Abstract
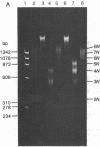
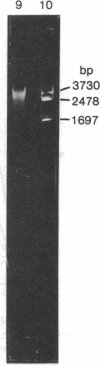
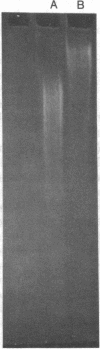
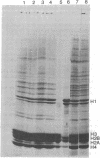
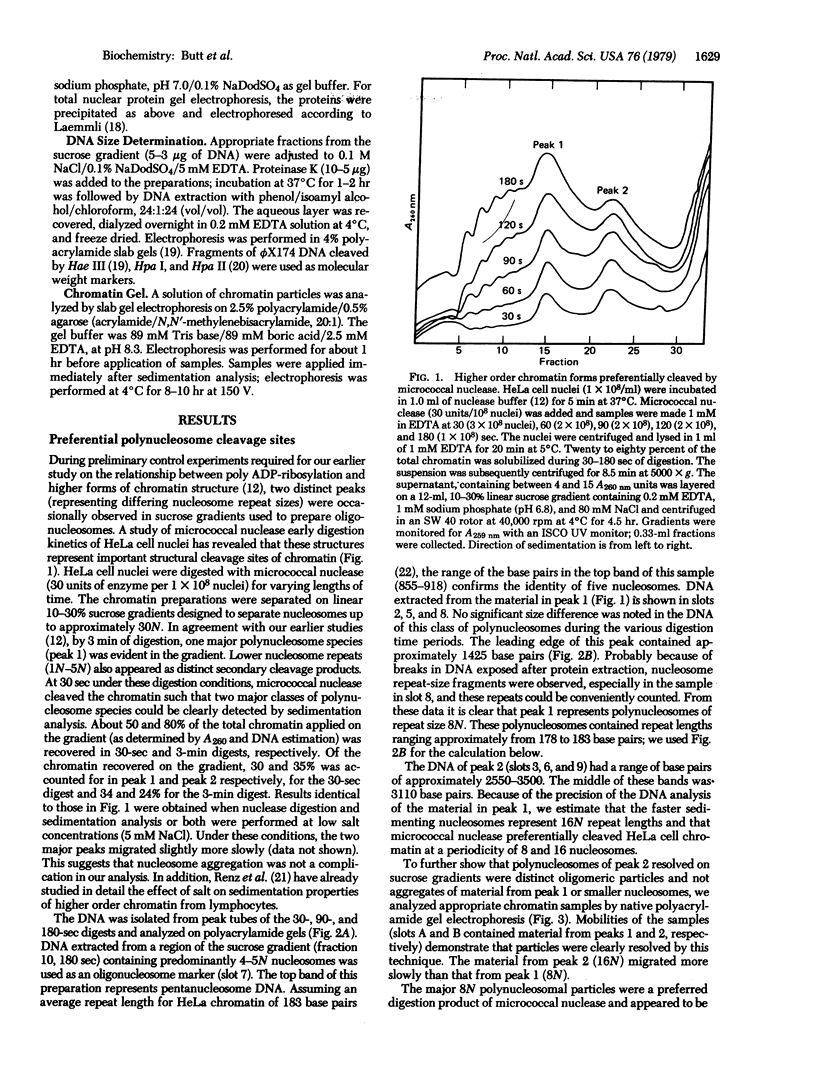
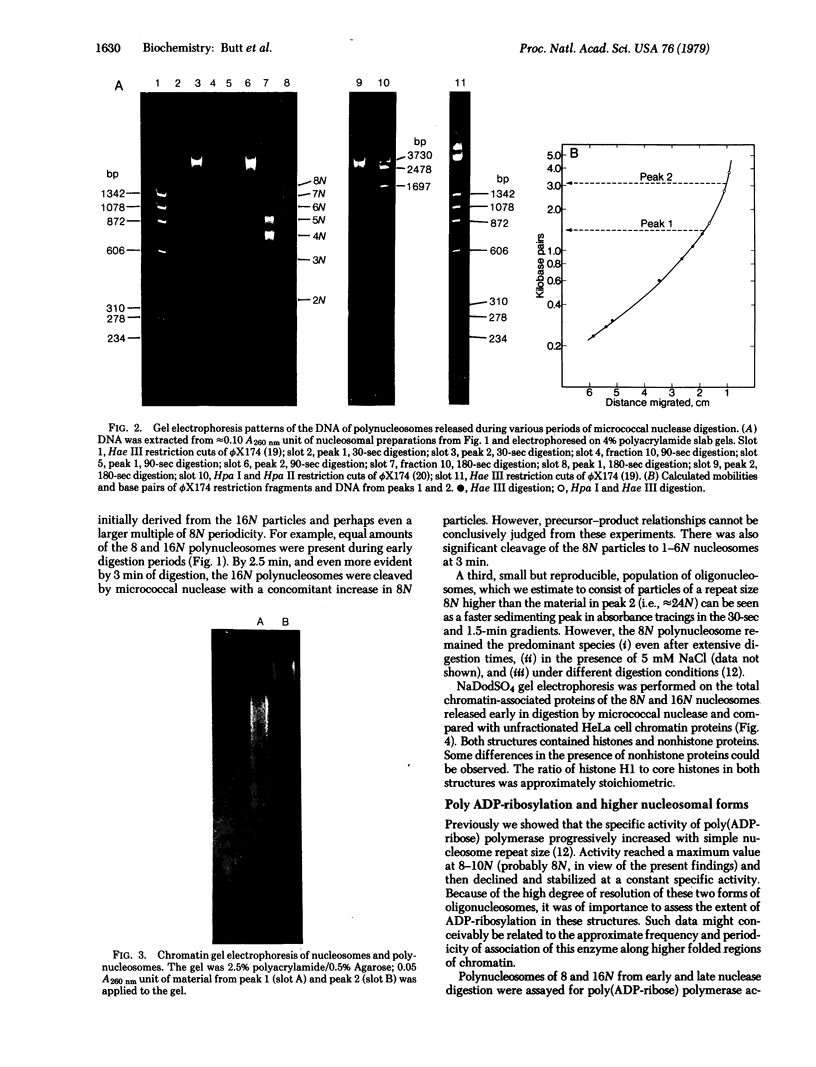
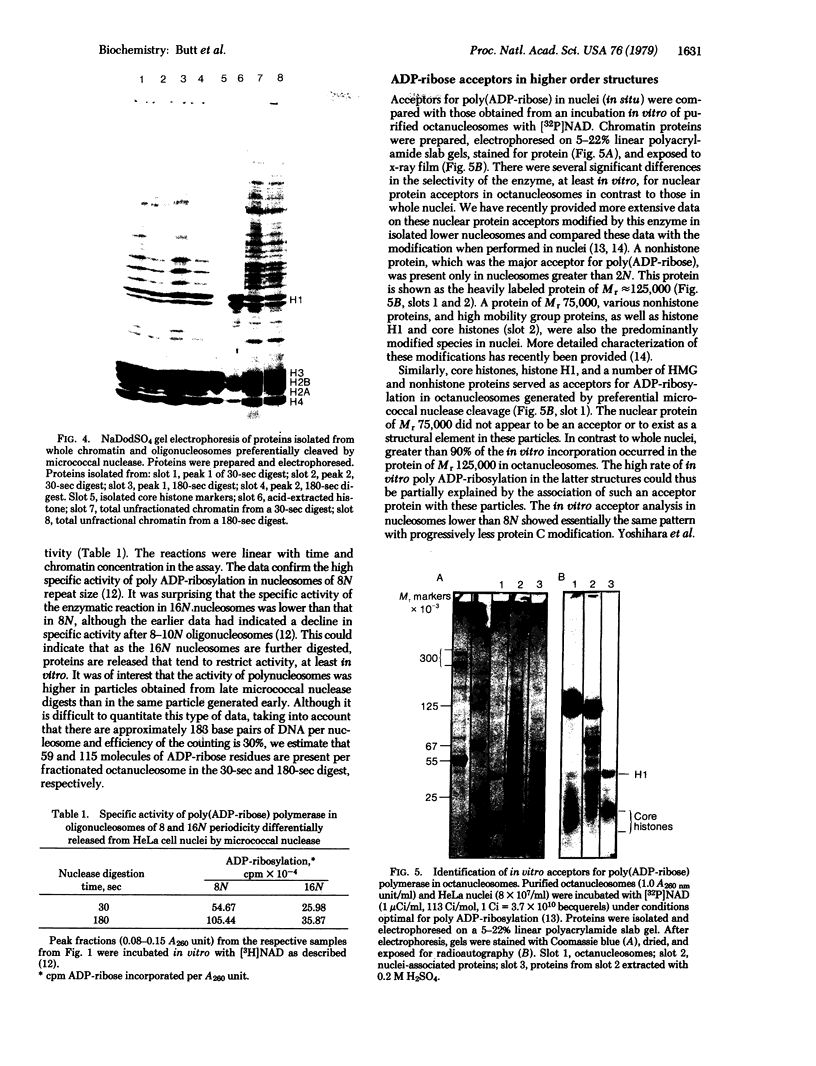
When HeLa cell nuclei were treated with micrococcal nuclease (nucleate 3-oligonucleotidohydrolase, EC 3.1.4.7), lysed, and centrifuged, the supernatant from early digests contained two predominant classes of polynucleosomes of repeat size 8N and 16N. With increasing digestion time, the 16 N polynucleosome appeared to be cleaved to the 8N species and finally to the basic subunit of chromatin. The size of the polynucleosomes has been determined by DNA analysis and on polyacrylamide electrophoretic gels of native chromatin particles. The 16N polynucleosome appears to be a unique higher ordered structural component of HeLa cell chromatin. Our recent report, showing that the nuclear protein-modifying enzyme poly(ADP-ribose) polymerase increases in specific activity progressively with increasing nucleosome repeat size up to 8-10N, has been extended in the present study. Activity was also elevated in the polynucleosomes of the 16N structure preferentially cleaved by micrococcal nuclease, although specific activity of the enzyme was highest in octanucleosomes. Acceptors for poly(ADP-ribose) have also been determined in these particles.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blakesley R. W., Wells R. D. 'Single-stranded' DNA from phiX174 and M13 is cleaved by certain restriction endonucleases. Nature. 1975 Oct 2;257(5525):421–422. doi: 10.1038/257421a0. [DOI] [PubMed] [Google Scholar]
- Bram S., Ris H. On the structure of nucleohistone. J Mol Biol. 1971 Feb 14;55(3):325–336. doi: 10.1016/0022-2836(71)90321-4. [DOI] [PubMed] [Google Scholar]
- Butt T. R., Brothers J. F., Giri C. P., Smulson M. E. A nuclear protein-modifying enzyme is responsive to ordered chromatin structure. Nucleic Acids Res. 1978 Aug;5(8):2775–2788. doi: 10.1093/nar/5.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B. G., Baldwin J. P., Bradbury E. M., Ibel K. Organisation of subunits in chromatin. Nucleic Acids Res. 1976 Jul;3(7):1739–1746. doi: 10.1093/nar/3.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Giri C. P., West M. H., Ramirez M. L., Smulson M. Nuclear protein modification and chromatin substructure. 2. Internucleosomal localization of poly(adenosine diphosphate-ribose) polymerase. Biochemistry. 1978 Aug 22;17(17):3501–3504. doi: 10.1021/bi00610a012. [DOI] [PubMed] [Google Scholar]
- Giri C. P., West M. H., Smulson M. Nuclear protein modification and chromatin substructure. 1. Differential poly(adenosine diphosphate) ribosylation of chromosomal proteins in nuclei versus isolated nucleosomes. Biochemistry. 1978 Aug 22;17(17):3495–3500. doi: 10.1021/bi00610a011. [DOI] [PubMed] [Google Scholar]
- Hozier J., Renz M., Nehls P. The chromosome fiber: evidence for an ordered superstructure of nucleosomes. Chromosoma. 1977 Jul 18;62(4):301–317. doi: 10.1007/BF00327030. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Mullins D. W., Jr, Giri C. P., Smulson M. Poly(adenosine diphosphate-ribose) polymerase: the distribution of a chromosome-associated enzyme within the chromatin substructure. Biochemistry. 1977 Feb 8;16(3):506–513. doi: 10.1021/bi00622a026. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Cotter R. I., Lilley D. M., Worcester D. L., Campbell A. M., Wooley J. C., Richards B. M. Scattering studies of chromatin subunits. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):11–22. doi: 10.1101/sqb.1978.042.01.004. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Histone H1 involvement in the structure of the chromosome fiber. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):245–252. doi: 10.1101/sqb.1978.042.01.026. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Seale R. L. Studies on the mode of segregation of histone nu bodies during replication in HeLa cells. Cell. 1976 Nov;9(3):423–429. doi: 10.1016/0092-8674(76)90087-8. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sporn M. B., Berkowitz D. M., Glinski R. P., Ash A., Stevens C. L. Irreversible inhibition of nuclear exoribonuclease by thymidine-3'-fluorophosphate and p-haloacetamidophenyl nucleotides. Science. 1969 Jun 20;164(3886):1408–1410. doi: 10.1126/science.164.3886.1408. [DOI] [PubMed] [Google Scholar]
- Stone P. R., Lorimer W. S., 3rd, Kidwell W. R. Properties of the complex between histone H1 and poly(ADP-ribose synthesised in HeLa cell nuclei. Eur J Biochem. 1977 Nov 15;81(1):9–18. doi: 10.1111/j.1432-1033.1977.tb11921.x. [DOI] [PubMed] [Google Scholar]
- Strätling W. H., Müller U., Zentgraf H. The higher order repeat structure of chromatin is built up of globular particles containing eight nucleosomes. Exp Cell Res. 1978 Dec;117(2):301–311. doi: 10.1016/0014-4827(78)90144-1. [DOI] [PubMed] [Google Scholar]
- Vengerov Y. Y., Popenko V. I. Changes in chromatin structure induced by EDTA treatment and partial removal of histone H1. Nucleic Acids Res. 1977 Sep;4(9):3017–3027. doi: 10.1093/nar/4.9.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M. H., Axel R. Nucleosomes in metaphase chromosomes. Nucleic Acids Res. 1976 Jun;3(6):1463–1471. doi: 10.1093/nar/3.6.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Yoshihara K., Hashida T., Yoshihara H., Tanaka Y., Ohgushi H. Enzyme-bound early product of purified poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1977 Oct 24;78(4):1281–1288. doi: 10.1016/0006-291x(77)91431-0. [DOI] [PubMed] [Google Scholar]