Abstract
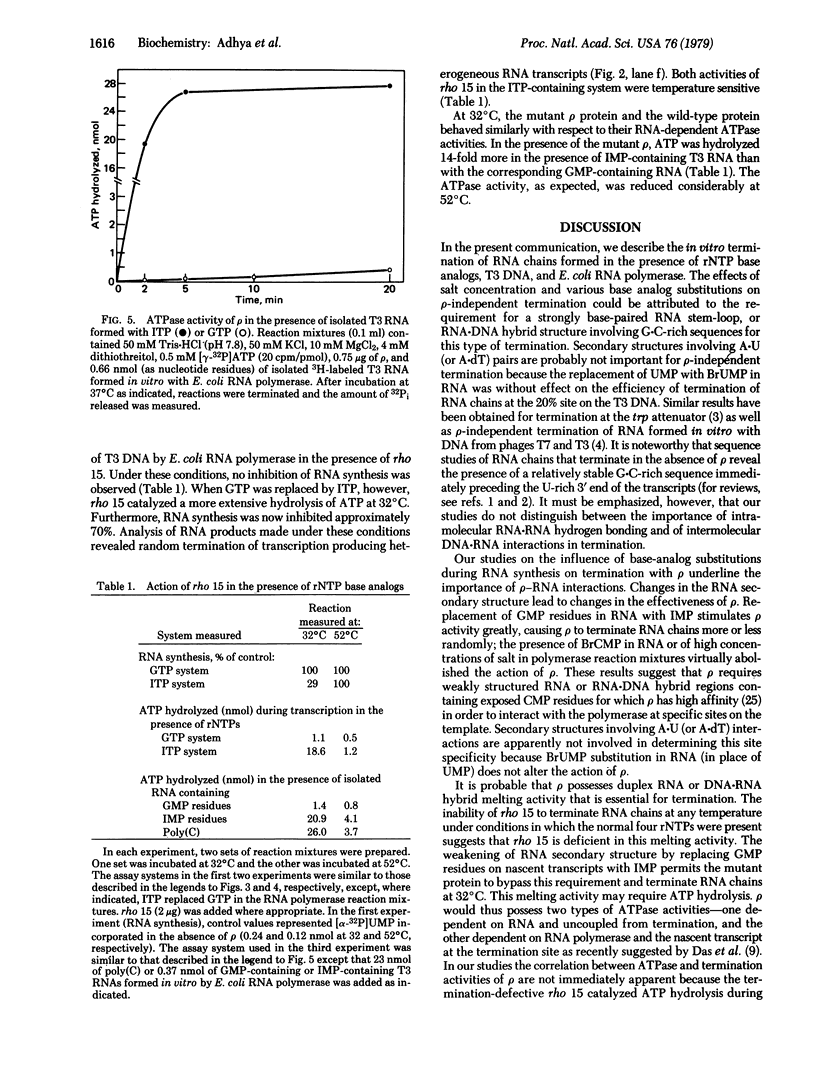
The effect of RNA secondary structure on ρ-independent and ρ-dependent termination of transcription of T3 DNA by Escherichia coli RNA polymerase has been studied by incorporating, into nascent transcripts, base analogs that lead to altered base-pairing properties. A guanine → hypoxanthine substitution, with attendant weakening of secondary structure, abolished the ρ-independent termination at 20% of the genome; in contrast, replacement of cytosine with 5-bromocytosine, which forms stronger pairs with guanine, enhanced termination at this site. ρ-Independent termination was not altered by replacing uracil with 5-bromouracil. There are two major ρ-dependent termination sites on the T3 DNA—at 8 and 15%. The termination activity of ρ in this system also depended on RNA secondary structure. The incorporation of 5-bromouracil instead of uracil into RNA did not alter the site specificity of ρ action but ρ was rendered inactive when cytosine was replaced by 5-bromocytosine. In contrast, replacement of GTP with ITP in the reaction increased ρ-dependent inhibition of RNA synthesis, caused production of heterogeneous-sized transcripts, and stimulated ρ-mediated ATP hydrolysis. The ρ-associated ATPase activity, in the presence of isolated T3 RNA, was also stimulated by inosine substitution. Furthermore, the temperature-sensitive ρ isolated from rho 15 mutant of E. coli, which does not terminate transcription in the presence of the common rNTPs, was active when GTP was replaced with ITP. These results suggest that strongly paired G·C-rich regions in RNA stem-loop structures or RNA·DNA hybrids are essential for ρ-independent termination, whereas ρ-dependent termination requires weakly paired cytosine residues for its action.
Keywords: rNTP base analog, rho ts15, ATP hydrolysis
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- CONWAY T. W., LIPMANN F. CHARACTERIZATION OF A RIBOSOME-LINKED GUANOSINE TRIPHOSPHATASE IN ESCHERICHIA COLI EXTRACTS. Proc Natl Acad Sci U S A. 1964 Dec;52:1462–1469. doi: 10.1073/pnas.52.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P. R., Bandyopadhyay P., Huang H. H., Maitra U. Fidelity of in vitro transcription of T3 deoxyribonucleic acid by bacteriophage T3-induced ribonucleic acid polymerase and by Escherichia coli ribonucleic acid polymerase. J Biol Chem. 1974 Nov 10;249(21):6901–6909. [PubMed] [Google Scholar]
- Chakraborty P. R., Salvo R. A., Majumder H. K., Maitra U. Further characterization of bacteriophage T3-induced ribonucleic acid polymerase. Studies on the size of in vitro transcripts and interaction of T3 RNA polymerase with T3 DNA. J Biol Chem. 1977 Sep 25;252(18):6485–6493. [PubMed] [Google Scholar]
- Chamberlin M. J. Comparative properties of DNA, RNA, and hybrid homopolymer pairs. Fed Proc. 1965 Nov-Dec;24(6):1446–1457. [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Merril C., Adhya S. Interaction of RNA polymerase and rho in transcription termination: coupled ATPase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4828–4832. doi: 10.1073/pnas.75.10.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnoff J. S., Maitra U. Characterization of the ribosome-dependent guanosine triphosphatase activity of polypeptide chain initiation factor IF 2. J Biol Chem. 1972 May 10;247(9):2876–2883. [PubMed] [Google Scholar]
- Dunn J. J., McAllister W. T., Bautz E. K. In vitro transcription of T3 DNA by Escherichia coli and T3 polymerases. Virology. 1972 Apr;48(1):112–125. doi: 10.1016/0042-6822(72)90119-5. [DOI] [PubMed] [Google Scholar]
- Franklin R. M. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B. H., de Crombrugghe B. ATPase activity required for termination of transcription by the Escherichia coli protein factor rho. J Biol Chem. 1976 Apr 25;251(8):2520–2524. [PubMed] [Google Scholar]
- Howard F. B., Frazier J., Miles H. T. Interaction of poly-5-bromocytidylic acid with polyinosinic acid. A study of helix stability and spectroscopic properties. J Biol Chem. 1969 Mar 10;244(5):1291–1302. [PubMed] [Google Scholar]
- INMAN R. B., BALDWIN R. L. Helix-random coil transitions in synthetic DNAs of alternating sequence. J Mol Biol. 1962 Aug;5:172–184. doi: 10.1016/s0022-2836(62)80082-5. [DOI] [PubMed] [Google Scholar]
- Issinger O. G., Hausmann R. Synthesis of bacteriophage-coded gene products during infection of Escherichia coli with amber mutants of T3 and T7 defective in gene 1. J Virol. 1973 Apr;11(4):465–472. doi: 10.1128/jvi.11.4.465-472.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller T., Sogo J. M., Bujard H. An electron microscopic method for studying nucleic acid-protein complexes. Visualization of RNA polymerase bound to the DNA of bacteriophages T7 and T3. Biopolymers. 1974 May;13(5):995–1009. doi: 10.1002/bip.1974.360130514. [DOI] [PubMed] [Google Scholar]
- Küpper H., Sekiya T., Rosenberg M., Egan J., Landy A. A rho-dependent termination site in the gene coding for tyrosine tRNA su3 of Escherichia coli. Nature. 1978 Mar 30;272(5652):423–428. doi: 10.1038/272423a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery-Goldhammer C., Richardson J. P. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974 May;71(5):2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesi termination factor p. I. Enzymatic properties and effects of inhibitors. J Biol Chem. 1977 Feb 25;252(4):1375–1380. [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. II. Influence of synthetic RNA homopolymers and random copolymers on the reaction. J Biol Chem. 1977 Feb 25;252(4):1381–1385. [PubMed] [Google Scholar]
- Maitra U., Hurwitz J. The role of deoxyribonucleic acid in ribonucleic acid synthesis. 13. Modified purification procedure and additional properties of ribonucleic acid polymerase from Escherichia coli W. J Biol Chem. 1967 Nov 10;242(21):4897–4907. [PubMed] [Google Scholar]
- Majumder H. K., Bishayee S., Chakraborty P. R., Maitra U. Ribonuclease III cleavage of bacteriophage T3RNA polymerase transcripts to late T3 mRNAs. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4891–4894. doi: 10.1073/pnas.74.11.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff N. F., Chamberlin M. J. Termination of transcription by Escherichia coli RNA polymerase in vitro is affected by ribonucleoside triphosphate base analogs. J Biol Chem. 1978 Apr 10;253(7):2455–2460. [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Hunter T. Sensitive methods for the detection and characterization of double helical ribonucleic acid. J Biol Chem. 1975 Jan 25;250(2):418–425. [PubMed] [Google Scholar]
- Rosenberg M., Court D., Shimatake H., Brady C., Wulff D. L. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978 Mar 30;272(5652):414–423. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]