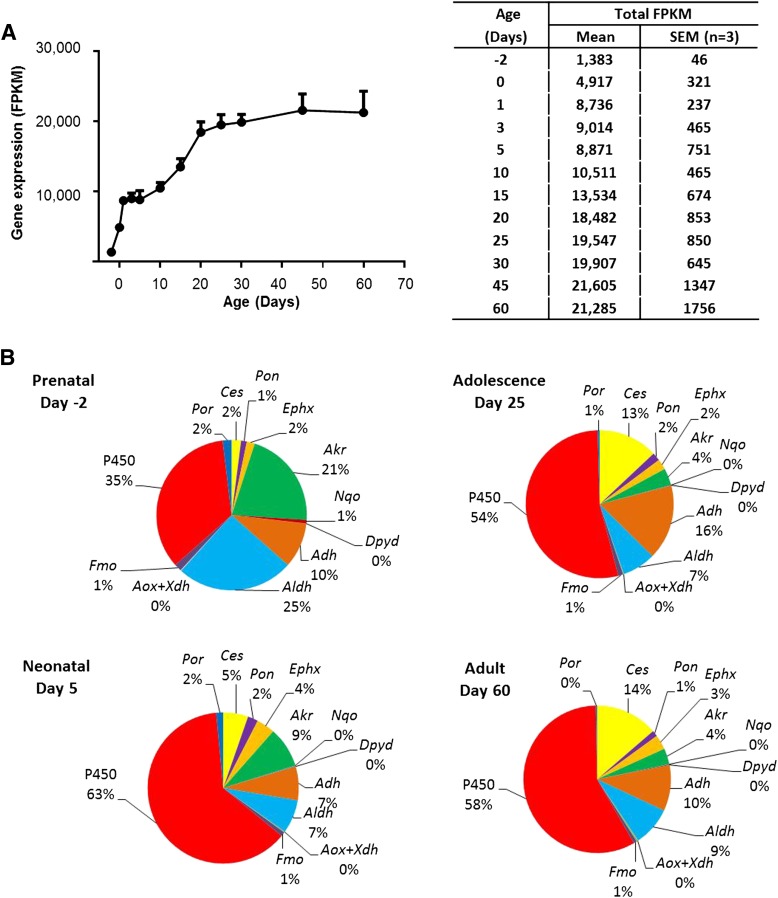
A significant change in expression of phase-I enzyme genes was observed around birth. The number of the expressed genes increased to 112 in the neonatal liver at day 5. The total expression levels of all phase-I mRNAs represented as cumulative FPKM values increased approximately 15-fold during postnatal liver maturation from day −2 (FPKM = 1383) to day 60 (21,285) with two surges (Fig. 1A), the first of which with a 6-fold increase from day −2 (1383) to day 1 (8736) and the other with 1.8-fold increase from day 10 (10,511) to day 20 (18,482), with each surge followed by a relatively stable value of cumulative FPKM. Figure 1B shows the composition of the phase-I families represented as percentages of the total FPKM value for each stage at prenatal (day −2), neonatal (day 5), adolescence (day 25), and adult (day 60).
Fig. 1.
(A) Total mRNA levels of the 186 mouse phase-I genes in the liver during postnatal maturation. RNA-Seq was done for liver mRNAs of male C57BL/6 mice at 12 ages from 2 days before birth to 60 days after birth. The FPKM values of all 186 phase-I genes at each age were added and plotted to show the developmental pattern of total phase-I mRNAs. Bars represent the mean ± S.E.M. of three individual animals. (B) Percentages of FPKM values of each phase-I family in prenatal (day −2), neonatal (day 5), adolescence (day 25), and adult (day 60) livers.
The major phase-I families expressed in the prenatal liver were P450 (35%), Aldh (25%), Akr (21%), and Adh (10%). After birth, there is a further increase in the relative proportions of the mRNAs of the P450s (from 35% prenatal levels to about 60% after birth). Other highly expressed phase-I families include Ces, Adh, and Aldh.
Ontogeny of Genes of Phase I Enzymes Involved in Hydrolysis Reactions
Carboxylesterases.
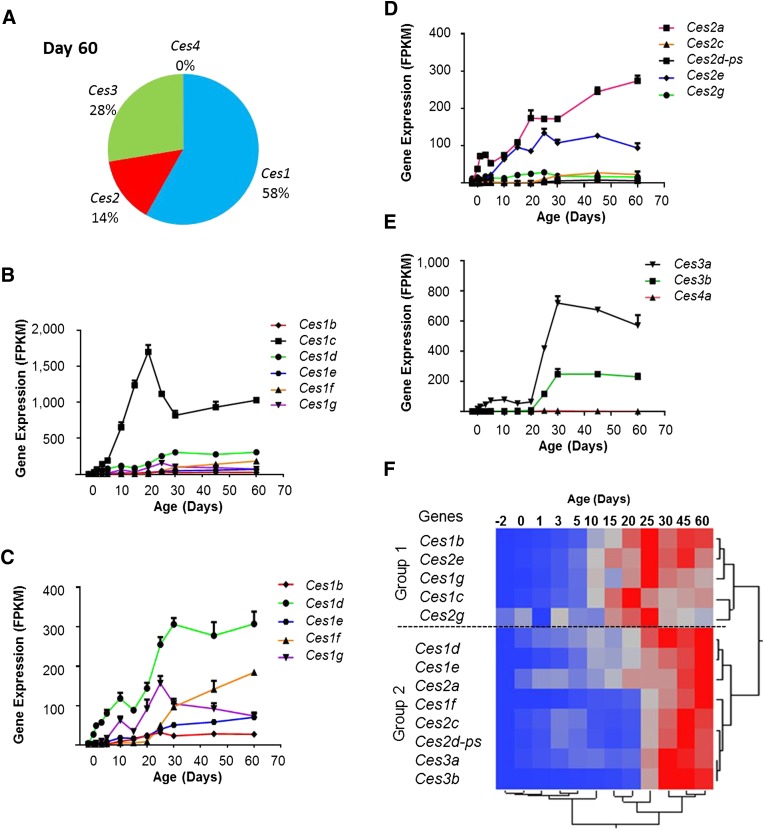
CESs are enzymes that participate in the hydrolysis and trans-esterification of a variety of esters and amides. Five families of Ces genes are annotated in the National Center for Biotechnology Information (NCBI) reference sequence database for mice. Expression of mRNA of four families of Ces (Ces1–Ces4) was detected in mouse liver samples. The most abundant family in the adult liver was Ces1, which comprised up to 58% of the total liver Ces mRNAs at age day 60 (Fig. 2A). Within the Ces family 1, Ces1c had much higher expression than other members at postnatal ages. The mRNA levels of Ces1 genes generally increased with age, but Ces1c and Ces1g showed a peak of expression around day 20 and day 25, respectively (Fig. 2B and 2C).
Fig. 2.
Expression of Ces mRNAs during liver development in male C57BL/6 mice. (A) mRNA proportion of each Ces family at the age of day 60. (B) Expression profiles of Ces1 gene family. (C) Expression profiles of Ces1 gene family without Ces1c. (D) Expression profiles of Ces2 gene family. (E) Expression profiles of Ces3 and Ces4 gene families. Data are expressed as mean FPKM and S.E.M. of three individual animals. (F) Hierarchical clustering of expression profiles for 13 differentially expressed Ces genes. The two trees describe the relationship between different gene expression profiles (right tree) and various ages (bottom tree). The dendrogram scale represents the correlation distances. Average FPKM values of three replicates per age are given by colored squares: red, relatively high expression; blue, relatively low expression. The dashed line categorizes the expression profiles into two major groups.
The Ces2 family was moderately expressed, with Ces2a and Ces2e being the major members. The expression of Ces2a gradually increased after birth till day 60, and Ces2e reached its adult levels at around day 25 (Fig. 2D).
The Ces3 family was also highly expressed in adult livers. Ces3a and Ces3b exhibited similar developmental patterns, with a rapid increase in expression between days 20 and 30 (Fig. 2E). The Ces4 family was expressed at a low level in the liver, and it did not show a significant difference in expression during development.
In total, 13 Ces genes had significant changes in expression during development. A two-way hierarchical clustering analysis revealed that the mRNA of these Ces were all low in prenatal and neonatal ages, but were markedly increased to adult levels around 20–30 days after birth (Fig. 2F).
Paraoxonases.
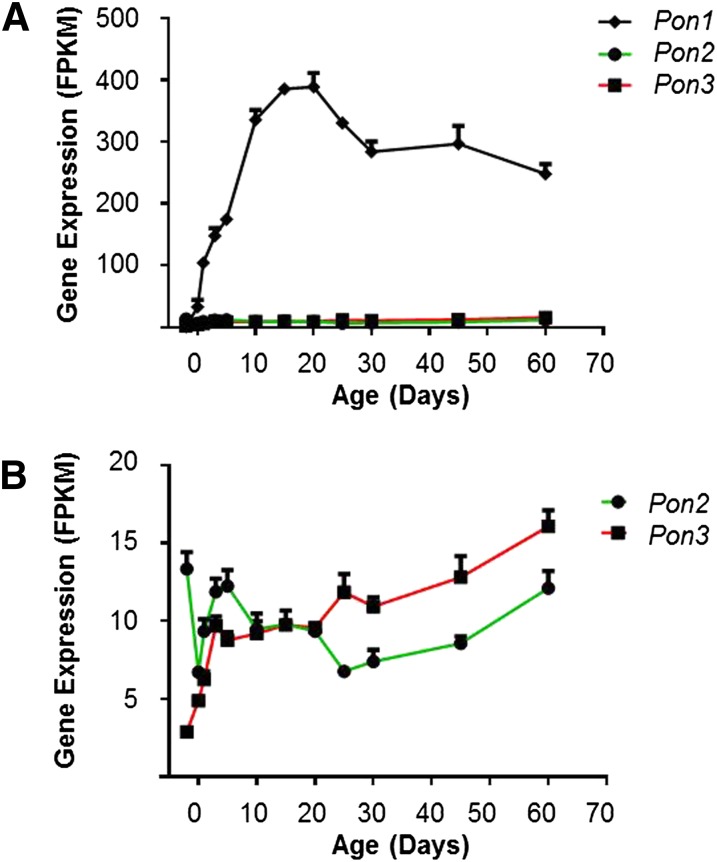
PON function in hydrolyzing a broad range of organophosphates, organophosphinites, aromatic carboxylic acid esters, cyclic carbonates, and lactones (Parkinson and Ogilvie, 2008). Mouse livers expressed all three Pons, with Pon1 being the most abundant form. Hepatic expression of Pon1 increased with age to a peak level around day 20 and then modestly decreased thereafter (Fig. 3A). At the prenatal age day −2, Pon2 mRNA level was higher than Pon1 and Pon3. After birth, the Pon2 mRNA expression first increased till day 5 then decreased till day 25, and then went up again to reach the adult level at day 60. Pon3 had an overall increased expression after birth with small fluctuations at adolescent ages (Fig. 3B).
Fig. 3.
Expression of Pon during liver development. (A) All three Pon ontogenic mRNA expression patterns. (B) Pon1 was removed to enlarge Pon2 and Pon3 expression patterns. Data are expressed as mean FPKM and S.E.M. of three individual animals.
Epoxide Hydrolases.
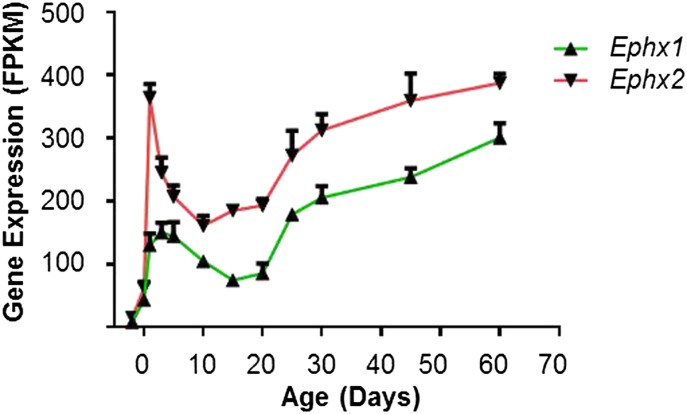
Ephxs are important in hydrolyzing and detoxifying electrophilic epoxides, which may otherwise cause cellular and genetic toxicity through binding to proteins and nucleic acids. Ephx1 and Ephx2 were the microsomal and soluble forms of these enzymes, respectively. Ephx2 had a higher expression at mRNA level than Ephx1, and they exhibited similar ontogenic patterns, with a sharp increase around birth, followed by a slight decrease through at least day 10, and then a gradual increase till adulthood (Fig. 4).
Fig. 4.
Expression patterns of Ephx1 and Ephx2 during liver development.
Ontogeny of Genes of Phase I Enzymes Involved in Reduction Reactions
Aldo-keto Reductases.
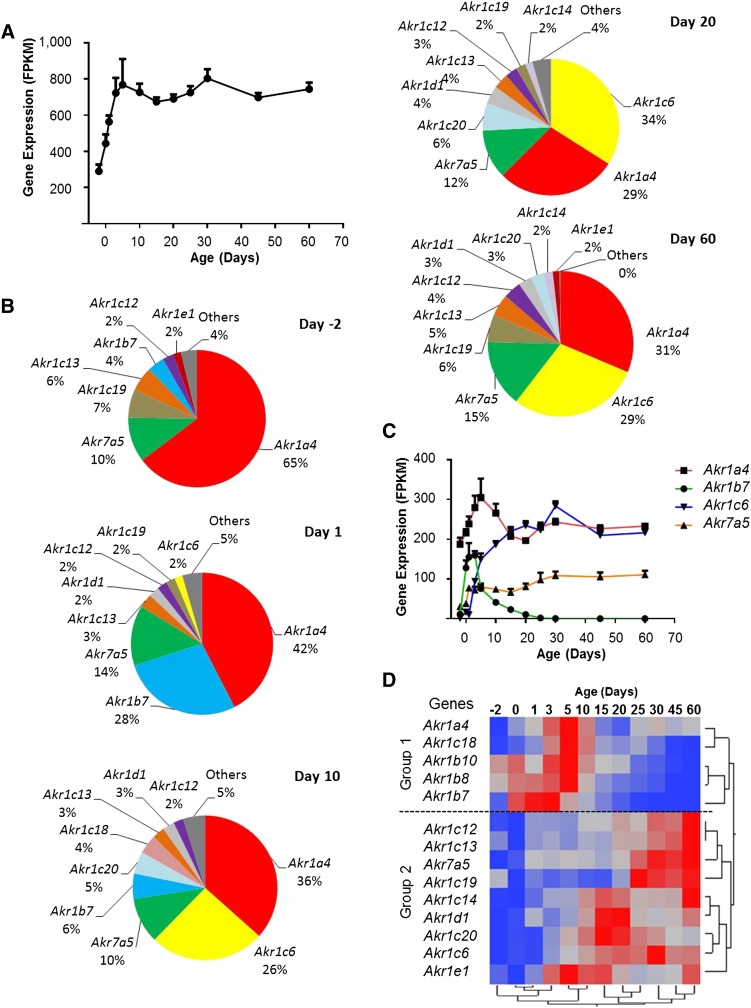
AKRs are a group of cytosolic enzymes that catalyze the reduction of aldehydes and ketones to primary and secondary alcohols, respectively (Jin and Penning, 2007). Mouse Akr genes consist of two families, Akr1 and Akr7, with a total of 16 genes. Akr1 is the larger family with five subfamilies (Akr1a-e) and 15 genes. The RNA-Seq data showed 14 out of the 16 mouse Akr genes were significantly expressed during liver maturation, and all the expressed Akrs exhibited significant differential expressions across the ages. The cumulative FPKM values of all expressed Akr mRNAs increased 2- to 3-fold during postnatal liver development (Fig. 5A).
Fig. 5.
(A) Total mRNA profile of Akr genes in the liver during development. The FPKM values of the 14 significantly expressed Akr genes are summed and plotted to show the developmental pattern of total Akr mRNAs. (B) Individual Akr mRNAs (shown as percentages of total Akr mRNAs) at 2 days before birth and 1, 10, 20, and 60 days after birth. Each gene is presented in a unique color for all ages. Only genes with mRNAs expressed at more than 1% at each age are listed, and the rest are grouped as “Others.” (C) Expression profile of the four highly expressed Akrs (>10% in B) in the liver during development. (D) Hierarchical clustering of expression profiles for the 14 differentially expressed Akr genes. The dendrogram scale represents the correlation distances. Average FPKM values of three replicates per age are given by colored squares: red, relatively high expression; blue, relatively low expression. The dashed line categorizes the expression profiles into two major groups.
The composition of Akr mRNA also changes with age, as evidenced by alterations in individual Akr genes. At day −2, Akr1a4 was the most abundant member, accounting for 65% of the total Akr mRNAs. After birth, Akr1a4 mRNA slightly increased, but its proportion in Akr mRNAs decreased (Fig. 5, B and C). Akr1b7 was highest during the neonatal stage. It accounted for 28% of the total Akr mRNA at day 1, but was undetectable after day 20. Akr1c6 was one of the major Akr genes in the liver; it was lowly expressed at birth, gradually increased to a peak at day 30, and then slightly decreased to adult levels around day 45. The mRNA expression profile of Akr7a5 was similar to the profile of total Akr mRNA, so the percentage of Akr7a5 mRNA was relatively constant during development (10%∼15%) (Fig. 5, B and C).
Two-way hierarchical clustering analysis of the differentially expressed Akrs revealed two major patterns (Fig. 5D). Unlike Ces genes, which all had low expression at the neonatal stage, some Akr genes displayed neonatal enriched expression (group 1). In group 2, Akr genes showed relatively low expression during the neonatal ages. These genes were a diverse group, with some genes enriched at the adolescent stage (e.g., Akr1c20) and some at the adult stage (e.g., Akr1c12).
Quinone Oxidoreductase.
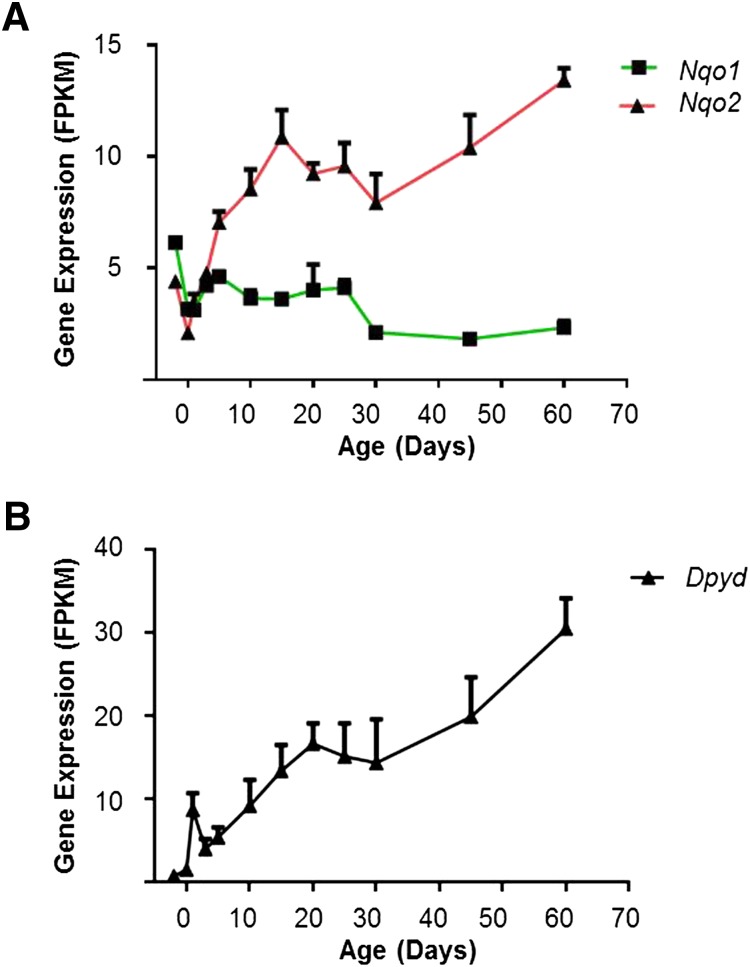
NQO1 and NQO2 perform two-electron reduction of quinones to hydroquinones. The RNA-Seq results demonstrated that the expression of Nqo1 was relatively stable during development, with two small drops around birth and day 25. Nqo2 mRNA was expressed higher than Nqo1 in mouse livers 5 days after birth. Nqo2 showed two periods of increased expression during development, one from day 0 to day 15, and the other from day 30 to day 60 (Fig. 6A).
Fig. 6.
(A) mRNA expression profile of Nqo gene in the liver during development. (B) mRNA expression profile of Dpyd in the liver during development.
Dihydropyrimidine Dehydrogenase.
DPYD is located mainly in liver cytosol. It catalyzes the reduction of 5-fluorouracil and related pyrimidines (Parkinson and Ogilvie, 2008). The mRNA expression of Dpyd showed a marked increase at day 1, dropped slightly till day 3, and then went up gradually to adult levels at day 60, with a small plateau between days 20 and 30 (Fig. 6B). The adult mRNA level of Dpyd was about 30 times higher than in newborns.
Ontogeny of Genes of Phase I Enzymes Involved in Oxidation Reactions
Alcohol Dehydrogenases.
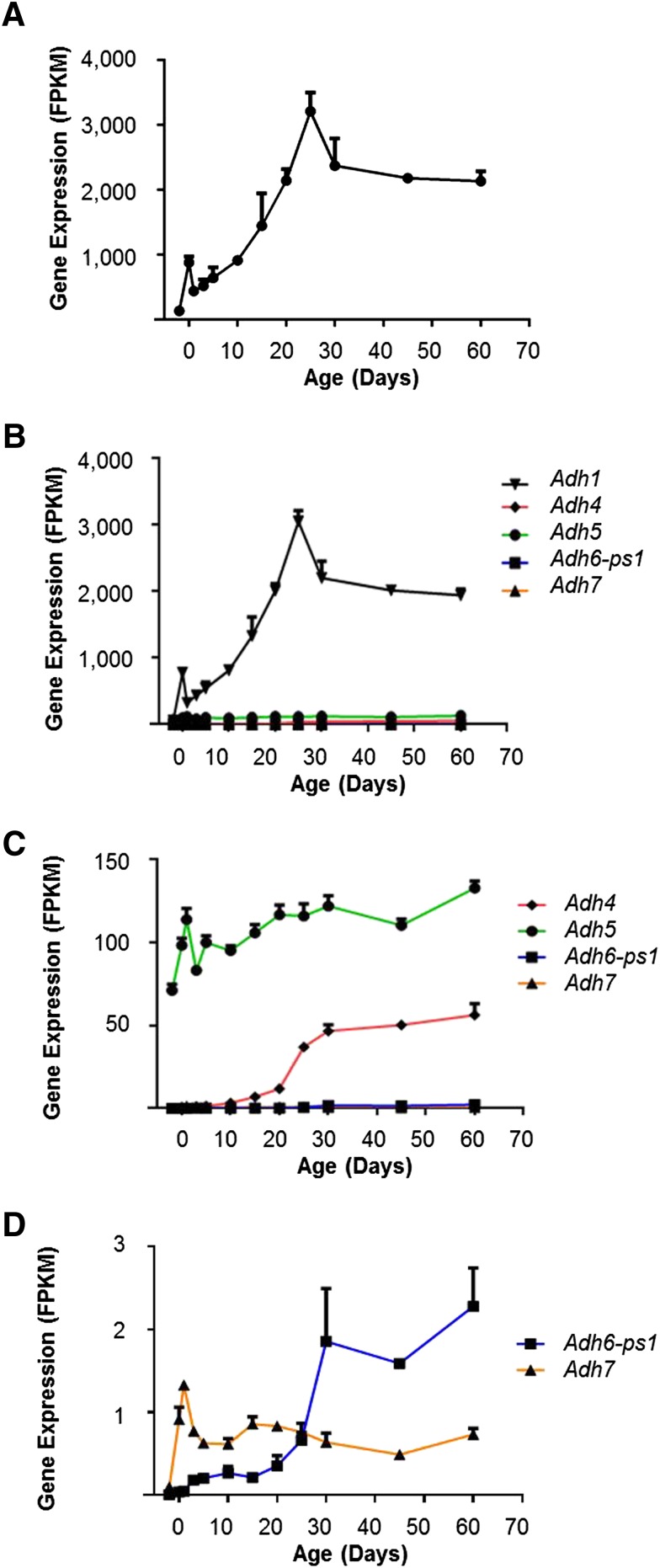
ADHs metabolize a wide spectrum of substrates, including ethanol, retinol, other aliphatic alcohols, hydroxysteroids, and lipid peroxidation products (Duester et al., 1999). Adhs generally had lower expression at younger ages and reached stable mature levels at day 30 or earlier (Fig. 7A). Five families of Adh genes exist in the mouse genome, and each family consists of only one member that is significantly expressed during liver development (Fig. 7, B–D). Adh1 mRNA was the most highly expressed, and was over 90% of all Adh mRNAs expressed in the adult liver (Fig. 7B), followed by Adh5 and then Adh4 (Fig. 7C). Adh6 and Adh7 were minimally expressed in the liver (Fig. 7D). Adh1 and Adh5 also showed a peak of expression around day 0 or day 1.
Fig. 7.
(A) Total mRNA profile of all five Adh genes during liver development. (B) Ontogenic mRNA expression patterns of individual Adh gene. (C) Adh1 removed to enlarge Adh4 and Adh5 expression patterns. (D) Adh4 and Adh5 removed to enlarge Adh6-ps1 and Adh7 expression patterns.
Aldehyde Dehydrogenase.
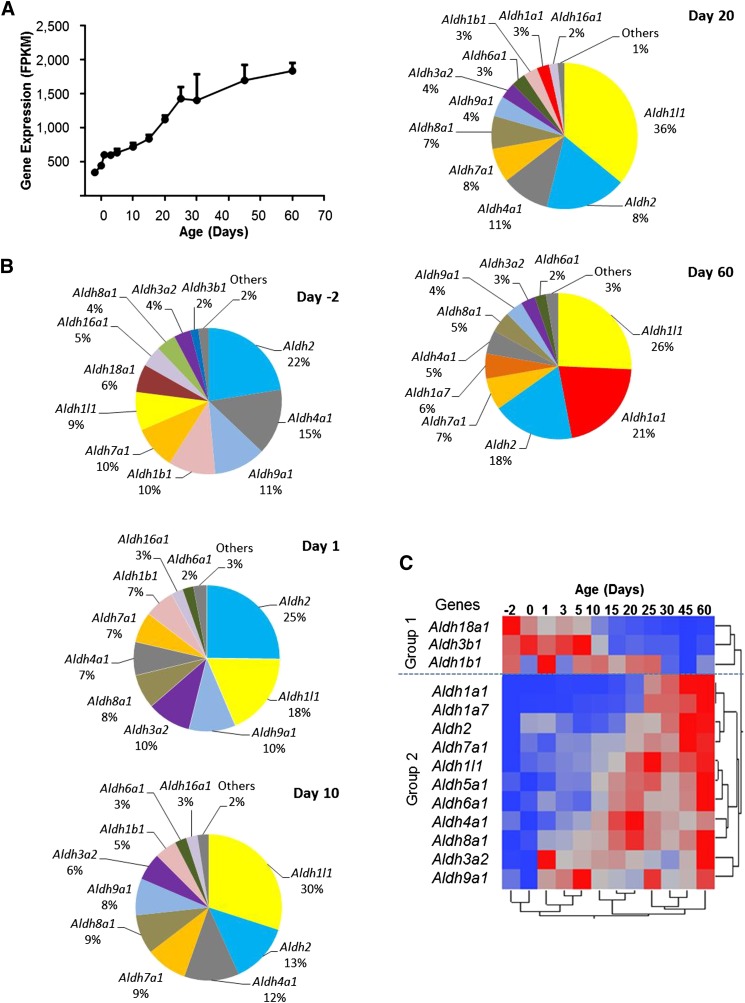
ALDHs are a group of enzymes that catalyze the oxidation and detoxification of aldehydes. There were total 20 Aldh genes in mouse genome, and 15 of these genes were significantly expressed at mRNA levels during liver maturation. Of these 15 genes, 14 showed differential expression across the ages. The total mRNA of Aldh genes increased almost linearly over 5-fold from 2 days before birth to 60 days after birth (Fig. 8A). Aldh1l1 and Aldh2 were the major Aldh genes expressed at all ages of liver development. Aldh1b1, Aldh4a1, and Aldh9a1 accounted for a high percentage of total Aldh mRNAs at the prenatal and adolescent stages, whereas Aldh1a1 matured later and became highly expressed only at the adult stage (Fig. 8B).
Fig. 8.
(A) Total mRNA profile of Aldh genes in the liver during development. The FPKM values of the 15 significantly expressed Aldh genes are summed and plotted to show the developmental pattern of total Aldh mRNAs. (B) Individual Aldh mRNAs (shown as percentages of total Aldh mRNAs) at 2 days before birth and 1, 10, 20, and 60 days after birth. Each gene is presented in a unique color for all ages. Only genes with mRNA expressed at more than 1% at each age are listed, and the rest are grouped as “Others.” (C) Hierarchical clustering of expression profiles for the 14 differentially expressed Aldh genes. The dendrogram scale represents the correlation distances. Average FPKM values of three replicates per age are given by colored squares: red, relatively high expression; blue, relatively low expression. The dashed line categorizes the expression profiles into two major groups.
Two-way hierarchical clustering analysis of the differentially expressed Aldhs has demonstrated diverse ontogenic patterns for the individual Aldhs. Similar to Akrs, group 1 Aldh genes had enriched expression at the perinatal stage, and genes in group 2 were expressed higher at the adult stage than at the perinatal stage (Fig. 8C).
Flavin Monooxygenases.
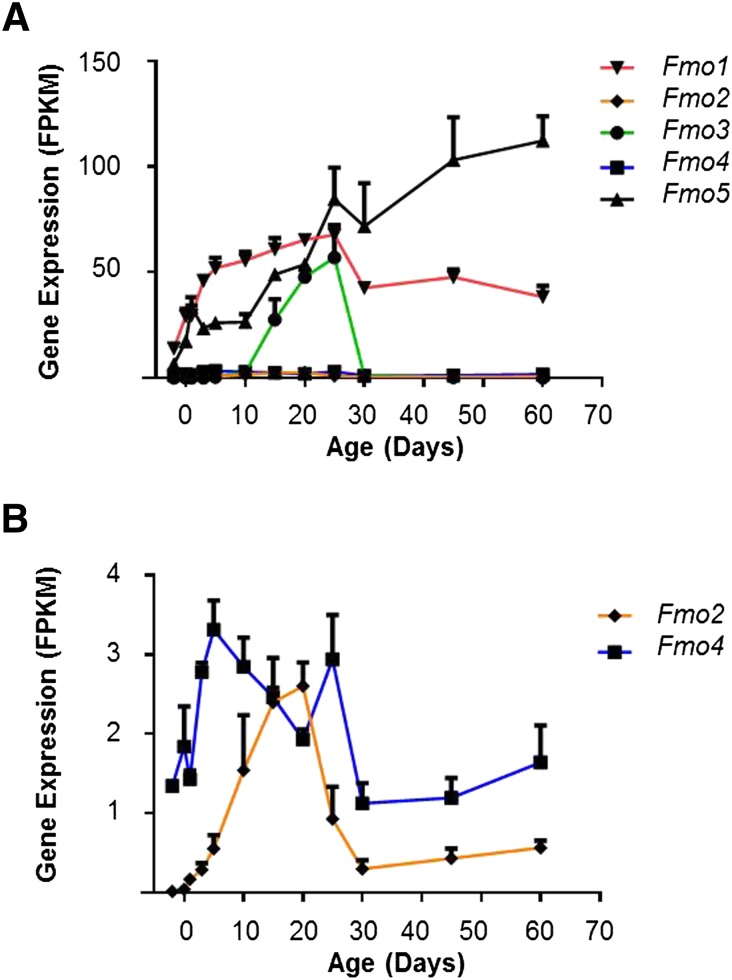
Like P450s, FMOs are microsomal enzymes that require NADPH and O2. They oxidize a variety of xenobiotics, including the nucleophilic nitrogen, sulfur, and phosphorus heteroatom (Parkinson and Ogilvie, 2008). Five families of Fmos are annotated in the mouse genome. The mRNA expression of Fmo5 increases gradually with age; it was the most abundant in adult males, followed by Fmo1, which showed a rapid increase of expression from birth to day 5, and then slightly increased to a peak at day 25 and decreased to adult expression levels at day 30. The expression of Fmo3 became detectable at 10 days after birth, increased to peak levels at day 25, and then dropped to negligible levels after day 30 in male mice (Fig. 9A). Fmo2 and Fmo4 were very lowly expressed in the liver during maturation, and Fmo2 was mainly detected at the adolescent stage from days 10 to 20 (Fig. 9B).
Fig. 9.
Expression of Fmo genes during liver development. (A) All five Fmo ontogenic mRNA expression patterns. (B) Fmo1, 3, 5 are removed to enlarge Fmo2 and Fmo4 expression patterns.
Molybdenum Hydroxylases.
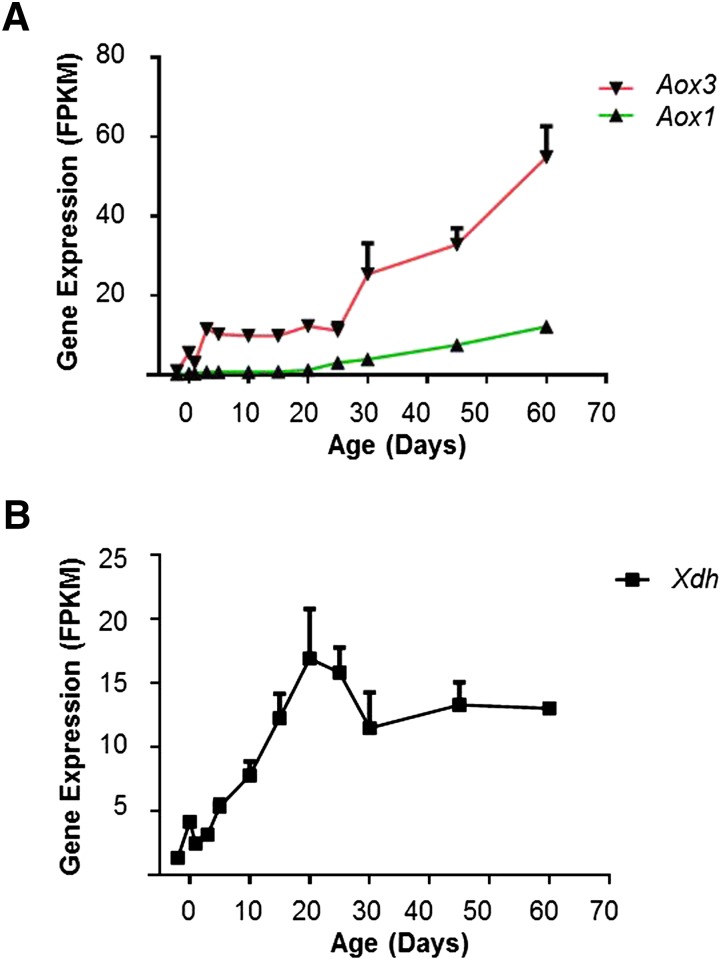
Mammalian molybdenum hydroxylases require flavin adenine dinucleotide (FAD) and molybdenum cofactor for their catalytic activity. There are two major molybdenum hydroxylases participating in the metabolism of xenobiotics: aldehyde oxidases (AOX) and xanthine oxidoreductase (XDH) (Parkinson and Ogilvie, 2008). Four Aox genes are annotated in the mouse genome, and two of them were expressed during liver maturation. Aox3 was the major Aox in mouse livers. Its mRNA level increased over 10-fold from day −2 to day 3, remained stable from day 3 to day 25, and then went up about 5-fold to adult level at day 60. Aox1 was expressed at a lower level than Aox3, and also showed an increase of expression after day 25 (Fig. 10A). Xdh mRNA expression gradually increased about 10-fold from days −2 to 20, and then remained relatively stable (Fig. 10B).
Fig. 10.
Expression of molybdenum hydroxylases during liver development. (A) mRNA ontogenic patterns of aldehyde oxidase Aox1 and Aox3. (B) mRNA ontogenic patterns of Xdh.
NADPH-Cytochrome P450 Oxidoreductase.
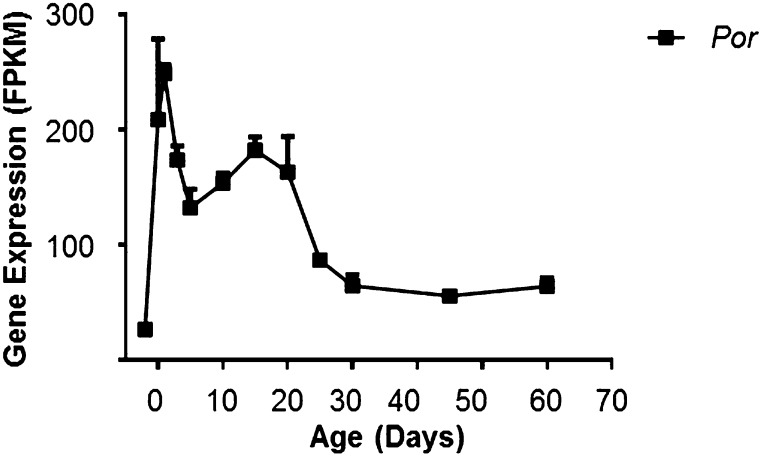
POR transfers electrons from NADPH to cytochrome P450s for their catalytic function. The expression of Por mRNA rose sharply around birth, and decreased to about half the level of day 1 by day 5. From days 5 to 20, Por expression was relatively stable. Then at day 25, it decreased again to about half the level of day 20, and it remained at that level till maturity (Fig. 11).
Fig. 11.
mRNA expression pattern of Por during liver development.
Discussion
The current study provides a comprehensive quantitative analysis of the developmental expression of major non-P450 phase-I enzymes on mRNA levels in mouse livers by RNA-Seq, including enzymes involved in hydrolysis, reduction, and oxidation. Compared with other commonly used methods for mRNA quantification, such as microarray, branched DNA, and real-time polymerase chain reaction, which detect mRNAs by probe hybridization and rely on hybridization specificity and efficiency, RNA-seq directly counts sequence reads of the nucleotide molecules in biologic samples. Thus, it can quantify the mRNA expression with minimal bias. The expression of a gene transcript is represented by FPKM, which normalizes sequencing depths between different samples and sizes between various genes, allowing direct comparison of mRNA abundance among various transcripts on a genomewide scale.
From perinatal through neonatal to adult livers, the total FPKM values of phase-I mRNAs increase 15-fold with two rapidly increasing stages (Fig. 1A), which may reflect the functional transition of the liver from a hematopoietic organ to a metabolic organ during liver maturation. The first surge occurs from 2 days before birth to 1 day after birth. During that period of time, the total FPKM values increase 8-fold, and the composition of each phase-I family also changes dramatically. P450s are the most changed phase-I enzymes during this perinatal surge, indicating an urgent need for the liver to deal with exposure to xenobiotics immediately after birth. The second surge occurs from day 10 to day 20, with a rapid increase in abundance of phase-I mRNAs. This period is also the most rapidly growing stage of postnatal liver maturation.
After day 25 (adolescence), the total FPKM levels and composition of the phase-I enzymes are consistent till day 60 (adult). These data indicate that during the developmental period from neonatal to adolescence, hepatic expression of many phase-I enzyme genes change dramatically. Expectedly, the ability to metabolize xenobiotics including drugs by phase-I enzymes should also change dramatically in this period of time. Determination of the mechanisms in regulation of the phase-I gene expression during this developmental period will provide insight for understanding drug metabolism in pediatric patients.
The ontogenic patterns of many phase-I genes we found in this study are consistent with the previous findings in mice and humans by other researchers. For example, the high expression of carboxylesterase 1 (Ces1) in our results was consistent with the literature that Ces1 was the major expressed carboxylesterase gene in the mouse liver (Holmes et al., 2010). Another study demonstrated that the expression of mouse Ces1 and Ces2 in the liver was markedly lower in newborns than in adults and increased gradually to levels of adult animals in 2 to 4 weeks (Zhu et al., 2009). Our data confirmed these observations and provides more details on changes with age and more information about individual members of each gene family (Fig. 2). Adult humans also express higher levels of CES1 and CES2 than children and fetuses (Yang et al., 2009). As a former study suggested that the CES genes from mouse and human had evolutionally conserved transcriptional regulatory mechanisms (Hosokawa et al., 2007), these ontogeny data in mice are expected to provide important resources for interpreting the developmental regulation of CESs in humans.
A study showed that lower levels of PON1 enzyme persisted in young children till at least 7 years of age (Huen et al., 2009), which was similar to the ontogenic pattern of Pon1 mRNA expression in mice. Another group found that the plasma PON1 activity in mice reached a plateau in 3 weeks after birth (Li et al., 1997), which is similar to our data of Pon1 mRNA expression in mouse livers (Fig. 3). A transgenic mouse strain that lacks endogenous Pon1 but has the human PON1 gene exhibited a similar developmental pattern of expression as wild-type mice, indicating conserved developmental regulatory elements between mouse and human PON1 (Cole et al., 2003), making our data helpful for mechanistic studies.
Quinones are highly reactive molecules. They can undergo one-electron reduction, commonly catalyzed by NADPH-cytochrome P450 reductase, and generate semiquinone radicals, which are reactive metabolites themselves and may cause oxidative stress by redox cycling. The resultant reactive oxygen species can lead to DNA damage, lipid peroxidation, membrane damage, cytotoxicity, and neoplasia. NQO1 and NQO2 compete for the above reaction and catalyze the two-electron reductive metabolism of quinones to produce stable hydroquinones, which are removed by glucuronidation or sulfonation (Long and Jaiswal, 2000; Parkinson and Ogilvie, 2008).
Quinones are ubiquitous in nature, and human exposure to quinones occurs through diet, airborne pollutants, and drugs (Monks and Jones, 2002). Therefore, quinone oxidoreductase has a highly important role in developmental toxicology. Our study is the first demonstration of the developmental expression of these genes. NQO1 and NQO2 are two closely related flavoproteins. Although they have overlapping substrate specificities, significant differences exist in relative affinities for various substrates (Das et al., 2006). Our results also revealed the differential expression of Nqo1 and Nqo2 mRNAs during development (Fig. 6A). The changes of expression during liver maturation may have a strong toxicity impact in children. Further studies are needed to address the ontogeny of NQO enzymes and their significance in human health.
Although FMO3 is the most highly expressed FMO family member in the adult human liver, it demonstrated gender-specific expression in mice and was not detectable in the male livers of mice (Falls et al., 1995). Our data only examined mRNA expression in male animals and are consistent with this result. Fmo3 was expressed equally in male and female mice, even at 4 weeks of age (Cherrington et al., 1998); after puberty, the gender difference appeared due to sex steroids (Falls et al., 1997). We also show the detailed time window when the shutdown of Fmo3 happened, which was between days 25 and 30 (Fig. 9A). Fmo1 in mice showed increased expression after birth, unlike humans in whom Fmo1 is most abundant in fetal livers and absent after birth (Hines, 2006). Interestingly, Fmo3 was not expressed immediately after birth in mice; it appeared after 10 days of age, which is similar in humans (Hines and McCarver, 2002). Thus, neonatal mice may serve as a model to study the mechanism of the delayed onset of FMO3 expression in humans.
AOX and XDH are important enzymes that catalyze the oxidation of electron-deficient carbon atoms, which are often found in nitrogen heterocycles such as purines and pyrimidines. These typically complement oxidations by cytochrome P450s, which catalyze the oxidation of carbon atoms with a high electron density. A broad range of xenobiotics are substrates for molybdoenzymes, including immunosuppressive drugs such as 6-mercaptopurine, antiviral drugs such as 6-deoxyacyclovir, and the antidepressant citalopram. They also perform important physiologic functions by metabolizing biogenic amines and catecholamines, and they may be related to neuron disease (Bendotti et al., 1997). The final electron acceptor of AOX and XDH is oxygen, so the reactions can generate reactive oxygen species and lead to oxidative stress and lipid peroxidation (Parkinson and Ogilvie, 2008). Although they have critical roles in biotransformation, the expression and regulation of molybdoenzymes are largely understudied, especially compared with other drug metabolizing enzymes. Our first report on the developmental expression pattern of these genes will facilitate drug metabolism and toxicity studies related to molybdoenzymes.
POR is the only electron donor for all microsomal P450s, and alteration in POR activity can affect P450-catalyzed drug oxidation (Hart et al., 2008). Our previous study revealed that the ontogenic mRNA expression of all P450s in the mouse liver and a large number of P450 genes had increased expression after birth (Peng et al., 2012). Here, we show hepatic mRNA expression of Por, which could influence P450 activities, actually decrease during postnatal development (Fig. 11). Thus, the level of Por needs to be taken into consideration when we study the developmental enzyme activity of P450s.
Protein levels and enzyme activities of phase-I genes were not determined in this study because of the extensive workload and technical limitations. Specific antibodies, substrates, and inhibitors for many individual mouse phase-I enzymes are not available. Yet compared with the few studies that have measured protein expression or enzyme activities of certain phase-I genes in the mouse liver during development, such as CES (Zhu et al., 2009), PON (Cole et al., 2003), EPHX (Rouet et al., 1984), and FMO (Cherrington et al., 1998), our mRNA expression patterns were fairly indicative of the protein or enzyme activity levels. But whether the mRNA expression can be extrapolated to the protein expression and enzymatic activities of the phase-I enzymes remains to be validated.
For future studies, it may be informative to apply certain pediatric drugs to mice of different developmental stages and to check the pharmacokinetics of these drugs to see if the metabolism patterns can be predicted by the current gene expression patterns. Technological breakthroughs in proteomics and metabolomics are essential to the study of ontogeny of phase-I enzymes on the protein contents and metabolite levels in the future.
In summary, our study provides the first knowledge regarding the true quantification of the mRNA ontogenic patterns of all major known non-P450 phase-I enzymes during mouse liver development. This knowledge will serve as a foundation for further understanding the regulation of gene expression and physiologic function of these enzymes in the liver during development and aid in a better understanding of the kinetics of xenobiotic metabolism during perinatal and postnatal maturation.
Supplementary Material
Acknowledgments
The authors thank Drs. Helen Renaud and Dan Li for help on sample collection, and Clark Bloomer from the KUMC genomic sequencing facilities for his technical assistance on A-Seq.
Abbreviations
- ADH or Adh
alcohol dehydrogenase
- AKR or Akr
aldo-keto reductase
- ALDH or Aldh
aldehyde dehydrogenase
- AOX or Aox
aldehyde oxidase
- CES or Ces
carboxylesterase
- DPYD or Dpyd
dihydropyrimidine dehydrogenase
- EPHX or Ephx
epoxide hydrolase
- FMO or Fmo
flavin monooxygenase
- FPKM
fragments per kilobase of exon per million reads mapped
- NQO or Nqo
quinone oxidoreductase
- P450
cytochrome P450
- PON or Pon
paraoxonase
- POR or Por
NADPH-cytochrome P450 oxidoreductase
- RNA-Seq
RNA sequencing
- XDH or Xdh
xanthine oxidoreductase
Authorship Contributions
Participated in research design: Zhong, Klaassen, Lu, Gunewardena, Peng, Cui, Yoo.
Conducted experiments: Peng, Cui.
Performed data analysis: Peng, Cui, Gunewardena, Yoo, Zhong.
Wrote or contributed to the writing of the manuscript: Zhong, Klaassen, Lu, Peng, Gunewardena, Yoo, Cui.
Footnotes
This study was supported by the National Institutes of Health National Institute for Environmental Health Sciences [Grant R01ES-019487] (to X.B.Z., C.D.K., and H.L.); the National Institutes of Health National Institute of General Medical Sciences [Grant R01GM-087376] (to X.B.Z.); and the National Institutes of Health National Institute for Environmental Health Sciences [Grant R01ES-009649] (to C.D.K.).
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Alnouti Y, Klaassen CD. (2008) Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101:51–64 [DOI] [PubMed] [Google Scholar]
- Bendotti C, Prosperini E, Kurosaki M, Garattini E, Terao M. (1997) Selective localization of mouse aldehyde oxidase mRNA in the choroid plexus and motor neurons. Neuroreport 8:2343–2349 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300 [Google Scholar]
- Blake MJ, Castro L, Leeder JS, Kearns GL. (2005) Ontogeny of drug metabolizing enzymes in the neonate. Semin Fetal Neonatal Med 10:123–138 [DOI] [PubMed] [Google Scholar]
- Cherrington NJ, Cao Y, Cherrington JW, Rose RL, Hodgson E. (1998) Physiological factors affecting protein expression of flavin-containing monooxygenases 1, 3 and 5. Xenobiotica 28:673–682 [DOI] [PubMed] [Google Scholar]
- Cole TB, Jampsa RL, Walter BJ, Arndt TL, Richter RJ, Shih DM, Tward A, Lusis AJ, Jack RM, Costa LG, et al. (2003) Expression of human paraoxonase (PON1) during development. Pharmacogenetics 13:357–364 [DOI] [PubMed] [Google Scholar]
- Croom EL, Stevens JC, Hines RN, Wallace AD, Hodgson E. (2009) Human hepatic CYP2B6 developmental expression: the impact of age and genotype. Biochem Pharmacol 78:184–190 [DOI] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Yoo B, Liu J, Renaud HJ, Lu H, Zhong XB, Klaassen CD. (2012a) RNA-Seq reveals different mRNA abundance of transporters and their alternative transcript isoforms during liver development. Toxicol Sci 127:592–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Renaud HJ, Klaassen CD. (2012b) Ontogeny of novel cytochrome P450 gene isoforms during postnatal liver maturation in mice. Drug Metab Dispos 40:1226–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Kole L, Wang LH, Barrios R, Moorthy B, Jaiswal AK. (2006) BALT development and augmentation of hyperoxic lung injury in mice deficient in NQO1 and NQO2. Free Radic Biol Med 40:1843–1856 [DOI] [PubMed] [Google Scholar]
- Duester G, Farrés J, Felder MR, Holmes RS, Höög JO, Parés X, Plapp BV, Yin SJ, Jörnvall H. (1999) Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem Pharmacol 58:389–395 [DOI] [PubMed] [Google Scholar]
- Falls JG, Blake BL, Cao Y, Levi PE, Hodgson E. (1995) Gender differences in hepatic expression of flavin-containing monooxygenase isoforms (FMO1, FMO3, and FMO5) in mice. J Biochem Toxicol 10:171–177 [DOI] [PubMed] [Google Scholar]
- Falls JG, Ryu DY, Cao Y, Levi PE, Hodgson E. (1997) Regulation of mouse liver flavin-containing monooxygenases 1 and 3 by sex steroids. Arch Biochem Biophys 342:212–223 [DOI] [PubMed] [Google Scholar]
- Hart SN, Cui Y, Klaassen CD, Zhong XB. (2009) Three patterns of cytochrome P450 gene expression during liver maturation in mice. Drug Metab Dispos 37:116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB. (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet Genomics 18:11–24 [DOI] [PubMed] [Google Scholar]
- Hines RN. (2006) Developmental and tissue-specific expression of human flavin-containing monooxygenases 1 and 3. Expert Opin Drug Metab Toxicol 2:41–49 [DOI] [PubMed] [Google Scholar]
- Hines RN. (2007) Ontogeny of human hepatic cytochromes P450. J Biochem Mol Toxicol 21:169–175 [DOI] [PubMed] [Google Scholar]
- Hines RN. (2008) The ontogeny of drug metabolism enzymes and implications for adverse drug events. Pharmacol Ther 118:250–267 [DOI] [PubMed] [Google Scholar]
- Hines RN. (2013) Developmental expression of drug metabolizing enzymes: impact on disposition in neonates and young children. Int J Pharm 452:3–7 [DOI] [PubMed] [Google Scholar]
- Hines RN, McCarver DG. (2002) The ontogeny of human drug-metabolizing enzymes: phase I oxidative enzymes. J Pharmacol Exp Ther 300:355–360 [DOI] [PubMed] [Google Scholar]
- Holmes RS, Wright MW, Laulederkind SJ, Cox LA, Hosokawa M, Imai T, Ishibashi S, Lehner R, Miyazaki M, Perkins EJ, et al. (2010) Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm Genome 21:427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M, Furihata T, Yaginuma Y, Yamamoto N, Koyano N, Fujii A, Nagahara Y, Satoh T, Chiba K. (2007) Genomic structure and transcriptional regulation of the rat, mouse, and human carboxylesterase genes. Drug Metab Rev 39:1–15 [DOI] [PubMed] [Google Scholar]
- Hrycay EG, Bandiera SM. (2009) Expression, function and regulation of mouse cytochrome P450 enzymes: comparison with human P450 enzymes. Curr Drug Metab 10:1151–1183 [DOI] [PubMed] [Google Scholar]
- Huen K, Harley K, Brooks J, Hubbard A, Bradman A, Eskenazi B, Holland N. (2009) Developmental changes in PON1 enzyme activity in young children and effects of PON1 polymorphisms. Environ Health Perspect 117:1632–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmohamed A, Hernandez D, Phillips IR, Shephard EA. (2004) Cell-, tissue-, sex- and developmental stage-specific expression of mouse flavin-containing monooxygenases (Fmos). Biochem Pharmacol 68:73–83 [DOI] [PubMed] [Google Scholar]
- Jin Y, Penning TM. (2007) Aldo-keto reductases and bioactivation/detoxication. Annu Rev Pharmacol Toxicol 47:263–292 [DOI] [PubMed] [Google Scholar]
- Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE. (2003) Developmental pharmacology—drug disposition, action, and therapy in infants and children. N Engl J Med 349:1157–1167 [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN. (2004) Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308:965–974 [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Simpson P, Yeung CK, Rettie AE, Hines RN. (2002) Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr Res 51:236–243 [DOI] [PubMed] [Google Scholar]
- Li WF, Matthews C, Disteche CM, Costa LG, Furlong CE. (1997) Paraoxonase (PON1) gene in mice: sequencing, chromosomal localization and developmental expression. Pharmacogenetics 7:137–144 [DOI] [PubMed] [Google Scholar]
- Long DJ, 2nd, Jaiswal AK. (2000) Mouse NRH:quinone oxidoreductase (NQO2): cloning of cDNA and gene- and tissue-specific expression. Gene 252:107–117 [DOI] [PubMed] [Google Scholar]
- Lu H, Cui JY, Gunewardena S, Yoo B, Zhong XB, Klaassen CD. (2012) Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing. Epigenetics 7:914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gunewardena S, Cui JY, Yoo B, Zhong XB, Klaassen CD. (2013) RNA-sequencing quantification of hepatic ontogeny and tissue distribution of mRNAs of phase II enzymes in mice. Drug Metab Dispos 41:844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, Oliver B. (2011) Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks TJ, Jones DC. (2002) The metabolism and toxicity of quinones, quinonimines, quinone methides, and quinone-thioethers. Curr Drug Metab 3:425–438 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- Muruganandan S, Sinal CJ. (2008) Mice as clinically relevant models for the study of cytochrome P450-dependent metabolism. Clin Pharmacol Ther 83:818–828 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson A, Ogilvie BW. (2008) Biotransformation of xenobiotics, in Casarett & Doull's Toxicology: The Basic Science of Poisons (Klaassen CD, ed) pp 161–304, McGraw-Hill, New York [Google Scholar]
- Peng L, Yoo B, Gunewardena SS, Lu H, Klaassen CD, Zhong XB. (2012) RNA sequencing reveals dynamic changes of mRNA abundance of cytochromes P450 and their alternative transcripts during mouse liver development. Drug Metab Dispos 40:1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lickteig AJ, Klaassen CD. (2013) Tissue distribution, ontogeny, and chemical induction of aldo-keto reductases in mice. Drug Metab Dispos 41:1480–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Dansette P, Frayssinet C. (1984) Ontogeny of benzo(a)pyrene hydroxylase, epoxide hydrolase and glutathione-S transferase in the brain, lung and liver of C57Bl/6 mice. Dev Pharmacol Ther 7:245–258 [DOI] [PubMed] [Google Scholar]
- Rowell M, Zlotkin S. (1997) The ethical boundaries of drug research in pediatrics. Pediatr Clin North Am 44:27–40 [DOI] [PubMed] [Google Scholar]
- Smith M, Hopkinson DA, Harris H. (1971) Developmental changes and polymorphism in human alcohol dehydrogenase. Ann Hum Genet 34:251–271 [DOI] [PubMed] [Google Scholar]
- Smolen TN, Smolen A, van de Kamp JL. (1990) Developmental profile of hepatic alcohol and aldehyde dehydrogenase activities in long-sleep and short-sleep mice. Alcohol 7:69–74 [DOI] [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ. (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307:573–582 [DOI] [PubMed] [Google Scholar]
- Stevens JC, Marsh SA, Zaya MJ, Regina KJ, Divakaran K, Le M, Hines RN. (2008) Developmental changes in human liver CYP2D6 expression. Drug Metab Dispos 36:1587–1593 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Pearce RE, Wang X, Gaedigk R, Wan YJ, Yan B. (2009) Human carboxylesterases HCE1 and HCE2: ontogenic expression, inter-individual variability and differential hydrolysis of oseltamivir, aspirin, deltamethrin and permethrin. Biochem Pharmacol 77:238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HJ, Appel DI, Jiang Y, Markowitz JS. (2009) Age- and sex-related expression and activity of carboxylesterase 1 and 2 in mouse and human liver. Drug Metab Dispos 37:1819–1825 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.