Abstract
Emiliania huxleyi, a key player in the global carbon cycle is one of the best studied coccolithophores with respect to biogeochemical cycles, climatology, and host-virus interactions. Strains of E. huxleyi show phenotypic plasticity regarding growth behaviour, light-response, calcification, acidification, and virus susceptibility. This phenomenon is likely a consequence of genomic differences, or transcriptomic responses, to environmental conditions or threats such as viral infections. We used an E. huxleyi genome microarray based on the sequenced strain CCMP1516 (reference strain) to perform comparative genomic hybridizations (CGH) of 16 E. huxleyi strains of different geographic origin. We investigated the genomic diversity and plasticity and focused on the identification of genes related to virus susceptibility and coccolith production (calcification). Among the tested 31940 gene models a core genome of 14628 genes was identified by hybridization among 16 E. huxleyi strains. 224 probes were characterized as specific for the reference strain CCMP1516. Compared to the sequenced E. huxleyi strain CCMP1516 variation in gene content of up to 30 percent among strains was observed. Comparison of core and non-core transcripts sets in terms of annotated functions reveals a broad, almost equal functional coverage over all KOG-categories of both transcript sets within the whole annotated genome. Within the variable (non-core) genome we identified genes associated with virus susceptibility and calcification. Genes associated with virus susceptibility include a Bax inhibitor-1 protein, three LRR receptor-like protein kinases, and mitogen-activated protein kinase. Our list of transcripts associated with coccolith production will stimulate further research, e.g. by genetic manipulation. In particular, the V-type proton ATPase 16 kDa proteolipid subunit is proposed to be a plausible target gene for further calcification studies.
Introduction
The prolific coccolithophore Emiliania huxleyi is distributed from sub-polar to tropical latitudes [1], [2] and often forms immense coastal and open ocean blooms that can cover more than 50.000 km2 [3], [4], [5]. These blooms can be detected via satellite imagery due to the reflection of the calcite platelets, the coccoliths [6], [7]. This makes E. huxleyi an important component within the global biogeochemical cycles of carbon and sulphur and one of the most important species on earth with respect to sediment formation and climate [8], [9]. Therefore, it is a key species for current studies on global biogeochemical cycles [10]. It is also of interest to scientists from fields as diverse as geology, biogeography, paleoclimatology, ecophysiology, material science, and medicine [11]. Whereas bloom formation is mainly stimulated by abiotic factors, bloom control and termination is often influenced by viral infection [12], [13]. A range of viruses specific to E. huxleyi (EhVs) have been isolated [14], [15] and were further analyzed for their phylogeny [16], [17], genome structure of Emiliania huxleyi virus 86 (EhV-86) [18], [19], [20], and ecological succession in mesocosm experiments [21], [22] and in nature [23]. Hence, it is one of the best studied eukaryotic phytoplankton host-virus systems to date [24], [25]. Previous studies have reported different genome sizes among morphotypes of E. huxleyi from different geographical regions via restriction fragment length polymorphism (RFLP) analysis [26], DNA microsatellites, [27], cox1b-atp4 genes [28], and tufa [29]. Results indicate the presence of different ecotypes of E. huxleyi with differences in genome organization as a response to environmental conditions or potential threats, such as viral infections. Furthermore, results show the lack of variation in coding (18S/16S) and non-coding (ITS/Rubisco) regions of E. huxleyi and its closest relative Gephyrocapsa oceanica, from which E. huxleyi separated about 270,000 years ago [30].
A genotype can develop multiple phenotypes depending on environmental conditions [31]. This so-called phenotypic plasticity is widespread in nature and can alter numerous interactions between organisms and their biotic and abiotic environments [32]. Furthermore, examples for a connection between genetic variation and virus susceptibility have been demonstrated [16], [33]. It was found that virus resistant strains of E. huxleyi display a higher dimethylsulfoniopropionate lyase (DMSP-lyase) activity than strains that are susceptible to virus infection. One reason for the different enzyme activities could be variations in expression or copy numbers of genes coding for DMSP lyase. Furthermore, Bidle and Kwityn [33] showed a connection between caspase activity and virus susceptibility. Resistant E. huxleyi strains were characterized by low caspase activity while sensitive strains had elevated caspase activity.A microbial genome can be partitioned into core and variable parts, both parts together making up the pan-genome [34]. The core genome is composed of genes which are present in all strains of a species and typically comprise genes necessary for basic metabolism [35], [36]. The variable genome consists of a set of genes which are detected in only some strains, and enable these strains to thrive in particular niches [36], [37], [38].
Over the last decade, comparative genomic hybridization (CGH) has been extensively utilized to elucidate genetic diversity between the genomes of closely related taxa, such as species and strains. CGH was mainly applied in bacterial systems, including Helicobacter pylori, Campylobacter jejuni, Francisella tularensis, and Escherichia coli among others [39], [40], [41], [42] but also in a eukaryote (yeast [39]). Results indicated that polymorphism in gene content is not uncommon, suggesting genetic adaptations to different ecological niches [39], [40], [41].
Here, we provide insights into the genetic diversity and genome evolution of a key player in marine phytoplankton, E. huxleyi, by CGH of 16 strains. We used a whole genome microarray comprising unique probes for predicted gene models of the sequenced strain E. huxleyi CCMP1516 (reference strain). This analysis gives information about a shared pool of genes (core genome) as well as about genes specific for the sequenced reference strain CCMP1516 (genome project conducted by JGI and lead by Betsy Read, http://genome.jgi-psf.org/Emihu1/Emihu1.home.html, [42]). Within the variable genome, we were able to identify candidate genes possibly involved in virus susceptibility and calcification.
Results and Discussion
Core Genome and Genetic Diversity of E. huxleyi
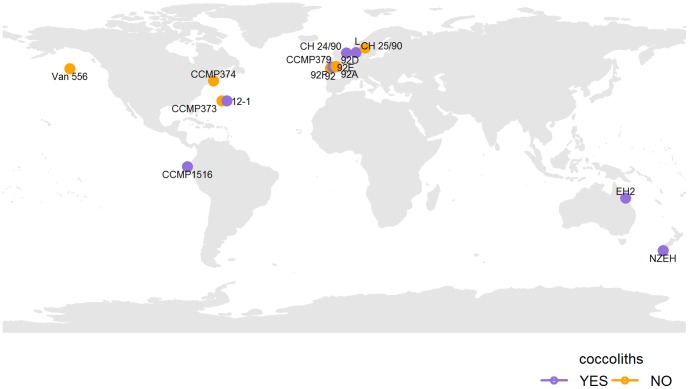
Genomic DNAs of 16 E. huxleyi strains from different geographic regions (Figure 1) were compared in order to identify genomic differences in terms of plasticity and possible relation to virus-susceptibility and calcification. Comparative genomic hybridization (CGH) was used to characterize 15 strains with respect to gene content similarities by using the sequenced virus susceptible and calcifying E. huxleyi strain CCMP1516 as the reference.
Figure 1. Isolation sites of the 16 E. huxleyi strains.
World map depicting the isolation sites of the 16 E. huxleyi strains and wether they possess coccoliths (blue) or not (orange).
Hybridization intensities were analysed statistically to determine either the presence, expressed as relative copy number of each gene transcript, or its absence or modification, indicated by the absence of cross-hybridisation. Gephyrocapsa oceanica and Isochrysis galbana, phylogenetically closely related taxa, were used as outgroup.
To check for possible unspecific probes, ANOVA was used (p<0.01). The obtained 31940 probes, representing gene models as identified in the genome by JGI, were classified as present or modified/absent genes by using the software GACK in its standard configuration [43]. By looking at the intersection of all genes showing 95% estimated probability of presence, we identified 14628 core genes in all 16 investigated strains (Figure 2, Table S1). As only slight modifications within a gene may result in absence of a hybridisation signal we regard this as a minimal number of core genes. Likely more genes belong to the core genome but this could only be clarified by sequencing of these strains.
Figure 2. Distribution of KOG annotations and comparison of the core and non-core (variable) genome of (16 strains of) E. huxleyi.
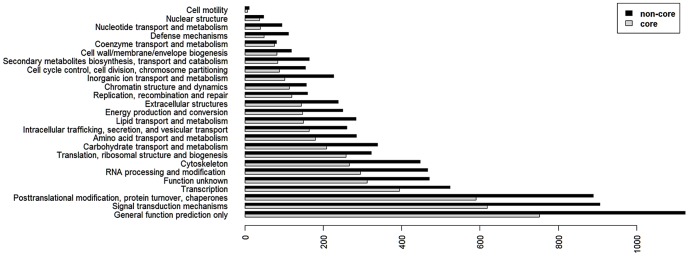
KOG annotations of the E. huxleyi genome strain CCMP1516 (JGI) were used to identify functional classes of the core and non-core genome.
We compared functional coverages of core and non-core genomes, respectively, using KOG statistics (Figure 2). Distribution of functions for both gene sets is quite similar, i.e. no KOG category is over-represented in either fraction. Surprisingly we found that genes of the variable genome (non-core) represented more KOG functions in all categories than core genes. This coincides with a higher number of annotated genes among the non-core genes versus the core genes. We expected this to be the opposite as the “core-genome” should contains house keeping genes necessary in all strains, and consequently genes better represented in the database and more easy to identify. For the strain specific genes we expected a high content of genes necessary only under particular conditions and as such less well known. The difference is not accounted for by different numbers of KOG hits per gene for core versus non-core genes. One possible exception is the category of coenzyme transport and metabolism where both group produced an almost equal number of KOG hits. The highest relative difference between the variable genome and the core genome is found in the category of defence mechanisms, implying a large variability of such genes among strains. Defence in general is “costly” and specific defence less often required than house keeping genes. Therefore, such genes are probably deleted more quickly from the core than others. Core genes among all analysed isolates are probably necessary for general survival in diverse environments where the species is found. The remaining specific set may reflect for the ‘niche’ adaptations of the different isolates, and for specific defence mechanisms.
Besides overlapping genes (i.e. those found in more than one strain) we identified 224 strain-specific probes representing 224 gene models for the reference genome from strain CCMP1516 (Table S2). This number could be an over-estimation as an oligo might not bind to an existing homologous gene only because 1–2 bases mismatch. Functions could be assigned to 74 genes as identified by the probes. According to KEGG classification (http://www.genome.jp/kegg/), these reflect proteins involved in metabolism, folding, sorting and degradation, replication and repair, cellular processes, transport and environmental information processing among the reference strain specific gene set. The remaining 150 genes had no significant similarity to sequences in the public sequence databases or were of unknown function.
A bootstrapped neighbour-joining consensus dendrogram of the 16 E. huxleyi strains and the out-group G. oceanica based on the CGH data is depicted in Figure 3. As expected, the out-group is separated from all E. huxleyi strains with 100% bootstrap (BT) support. Interestingly, the E. huxleyi strain EH2 also is separated from other E. huxleyi strains by BT support. The remaining 15 E. huxleyi strains grouped into two main clusters. The strains Van556, 92D, 92F and CCMP373 showed the highest degree of divergence to all other E. huxleyi strains and clustered into one of the main groups. The second main cluster can be subdivided into sub-groups on different scales. Strain 12-1 was most similar to the reference strain (89.1% identity in gene contents) and clustered into one of the five sub-groups.
Figure 3. Bootstrapped neighbour-joining consensus dendogram of the 16 E. huxleyi strains and G. oceanica.
Dendrogram with bootstrap values obtained from 1000 replicates of 16 E. huxleyi strains and G. oceanica as outgroup. Distances were computed from a matrix of ones (gene present with probability >95%) and zeros (rest).
The analysis of hybridization patterns showed that the gene dissimilarity between strains ranged from 10,9% to 30,1% (3015 genes differed, compared to 31468 positive reference hybridization signals). The variation exhibited in these genes is probably associated with (partial) gene deletion, nucleotide sequence divergence or gene duplication. When comparisons were made between the outgroup G. oceanica and the reference, the genetic distance increased only slightly up to 30,4% (9232 genes, summarized in Table S1).
So far, genome comparative studies of microorganisms using CGH were restricted to bacteria such as E. coli [44], Bacillus subtilis [45], and Streptococcus [36] and to yeast [39]. Fewer studies exist of genome comparisons in marine environments (Vibrio cholerae [46], Ectocarpus siliculosus [47]). Genome comparisons of strains for a given species in these studies showed differential hybridization between 0.17 and 16.7 per cent of the gene transcripts.
In contrast, our results revealed between 10.9 and 30.1% gene variation within the species and up to 30.4% compared to the genus G. oceanica, their last common ancestor. As E. huxleyi has evolved from G. oceanica only 268.000 years ago [30] and became dominant around 70.000 years ago, this gene diversity indicates that E. huxleyi is undergoing a more rapid evolutionary radiation than other species and is better described as a species-morpho-complex than as a single species. This could also explain its enormous phenotypic plasticity described in the literature [48], [49], [50].
The highest variability amongst the strains was observed for strain 1516 against strain EH2 (30.1%), almost as much as between the genera Gephyrocapsa and Emiliania (30.4%). Strain EH2 is virus susceptible and can produce coccoliths as the reference strain (CCMP1516). The reasons for the high genomic deviation from the reference could be many fold, including its different geographic origin, ecological niche, and predation (grazing, virus infection etc.) The reference strain was isolated near the coast of Ecuador whereas EH2 was obtained from the Great Barrier Reef (Table 1, Figure 1). Clearly differences in the ecological and life cycle strategies of E. huxleyi strains (e.g. bloom dynamics) could also cause these gene differences [51]. Results of the recent study of Cook et al (2011) using tufA as a molecular marker support our study showing two distinct clades (Bayesian analysis): one of the southern hemisphere and one showing G. oceanica and E. huxleyi together suggesting inter-breeding between the two genera (Linda Medlin, personal comment) which could be hypothesized for our case.
Table 1. Isolation sites and date of the 16 E. huxleyi strains and whether they possess coccoliths (Y = yes) or not (N).
| Emiliania huxleyi strain | Coccoliths | Collection site | Isolation date |
| 92 (English Channel) | N | 49°19N 07°26W | 1950 |
| 92A (English Channel) | N | 50°10N 4°15W 1mile west of Eddystone | 1957 |
| 92D (English Channel) | Y | 50°02N 4°22W | 1975 |
| 92E (English Channel) | Y | 49°52N 06°12W 2m depth | 1992 |
| 92F (English Channel) | Y | 49°52N 06°12W 2m depth | 1992 |
| CCMP379 ( = 92A, according to CCMP) | N | 50°10N 4°154W 1mile west of Eddystone | 1992 |
| CCMP374 (Gulf of Maine) | N | 42°30N 69°W Gulf of Maine (5 meters) | 1989 |
| CCMP373 (Sargasso Sea) | N | 32°10N 64°30W | 1960 |
| 12-1 ( = CCMP371) (Sargasso Sea) | Y | 32°00N 62°00W (50 meter depth) | 1987 |
| CCMP1516 (South Pacific) | Y | 2°40S 82°43W (surface) | 1991 |
| Van 556 (North Pacific) | N | 49°05N 144°40W | 1984 |
| CH 24/90 (North Atlantic) | Y | 57°20N 01°09E | 1990 |
| CH 25/90 (North Atlantic) | Y | 57°26N 6°13E | 1990 |
| L (Oslo Fjord) | N | 60°N 11°E | 1959/68/80 |
| NZEH (South Pacific) | Y | Big Glory Bay, NZ | 1992 |
| EH2 (South Pacific) | Y | Great Barrier Reef | 1990 |
Other reasons for the genetic distance between these two strains include different genome size. Read et al. [42] report on genome sizes ranging from 99 to 133 Mb (haploid) for different strains of E. huxleyi (Supplementary material to [42]: Table 6). They also estimate numbers of missing genes, compared to the reference. This documents that genome size is very likely to show a signature in genetic distance in terms of gene function repertoire.
As highlighted above, E. huxleyi is an important calcifier in the ocean, and at the same time non-calcifying strains exist. Its immense blooms are often terminated by viruses. Therefore, within the variable genome, we searched for specific genes which may be involved in virus susceptibility and coccolith production. The 15 compared E. huxleyi strains were divided into the following groups: virus susceptible vs. virus resistant (Table 2) and possession of coccoliths vs. no possession of coccoliths (Table 1), genes specific for either group were identified.
Table 2. Virus susceptibility and resistance of E. huxleyi strains derived from Allen et al. [18].
| Emiliania huxleyi virus (EhV) strain | |||||||||
| Emiliania huxleyi host strain | 86 | 84 | 88 | 163 | 201 | 205 | 202 | 208 | 207 |
| 92 (English Channel) | – | – | – | – | – | – | – | – | – |
| 92A (English Channel) | – | – | – | – | – | – | – | – | – |
| 92D (English Channel) | – | – | – | – | – | – | – | – | – |
| 92E (English Channel) | – | – | – | – | – | – | – | – | – |
| 92F (English Channel) | + | + | + | + | + | + | + | + | + |
| CCMP379 (Unknown) | – | – | – | – | – | – | – | – | – |
| CCMP374 (Gulf of Maine) | + | + | + | + | + | + | + | + | + |
| CCMP373 (Sargasso Sea) | – | – | – | – | – | – | – | – | – |
| 12-1 (Sargasso Sea) | – | – | + | + | + | + | + | + | + |
| CCMP1516 (South Pacific) | + | + | + | – | + | + | + | + | + |
| Van 556 (North Pacific) | – | – | – | – | – | – | – | – | – |
| CH 24/90 (North Atlantic) | – | – | – | – | – | – | – | + | – |
| CH 25/90 (North Atlantic) | – | – | – | – | – | – | – | – | – |
| L (Oslo Fjord) | + | + | + | + | + | – | + | + | + |
| NZEH (South Pacific) | – | – | – | – | – | – | – | – | – |
| EH2 (South Pacific) | + | + | + | – | + | + | + | + | + |
+, culture lysis; –, no evidence of lysis after 14 days of viral infection cultures were not lysed and considered to be non-susceptible to the virus strain [18].
Genes involved in virus susceptibility
The E. huxleyi EhV system has emerged as an excellent model to understand the biochemical “arms race” in the oceans (summarized by [25]). It involves infection-induced ROS production [52] and subsequent caspase-induced programmed cell death (PCD) controlled by the virus [24]. E. huxleyi possibly counter-acts by dimethylsulfoniopropionate (DMS) and acrylic acid release which can scavenge oxygen radicals [53]. Glycosphingolipids (GSL) seem to play an important role in the process in that the virus controls synthesis of viral myristoyl-based GSL, different from the host palmitoyl-based SPL, which in turn control host metabolism and EhV production; viral GSL can even induce PCD [23]. Viral GSL are likely also present in the viral envelope which could be used for a fusion mechanism to enter the host cell, similar to the situation in many animal viruses.
The analysis of gene contents correlated to virus susceptibility yielded 94 genes (Table S3) linked to this trait. More than half of these (54) showed no similarity to any sequences in public sequence databases or were of unknown function. According to KEGG (http://www.genome.jp/kegg/) classification we found proteins involved in metabolism (12), transcription and translation (7), transport (5), cellular processes and signalling (4), carbohydrate and lipid metabolism (4), genetic information processing (2), signal transduction (4), and folding, sorting, and degradation (2). Within these groups, a Bax inhibitor-1 like protein (BI-1) and ten different protein kinases, including three leucine-rich repeat (LRR) receptor-like protein kinases and one mitogen-activated protein kinase (MAPK) were identified and might be of particular interest.
The identification of BI-1 (Figure S1) is rather plausible, as this protein might serve as a cell death regulator protein that inhibits Bax induced cell death as has been shown by Hückelhoven [54], [55]. BI-1 is a conserved protein found in all organisms including plants and fungi. Even viruses code for proteins with a domain architecture similar to BI-1 indicating that BI-1 has been corrupted during evolution by pathogens to reprogram a living host cell [54]. Eukaryotic cells permanently have to cope with environmental cues and to integrate developmental signals. Cell survival or death is the possible outcome [56]. Likely most E. huxleyi blooms are terminated by viral infection via virus-induced apoptosis, as a form of (PCD) [24]. Indeed, as Bidle et al (2007) showed the virus rather makes use of and induces PCD in E. huxleyi for its benefit and proliferation rather than the host inducing PCD to terminate virus proliferation. Recently, it has been shown that BI-1 is required for full susceptibility of barley to powdery mildew, suppressing the defense response of the host [57]. Accordingly BI-1 could therefore well be a susceptibility factor of E. huxleyi strains and involved in virus-induced cells apoptosis.
LRR motifs are found in many plant and animal proteins and are usually involved in protein-protein interactions and ligand binding [58]. Receptor-ligand interactions are very sensitive to point mutations of the DNA-sequence, which can lead to viral resistance/or can allow pathogens to avoid recognition [59]. LRRs are also found in the human Interleukin-1 and Toll-like receptors, which participate in the regulation of immune responses [60], [61]. The identification of MAPK is consistent with recent observations of Marchant et al. [60], which have shown that MAPK is a determinant of virus infection even knowing that the MAPK pathway is involved in many substantial regulative processes (see [62]). It has furthermore been shown, that the vaccinia virus replication requires the MAPK pathway [63].
Animal dsDNA enveloped viruses like herpes simplex virus (HSV) and vaccinia virus enter their host either via clathrin-mediated endocytosis or by fusion with the plasma membrane [64]. Both processes involve the fusion of the virion envelope with a cell membrane, either the plasma membrane or a vesicle membrane. In general, the first step of virus infection involves attachment of virus particles to host-specific cell surface receptors [65], [66] prior to entering the host cell. Once inside the host cell, viruses utilize the host machinery in order to enhance the efficiency of its replication process. This is of particular importance for EhV86 which encodes hundreds of genes as compared to only a handful in ssRNA animal viruses. Consequently, the expression of a receptor on the outer surface of the host is a major determinant of the route of entry of the virus into the host and of the patterns of virus spread and pathogenesis in the host [65]. Viruses have evolved to exploit these receptors to gain entry into cells. As each virus is looking for only one specific receptor that fits its attachment protein, the host receptor will, in part, determine the susceptibility of different hosts to the same virus. Previous studies have demonstrated that the lack of receptor expression restricts virus entry [67], [68], [69] and that protein kinases influence virus entry and infectivity [70], [71], suggesting that the LRR receptor-like protein kinase as well as MAPK and serine/threonine protein kinase could be involved in virus susceptibility, infection or induced defence mechanisms. We have identified three leucine-rich repeat (LRR) receptor-like protein kinases and MAPK among the genes present only in virus-susceptible E. huxleyi strains. These genes suggest the route of entry of the virus into the host for the virus-susceptible group and may be similar to above described animal virus infection pathways. It also shows that the lack of them due to possible point mutations tends to lead to resistance to such viruses, possibly by avoidance of recognition.
Our analysis is limited by the availability of only one sequenced E. huxleyi genome which is from a virus susceptible strain. Therefore we can not identify possible resistance genes in the resistant strains due to the limitation of our microarray design. We therefore regard it as very likely that virus susceptibility of E. huxleyi may be dependent on the expression of other genes or factors for viral entry. However, differences in copy numbers or point-mutations of coding sequences of the identified receptor-like protein kinases could be an indication for differences in virus susceptibility, making them suitable targets for further studies.
Genes involved in calcification
A total of only 11 genes were identified as possibly associated with calcification (Table S4). Three of them showed no similarity to sequences in the public sequence databases or were of unknown function. We identified a kelch-like protein, one activator of 90 kDa heat shock protein ATPase homolog, and one uncharacterized oxidoreductase. Moreover, we identified one arylsulfatase which binds one Ca2+ ion per subunit suggesting a potential role in calcification. A long-chain-fatty-acid-CoA ligase was identified, which is involved in the lipid metabolism. The identification of two V-type proton ATPase 16 kDa proteolipid subunits is of high relevance as these are involved in transport and were also found in the calcifying coccolithophore Pleurochrysis carterae [72]. Studies of calcification in the coccolithophore P. carterae have previously localized a vacuolar H+-ATPase (V-ATPase) in the coccolith vesicle mediating Ca2 +/H+ exchange [72], [73], [74]. Moreover, recent transcriptome analyses have shown an over-expression of numerous V-ATPase subunits in diploid calcifying cells [75], [76] indicating putative roles in the calcification process as reviewed by Mackinder et al. [77].
Coccolith formation and structure has been extensively studied in the species Emiliania huxleyi [77], [78], Pleurochrysis carterae [79] and Coccolithus pelagicus [80]. Formation of coccoliths takes place in a Golgi-derived intracellular vesicle [81], [82] before transport to the cell cortex and secretion to the cell surface in a single exocytotic extrusion event [80]. In Pleurochrysis coccolith precursors are mediated by acidic polysaccharides [81], [83]. However, despite the existence of several hypotheses (for an overview see Young [84] and Mackinder et al. [77]) and the discovery of novel genes possibly involved in calcification and coccolithogenesis by using EST approaches, suppression subtractive hybridization, long serial analysis of gene expression, microarrays for gene expression analysis and quantitative RT-PCR [11], [75], [76], [85], [86], [87], [88], [89], the details of the process of and the genes involved in coccolith formation in E. huxleyi are still unknown.
Genes potentially involved in calcification like carbonic anhydrase or the calcium-binding glycoprotein with a high glutamic acid, proline, and alanine content (GPA) [88], [90], [91] were not detected as calcifying factors in our study. This indicates that these two genes might be regulated at the transcript level or they fulfill cell-biological tasks in the non-calcifying life-cycle stage as well, as also indicated recently by Dassow et al. [75] and Rokitta et al. [76].
Emiliania huxelyi is known for its flexible responses in eco-physiological studies [49]. In particular, recent studies on carbonate chemistry changes showed strain-specific sensitivities to acidification of seawater [49], [50] which might be due to genetic variability described here. However, even with the same strain the diploid stage 1916 and the haploid 1917 exhibit different strategies and gene sets to acclimate to changing environmental conditions [89].
The genes possibly involved in virus susceptibility and calcification identified in this study provide targets for future studies on their expression, e.g. under virus attack, and for gene knock-out experiments.
Methods
Strains and culture conditions
Emiliania huxleyi strains (Table 1) and Gephyrocapsa oceanica were cultured in f/2 medium and Isochrysis galbana in K media at 15°C with a 16:8 light-dark cycle and 150 µE m−2 s−1. Strains EH2 and NZEH were treated with 1000 µg/mL Kanamycin because they were too sensitive against the antibiotic mixture. All other cultures were treated with a mixture of Ampicillin, Gentamycin, Streptomycin, Chloramphenicol and Ciprofloxacin (Table 3). Antibiotic treatment took place over 10–12 days. After 5–6 days cultures grown in 200 mL treated with antibiotics were transferred to 800 mL antibiotic treated f/2 media. Five to six days later cells were harvested on 1.2 µm RTTP ISOPORE filters Millipore. Cultures were checked against bacteria with acridine-orange staining. Only samples with no observed bacteria were used for analysis, although we cannot reduce a highly reduced bacterial background.
Table 3. Antibiotic treatment mixture.
| Antibiotic | Concentration in culture [mg/mL] |
| Ampicillin | 0.05 |
| Gentamycin | 0.003 |
| Streptomycin | 0.025 |
| Chloramphenicol | 0.001 |
| Ciprofloxacin | 0.010 |
Genomic DNA labelling
All steps were performed in technical triplicates in order to avoid methodological errors in the hybridisation patterns interpretation. Genomic DNA was isolated from the samples using Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, D) and then subjected to amplification according to Agilent’s protocol for oligonucleotide array-based CGH for genomic DNA (version 5.0, June 2007). Restriction digestion was performed with 200 ng of genomic DNA for 8 h at 37°C. Digested DNA from each test strain and species was labelled with Cy5-dUTP whereas E. huxleyi strain CCMP1516 was labelled with Cy3 as reference. Labelled DNA sample yields and dye incorporation efficiencies were assessed photometrical (Nanodrop ND-1000, PecLab). Specific activity (pmol dyes perµg genomic DNA) were calculated as [pmol perµL dye/µg perµL genomic DNA] from the results of photometry.
Microarray hybridizations
Labelled samples were then co-hybridized with DNA of the reference E. huxleyi strain CCMP1516 in triplicates to Agilent oligonucleotide-based 44k custom-made microarrays. One Array contained 37880 different transcripts derived from the E. huxleyi CCMP1516 genome project conducted by the U.S. department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) using the best gene model for each locus. Microarrays were designed with Agilent’s eArray online application tool version 5.0 (accession number of the array design at ArrayExpress: A-MEXP-1696; http://www.ebi.ac.uk/arrayexpress).
A self-versus-self hybridization was performed in triplicates for determining probe specificity, array reproducibility, and microarray feature uniformity. Hybridizations were done for 24 h with 20 rpm using a hybridization chamber (Agilent technologies). After hybridization, the microarrays were washed according to the manufacturer’s instructions (Wash with Stabilization and Drying Solution, Agilent Technologies).
Data acquisition and analysis
Microarrays were scanned using a G25655B Agilent microarray scanner with 100% photomultiplier tube (PMT) settings for both channels and 5 µm scan resolution. Signal intensities were detected and normalized by Feature Extraction software version 9.5 (Agilent Technologies) using the GE protocol and matrix. Spots which were not well above background in the self-self hybridization were removed before further analysis. Results were first analyzed using the MeV software package from TIGR [92]. An ANOVA test was performed for all groups with an alpha-level of 0.01 and a standard Bonferroni correction for multiple testing. The average intensity from the significant genes of the triplicates was used for further analysis. Probes which did not hybridize in the self-self hybridisation of the reference strain were excluded from the analysis. The software GACK was used to determine the cut-off on a strain-by-strain basis, accounting for variation in strain composition and hybridization quality [43]. GACK uses an estimated probability of presence (EPP) as an assignment scale. The hybridization against I. galbana was excluded from the following analyses due to problems in cut-off determination (visually misplaced Gaussian, data not shown). The criterion for the presence of genes in the non-reference strains hybridizations was EPP>95%. A consensus dendrogram (neighbour-joining) from the matrix of assumed presence of genes, including bootstrap analyses, was computed with the ape-package [93] in the data analysis software R [94].
Identification of genes regarding virus susceptibility and calcification
The reference strain (CCMP1516) and the two out-groups G. oceanica and I. galbana were excluded in this analysis. Strains were grouped according to their virus susceptibility (Table 2) and possession of coccoliths (Table 1). A lower limit for the estimated probability of “gene present” >95% (EPP>95%) was used to elucidate whether a lack of certain genes, copy number changes or sequence divergence between reference and tester strain may explain the different biological properties of virus susceptibility or possession of coccoliths. To identify genes involved in virus susceptibility and calcification, we filtered for combinations of present genes in the susceptible (or calcifying, respectively) group of species versus absent genes in the resistant (or non-calcifying, respectively) group. We relaxed the criterion of perfect matches to allow for one false negative or one false positive, respectively. The resulting genes were manually analysed using BLASTP [95], [96] similarity searches version 2.2.24+ against the NCBI non-redundant protein database and the SwissProt database and were compared with matches of Pfam families [97]. All similarity search programs were applied with default parameters. Original data files for all arrays were uploaded in MIAME format to ArrayExpress with accession number E-MEXP-2388 (http://www.ebi.ac.uk/arrayexpress).
Supporting Information
Multiple alignment of Bax Inhibitor 1-like protein (BI-1) using Clustal W in BioEdit. The protein sequence of E. huxleyi (ID 198434) was analysed using blastp [95], [96] similarity searches version 2.2.26 + against the SwissProt database in its standard configurations. The alignment was done with the four hits Q94A20 (Arabidopsis thaliana), Q49P94 (Vaccinia virus Lister), Q9DA39 (Mus musculus), and O7488 (Schizosaccharomyces pombe 972h) with ClustalW [98] in BioEdit. BLOSUM 62 was used as similarity Matrix.
(TIFF)
Table of pair wise overlaps of all E. huxleyi strains and G. oceanica in terms of common present genes (EPP>95%). Excel-file including strain name and number of overlaps between each strain, including the reference strain.
(XLSX)
Table of strain-specific genes for the reference strain E. huxleyi CCMP1516. Excel-file including array-, gene- and protein-ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, and the description and function of the identified genes.
(XLS)
Identified genes of E. huxleyi in respect to virus susceptibility. Excel-file including array, gene and protein- ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, description and function of the identified genes of the 16 E. huxleyi strains.
(XLS)
Identified genes of E. huxleyi related to the production of coccoliths. Excel-file including array, gene and protein- ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, description and function of the identified genes of the 16 E. huxleyi strains.
(XLS)
Acknowledgments
E. huxleyi genome sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the International E. huxleyi Genome Sequencing Consortium community. We would like to thank Betsy Read and the JGI for giving us the opportunity to start working with the sequences of E. huxleyi CCMP1516 before submission. William H. Wilson and Michael J. Allen are acknowledged for sharing E. huxleyi cultures.
Funding Statement
Financial support was partly provided by the PACES research program of the Alfred Wegener Institute, within the Helmholtz Foundation Initiative in Earth and Environment, and by Marine Genomics Europe (EU Contract n° 505403). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Balch WM, Holligan PM, Kilpatrick KA (1992) Calcification, photosynthesis and growth of the bloom-forming coccolithophore, Emiliania huxleyi . Continental Shelf Research 12: 1353–1374. [Google Scholar]
- 2. Brown CW, Yoder JA (1993) Blooms of Emiliania huxleyi (Prymnesiophyceae) in surface waters of the Nova Scotian Shelf and the Great Bank. Journal of Plankton Research 15: 1429–1438. [Google Scholar]
- 3.Winter A, Jordan RW, Roth PH (1994) Biogeography of living coccolithophores in ocean waters. In: Winter A, Siesser WG, editors. Coccolithophores. Cambridge: Cambridge University Press. pp. 161–177.
- 4. Holligan PM, Fernández E, Aiken J, Balch WM, Boyd P, et al. (1993) A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Global Biogeochemical Cycles 7: 879–900. [Google Scholar]
- 5. Sukhanova IN, Flint MV (1998) Anomalous blooming of coccolithophorids over the Eastern Bering Sea Shelf. Oceanology 38: 502–505. [Google Scholar]
- 6. Holligan PM, Viollier M, Harbour DS, Camus P, Champagne-Philippe M (1983) Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature 304: 339–342. [Google Scholar]
- 7. Balch WM, Holligan PM, Ackleson SG, Voss KJ (1991) Biological and optical properties of mesoscale coccolithophore blooms in the Gulf of Maine. Limnology and Oceanography 36: 629–643. [Google Scholar]
- 8.Malin G, Liss PS, Turner SM (1994) Dimethyl sulphide: production and atmospheric consequences. In: Green JC, Leadbeater BSC, editors. The Haptophyte Algae. Oxford: Clarendon Press. pp. 303–320.
- 9. Westbroek P, Brown CW, Bleijswijk J, Brownlee C, Brummer GJ, et al. (1993) A model system approach to biological climate forcing. The example of Emiliania huxleyi . Global and Planetary Change 8: 27–46. [Google Scholar]
- 10.Westbroek P, Hinte JE, Brummer G-J, Veldhuis M, Brownlee C, et al. (1994) Emiliania huxleyi as a key to biosphere-geosphere interactions. In: Green JC, Leadbeater BSC, editors. The Haptophyte Algae. Oxford: Clarendon Press. pp. 321–334.
- 11. Nguyen B, Bowers RM, Wahlund TM, Read BA (2005) Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Applied and Environmental Microbiology 71: 2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jacquet S, Heldal M, Iglesias-Rodriguez D, Larsen A, Wilson W, et al. (2002) Flow cytometric analysis of an Emiliana huxleyi bloom terminated by viral infection. Aquatic Microbial Ecology 27: 111–124. [Google Scholar]
- 13. Bratbak G, Egge JK, Heldal M (1993) Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Marine Ecology Progress Series 93: 39–48. [Google Scholar]
- 14. Castberg T, Thyrhaug R, Larsen A, Sandaa R-A, Heldal M, et al. (2002) Isolation and characterization of a virus that infects Emiliania huxleyi (Haptophyta). Journal of Phycology 38: 767–774. [Google Scholar]
- 15. Wilson WH, Tarran GA, Schroeder D, Cox M, Oke J, et al. (2002) Isolation of viruses responsible for the demise of an Emiliania huxleyi bloom in the English Channel. J Mar Biolog Assoc UK 82: 369–377. [Google Scholar]
- 16. Schroeder DC, Oke J, Malin G, Wilson WH (2002) Coccolithovirus (Phycodnaviridae): Characterisation of a new large dsDNA algal virus that infects Emiliana huxleyi . Archives of Virology 147: 1685–1698. [DOI] [PubMed] [Google Scholar]
- 17. Allen MJ, Schroeder DC, Wilson WH (2006) Preliminary characterisation of repeat families in the genome of EhV-86, a giant algal virus that infects the marine microalga Emiliania huxleyi . Archives of Virology 151: 525–535. [DOI] [PubMed] [Google Scholar]
- 18. Allen MJ, Martinez-Martinez J, Schroeder DC, Somerfield PJ, Wilson WH (2007) Use of microarrays to assess viral diversity: from genotype to phenotype. Environmental Microbiology 9: 971–982. [DOI] [PubMed] [Google Scholar]
- 19. Allen MJ, Schroeder DC, Holden MTG, Wilson WH (2006) Evolutionary History of the Coccolithoviridae. Molecular Biology and Evolution 23: 86–92. [DOI] [PubMed] [Google Scholar]
- 20. Wilson WH, Schroeder DC, Allen MJ, Holden MTG, Parkhill J, et al. (2005) Complete Genome Sequence and Lytic Phase Transcription Profile of a Coccolithovirus. Science 309: 1090–1092. [DOI] [PubMed] [Google Scholar]
- 21. Schroeder DC, Oke J, Hall M, Malin G, Wilson WH (2003) Virus succession observed during an Emiliania huxleyi bloom. Applied and Environmental Microbiology 69: 2484–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinéz JM, Schroeder DC, Larsen A, Bratbak G, Wilson WH (2007) Molecular Dynamics of Emiliania huxleyi and Cooccurring Viruses during Two Separate Mesocosm Studies. Applied and Environmental Microbiology 73: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, et al. (2009) Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Science 326: 861–865. [DOI] [PubMed] [Google Scholar]
- 24. Bidle KD, Haramaty L, Barcelos e Ramos J, Falkowski P (2007) Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi . Proceedings of the National Academy of Sciences 104: 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bidle KD, Vardi A (2011) A chemical arms race at sea mediates algal host-virus interactions. Current Opinion in Microbiology 14: 449–457. [DOI] [PubMed] [Google Scholar]
- 26. Medlin LK, Barker GLA, Campbell L, Green JC, Hayes PK, et al. (1996) Genetic characterisation of Emiliania huxleyi (Haptophyta). Journal of Marine Systems 9: 13–31. [Google Scholar]
- 27. Iglesias-Rodriguez MD, Saez AG, Groben R, Edwards KJ, Batley J, et al. (2002) Polymorphic microsatellite loci in global populations of the marine coccolithophorid Emiliania huxleyi . Molecular Ecology Notes 2: 495–497. [Google Scholar]
- 28. Hagino K, Bendif EM, Young JR, Kogame K, Probert I, et al. (2011) New evidence for morphological and genetic variation in the cosmopolitan coccolithophore Emiliania huxleyi (Prymnesiophyceae) from the cox1b-atp4 genes. Journal of Phycology 47: 1164–1176. [DOI] [PubMed] [Google Scholar]
- 29. Cook SS, Whittock L, Wright SW, Hallegraeff GM (2011) Photosynthetic pigment and genetic differences between two southern ocean morphotypes of Emiliania huxleyi (Haptophyta). Journal of Phycology 47: 615–626. [DOI] [PubMed] [Google Scholar]
- 30. Thierstein HR, Geitzenauer KR, Molfino B, Shackleton NJ (1977) Global synchronicity of late Quarternary coccolith datum levels: Validation by oxygen isotopes. Geology 5: 400–404. [Google Scholar]
- 31. Bradshaw AD (1965) Evolutionary significance of phenotypic plasticity in plants. Adv Genet 13: 115–155. [Google Scholar]
- 32. Miner B, Sultan S, Morgan S, Padilla D, Relyea R (2005) Ecological consequences of phenotypic plasticity. Trends in Ecology & Evolution 20: 685–692. [DOI] [PubMed] [Google Scholar]
- 33. Bidle KD, Kwityn CJ (2012) Assessing the role of caspase activity and metacaspase expression on viral susceptibility of the coccolithophore, Emiliania huxleyi (Haptophyta). Journal of Phycology 48: 1079–1089. [DOI] [PubMed] [Google Scholar]
- 34. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. PNAS 102: 13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R (2005) The microbial pan-genome. Current Opinion in Genetics & Development 15: 589–594. [DOI] [PubMed] [Google Scholar]
- 36. Rasmussen TB, Danielsen M, Valina O, Garrigues C, Johansen E, et al. (2008) Streptococcus thermophilus Core Genome: Comparative Genome Hybridization Study of 47 Strains. Appl Environ Microbiol 74: 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ (2009) Biogeography of the Sulfolobus islandicus pan-genome. Proceedings of the National Academy of Sciences 106: 8605–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lefébure T, Stanhope MJ (2007) Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biology 8: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watanabe T, Murata Y, Oka S, Iwahashi H (2004) A new approach to species determination for yeast strains: DNA microarray-based comparative genomic hybridization using a yeast DNA microarray with 6000 genes. Yeast 21: 351–365. [DOI] [PubMed] [Google Scholar]
- 40. Riley M, Serres MH (2000) Interim report on genomics of Escherichia coli . Annual Review of Microbiology 54: 341–411. [DOI] [PubMed] [Google Scholar]
- 41. Pearson BM, Pin C, Wright J, I'Anson K, Humphrey T, et al. (2003) Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Letters 554: 224–230. [DOI] [PubMed] [Google Scholar]
- 42. Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, et al. (2013) Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499: 209–213. [DOI] [PubMed] [Google Scholar]
- 43. Kim CC, Joyce EA, Chan K, Falkow S (2002) Improved analytical methods for microarray-based genome-composition analysis. Genome Biology 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fukiya S, Mizoguchi H, Tobe T, Mori H (2004) Extensive Genomic Diversity in Pathogenic Escherichia coli and Shigella Strains Revealed by Comparative Genomic Hybridization Microarray. Journal of Bacteriology 186: 3911–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Earl AM, Losick R, Kolter R (2007) Bacillus subtilis Genome Diversity. J Bacteriol 189: 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keymer DP, Miller MC (2007) Schoolnik GK, Boehm AB (2007) Genomic and Phenotypic Diversity of Coastal Vibrio cholerae Strains Is Linked to Environmental Factors. Applied and Environmental Microbiology 73: 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dittami SM, Proux C, Rousvoal S, Peters AF, Cock JM, et al. (2011) Microarray estimation of genomic inter-strain variability in the genus Ectocarpus (Phaeophyceae). BMC Molecular Biology 12: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lakeman MB, von Dassow P, Cattolico RA (2009) The strain concept in phytoplankton ecology. Harmful Algae 8: 746–758. [Google Scholar]
- 49. Langer G, Nehrke G, Probert I, Ly J, Ziveri P (2009) Strain-specific responses of Emiliania huxleyi to changing seawater carbonate chemistry. Biogeosciences 6: 2637–2646. [Google Scholar]
- 50. Hoppe CJM, Langer G, Rost B (2011) Emiliania huxleyi shows identical responses to elevated pCO2 in TA and DIC manipulations. Journal of Experimental Marine Biology and Ecology 406: 54–62. [Google Scholar]
- 51. Thyrhaug R, Larsen A, Brussaard CPD, Bratbak G (2002) Cell cycle dependent virus production in marine phytoplankton. Journal of Phycology 38: 338–343. [Google Scholar]
- 52. Evans C, Malin G, Mills GP, Wilson WH (2006) Viral infection of Emiliania huxleyi (Prymnesiophyceae) leads to elevated production of reactive oxygen species. Journal of Phycology 42: 1040–1047. [Google Scholar]
- 53. Evans C (2007) The relative significance of viral lysis and microzooplankton grazing as pathways of dimethylsulfoniopropionate (DMSP) cleavage: An Emiliania huxleyi culture study. Limnology and Oceanography 52: 1036–1045. [Google Scholar]
- 54. Hückelhoven R (2004) BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9: 299–307. [DOI] [PubMed] [Google Scholar]
- 55. Hückelhoven R, Dechert C, Kogel K-H (2003) From the Cover: Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis . PNAS 100: 5555–5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cacas J-L (2010) Devil inside: does plant programmed cell death involve the endomembrane system? Plant, Cell & Environment 33: 1453–1473. [DOI] [PubMed] [Google Scholar]
- 57. Eichmann R, Bischof M, Weis C, Shaw J, Lacomme C, et al. (2010) BAX INHIBITOR-1 Is Required for Full Susceptibility of Barley to Powdery Mildew. Molecular Plant-Microbe Interactions 23: 1217–1227. [DOI] [PubMed] [Google Scholar]
- 58. Jones DA, Jones JDG (1997) The Role of Leucine-Rich Repeat Proteins in Plant Defences. Advances in Botanical Research 24: 89–167. [Google Scholar]
- 59. Ellis J, Dodds P, Pryor T (2000) Structure, function and evolution of plant disease resistance genes. Current Opinion in Plant Biology 3: 278–284. [DOI] [PubMed] [Google Scholar]
- 60. Marchant D, Singhera GK, Utokaparch S, Hackett TL, Boyd JH, et al. (2010) Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism. Journal of Virology 84: 11359–11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Compton T (2004) Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends in Cell Biology 14: 5–8. [DOI] [PubMed] [Google Scholar]
- 62. Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, et al. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22: 153–183. [DOI] [PubMed] [Google Scholar]
- 63. Andrade AA, Silva PN, Pereira AC, De Sousa LP, Ferreira PC, et al. (2004) The vaccinia virus-stimulated mitogen-activated protein kinase (MAPK) pathway is required for virus multiplication. The Biochemical Journal 381: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mudhakir D, Harashima H (2009) Learning from the Viral Journey: How to Enter Cells and How to Overcome Intracellular Barriers to Reach the Nucleus. 11: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Norkin LC (1995) Virus receptors: implications for pathogenesis and the design of antiviral agents. Clin Microbiol Rev 8: 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baranowski E, Ruiz-Jarabo CM, Domingo E (2001) Evolution of Cell Recognition by Viruses. Science 292: 1102–1105. [DOI] [PubMed] [Google Scholar]
- 67.Ejrnaes M, Filippi CM, Martinic MM, Ling EM, Togher LM, et al.. (2006) Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. [DOI] [PMC free article] [PubMed]
- 68. Erbar S, Diederich S, Maisner A (2008) Selective receptor expression restricts Nipah virus infection of endothelial cells. Virology Journal 5: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ren RB, Costantini F, Gorgacz EJ, Lee JJ, Racaniello VR (1990) Transgenic mice expressing a human poliovirus receptor: a new model for poloimyelitis. Cell 63: 353–362. [DOI] [PubMed] [Google Scholar]
- 70. Monick MM, Staber JM, Thomas KW, Hunninghake GW (2001) Respiratory Syncytial Virus Infection Results in Activation of Multiple Protein Kinase C Isoforms Leading to Activation of Mitogen-Activated Protein Kinase. J Immunol 166: 2681–2687. [DOI] [PubMed] [Google Scholar]
- 71. Farquhar MJ, Harris HJ, Diskar M, Jones S, Mee CJ, et al. (2008) Protein Kinase A-Dependent Step(s) in Hepatitis C Virus Entry and Infectivity. Journal of Virology 82: 8797–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Corstjens PLAM, Araki Y, Westbroek P, Gonzalez EL (1996) A Gene Encoding the 16 kD Proteolipid Subunit of a Vacuolar-Type H(+)- ATPase from Pleurochrysis carterae strain 136 (Accession No. U48365 and U53182) (PGR 96-038). Plant Physiology 111: 652. [Google Scholar]
- 73. Corstjens PLAM, Araki Y, González EL (2001) A Coccolithophorid calcifying vesicle with a vacoular-type ATPase proton pump: cloning and immunolocalizatio of the V0 subunit c. Journal of Phycology 37: 71–78. [Google Scholar]
- 74. Araki Y, González EL (1998) V- and P-Type Ca2+-Stimulated Atpases in a Calcifying Strain of Pleurochrysis Sp. (Haptophyceae). Journal of Phycology 34: 79–88. [Google Scholar]
- 75. von Dassow P, Ogata H, Probert I, Wincker P, Da Silva C, et al. (2009) Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. 10: R114–R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rokitta SD, De Nooijer L, Trimborn S, De Vargas C, Rost B, et al. (2011) Transcriptome analyses reveal differential gene expression patterns between life-cycle stages of Emiliania huxleyi (Haptophyta) and reflect specialization to different ecological niches. Journal of Phycology 47: 829–838. [DOI] [PubMed] [Google Scholar]
- 77. Mackinder L, Wheeler G, Schroeder D, Riebesell U, Brownlee C (2010) Molecular Mechanisms Underlying Calcification in Coccolithophores. Geomicrobiology Journal 27: 585. [Google Scholar]
- 78.Read BA, Wahlund TM (2007) Molecular Approaches to Emiliania huxleyi Coccolith Formation. In: Bäuerlein E, editor. Handbook of Biomineralization. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA. pp. 227–241.
- 79.Marsh ME (2007) Regulation of Coccolith Calcification in Pleurochrysis carterae. In: Bäuerlein E, editor. Handbook of Biomineralization. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA. pp. 211–226.
- 80.Taylor AR, Russell MA, Harper GM, Collins TT, Brownlee C (2007) Dynamics of formation and secretion of heterococcoliths by Coccolithus pelagicus ssp. braarudii European Journal of Phycology 42: : 125 – 136. [Google Scholar]
- 81. Marsh ME (2003) Biochemistry and Molecular Biology : Regulation of CaCO3 formation in coccolithophores. Comparative Biochemistry and Physiology Part B 136: 743–754. [DOI] [PubMed] [Google Scholar]
- 82. Paasche E (2002) A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia 40: 503–529. [Google Scholar]
- 83. Ozaki N, Sakuda S, Nagasawa H (2007) A novel highly acidic polysaccharide with inhibitory activity on calcification from the calcified scale "coccolith" of a coccolithophorid alga, Pleurochrysis haptonemofera . Biochemical and Biophysical Research Communications 357: 1172–1176. [DOI] [PubMed] [Google Scholar]
- 84.Young JR (1994) Functions of coccoliths. In: Winter A, Siesser WG, editors. Coccolithophores. Cambridge: Cambridge University Press. pp. 63–82.
- 85. Dyhrman ST, Haley ST, Birkeland SR, Wurch LL, Cipriano MJ, et al. (2006) Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi . Applied and Environmental Microbiology 72: 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Quinn P, Bowers RM, Zhang YY, Wahlund TM, Fanelli MA, et al. (2006) cDNA Microarrays as a Tool for Identification of Biomineralization Proteins in the Coccolithophorid Emiliania huxleyi (Haptophyta). Applied and Environmental Microbiology 72: 5512–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wahlund TM, Hadaegh AR, Clark R, Nguyen B, Fanelli M, et al. (2004) Analysis of Expressed Sequence Tags from Calcifying Cells of Marine Coccolithophorid (Emiliania huxleyi). Marine Biotechnology 6: 278–290. [DOI] [PubMed] [Google Scholar]
- 88. Richier S, Kerros M-E, de Vargas C, Haramaty L, Falkowski PG, et al. (2009) Light-Dependent Transcriptional Regulation of Genes of Biogeochemical Interest in the Diploid and Haploid Life Cycle Stages of Emiliania huxleyi . Applied and Environmental Microbiology 75: 3366–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rokitta SD, Rost B (2012) Effects of CO2 and their modulation by light in the life-cycle stages of the coccolithophore Emiliania huxleyi Limnology and Oceanography. 57: 607–618. [Google Scholar]
- 90. Corstjens PLAM, van der Kooij A, Linschooten C, Brouwers G-J, Westbroek P, et al. (1998) GPA, a calcium-binding protein in the coccolithophorid Emiliania huxleyi (Prymnesiophyceae). Journal of Phycology 34: 622–630. [Google Scholar]
- 91. Soto AR, Zheng H, Shoemaker D, Rodriguez J, Read BA, et al. (2006) Identification and Preliminary Characterization of Two cDNAs Encoding Unique Carbonic Anhydrases from the Marine Alga Emiliania huxleyi . Applied and Environmental Microbiology 72: 5500–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Saeed AI, Sharov V, White J, Li J, Liang W, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- 93. Paradis E, Claude J, Strimmer K (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- 94.Team TRDC (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing 2: ISBN 3-900051-900007-900050.
- 95. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, et al. (2005) Protein database searches using compositionally adjusted substitution matrices. The FEBS Journal 272: 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, et al. (2002) The Pfam Protein Families Database. Nucleic Acids Research 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignment of Bax Inhibitor 1-like protein (BI-1) using Clustal W in BioEdit. The protein sequence of E. huxleyi (ID 198434) was analysed using blastp [95], [96] similarity searches version 2.2.26 + against the SwissProt database in its standard configurations. The alignment was done with the four hits Q94A20 (Arabidopsis thaliana), Q49P94 (Vaccinia virus Lister), Q9DA39 (Mus musculus), and O7488 (Schizosaccharomyces pombe 972h) with ClustalW [98] in BioEdit. BLOSUM 62 was used as similarity Matrix.
(TIFF)
Table of pair wise overlaps of all E. huxleyi strains and G. oceanica in terms of common present genes (EPP>95%). Excel-file including strain name and number of overlaps between each strain, including the reference strain.
(XLSX)
Table of strain-specific genes for the reference strain E. huxleyi CCMP1516. Excel-file including array-, gene- and protein-ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, and the description and function of the identified genes.
(XLS)
Identified genes of E. huxleyi in respect to virus susceptibility. Excel-file including array, gene and protein- ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, description and function of the identified genes of the 16 E. huxleyi strains.
(XLS)
Identified genes of E. huxleyi related to the production of coccoliths. Excel-file including array, gene and protein- ID, html-link to the genome website of the reference strain E. huxleyi CCMP1516, description and function of the identified genes of the 16 E. huxleyi strains.
(XLS)