Abstract
Toxicoproteomics integrates the proteomic knowledge into toxicology by enabling protein quantification in biofluids and tissues, thus taking toxicological research to the next level. Post-translational modification (PTM) alters the three-dimensional (3D) structure of proteins by covalently binding small molecules to them and therefore represents a major protein function diversification mechanism. Because of the crucial roles PTM plays in biological systems, the identification of novel PTMs and study of the role of PTMs are gaining much attention in proteomics research. Of the 300 known PTMs, protein acylation, including lysine formylation, acetylation, propionylation, butyrylation, malonylation, succinylation, and crotonylation, regulates the crucial functions of many eukaryotic proteins involved in cellular metabolism, cell cycle, aging, growth, angiogenesis, and cancer. Here, I reviewed recent studies regarding novel types of lysine acylation, their biological functions, and their applicationsin toxicoproteomics research.
Keywords: Lysine acylation, Post-translational modification, Toxicoproteomics
INTRODUCTION
Post-translational modification (PTM) provides an option for expanding protein functionally in a cell or organism (1). PTMs involve protein backbone cleavages or the covalent binding of small molecules to protein residues in order to change the properties of proteins (2). In particular, covalent binding efficiently increases the diversity of proteins and changes 3D protein structures (1). As many as 300 protein PTMs have been described and found to possess fundamental biological roles (1,3). For examples, protein phosphorylation is the most intensively studied, and involves the attachment of phosphate moieties to serine, threonine or tyrosine residues by protein kinases (4). Reversible protein phosphorylation regulates most crucial cellular processes including the cell cycle, apoptosis, metabolism, signal transduction, proliferation and development (5-7).
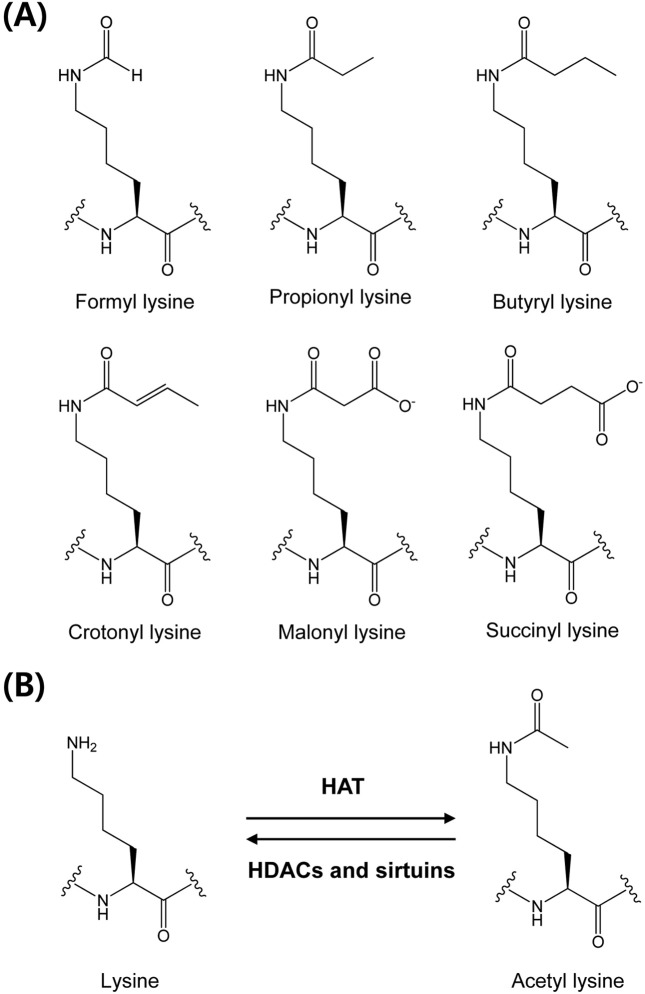
As well as phosphorylation, acylations at lysine residues include formylation, acetylation, propionylation, butyrylation, malonylation, succinylation, and crotonylation, and these processes are crucial for functional regulations of many eukaryotic proteins (Fig. 1A). Lysine acetylation was first discovered as a post-translational modification of histones in 1964 (4). A role of histone acetylation is crucial chromatin remodeling for gene transcription since its discovery for the first 30 years (8). During the past 30 years, the biological roles of lysine acetylation have been developed in nonhistone proteins. In particular, to identify protein acetylation involvement in complex biological process, the acetylome study has been develop to global analysis (8). In 2006, Kim et al., developed a method to study global protein acetylation using antibodies that selectively bind to acetylated lysine, and reported about 400 lysine acetylation sites in almost 200 proteins (9). The study revealed that > 20% of mitochondrial proteins are commonly acetylated, and the authors suggested the regulation of mitochondrial function and metabolism by reversible acetylation. Choudhary et al., identified over 3500 acetylation sites in about 1700 acetylated protein, and increased the size of the acetylome to near that of phosphorylation, the most dominant PTM (10). Thus lysine acetylation has emerged as a key PTM in cellular metabolism, cell cycle, aging, growth, angiogenesis and cancer (11-16).
Fig. 1. Protein lysine acetylation and deacetylation are controlled by acetyltransferases (HATs) and deacetylase (HDACs and sirtuins) (A) and chemical structures of lysine acylation (B).
Several other PTMs, such as, propionylation, butyrylation, malonylation, succinylation, and crotonylation, at lysine residues have been discovered in the past few years (17-19). The biological functions of these novel PTMs are uncertain, and much work is being done to identify their roles in cellular regulation. This review introduces biological roles of lysine acylation in toxic response and discusses recent advances in this active topic in the field of toxicology.
Lysine acetylation. The positively charged lysine residue plays an important role in protein folding and function. Neutralization of the charge often has a profound impact on substrate proteins. Lysine acetylation is an abundant, reversible, and highly regulated post-translational modification, which plays important roles in diverse cellular processes, such as, apoptosis, metabolism, transcription, and stress response (9). Lysine acetylation is known to be controlled by two opposing types of enzymes, acetyltransferases and deacetylase (11) (Fig. 1B). In case of fasting, toxicants exposure, and infections, the disruption of balance between two enzymatic reactions may trigger the potent toxic reaction (20). For historical reasons, the protein lysine acetyltransferases are called histone acetyltransferases (HATs), and protein lysine deacetylases is consist of histone deacetylases (HDACs) and sirtuins (21). There are three major groups of HATs: Gcn5-related N-acetyltransferases (GNATs), E1A-associated protein of 300 kDa (p300)/CREB-binding protein (CBP), and MYST proteins (22). Known HDACs are divided into Rpd3/Hda1 and sirtuin families. In humans, the former can be divided to three classes as follows: HDAC 1-3, and 8 (class I); HDAC 4-7, and 9-10 (class II); and HDAC 11 (class IV) (21). The mammalian sirtuin family comprises seven proteins (SIRT 1-7) (14). Sirtuins target a wide range of cellular proteins in nuclei, cytoplasm, and mitochondria for post-translational modification by acetylation (SIRT 1, 2, 3, and 5) or ADP-ribosylation (SIRT 4 and 6).
Alcohol-induced protein hyperacetylation: Recent studies have indicated that ethanol exposure induces global protein hyperacetylation (23). Mitochondrial protein hyperacetylation is a known consequence of sustained ethanol consumption and has been proposed to play a role in the pathogenesis of alcoholic liver disease (24). The mechanism is underlying acetylome alterations in lipid and fatty acid metabolism, antioxidant response, amino acid biosynthesis, and in the electron transport chain pathways. Chronic ethanol consumption substantially down-regulated hepatic SIRT 1 in mice, and was associated with an increase in the acetylated active nuclear form of sterol regulatory element-binding protein 1 in the livers from ethanol fed mice (25). Thus, alcohol consumption changes hepatic lipid metabolism and originates the development of alcoholic fatty liver.
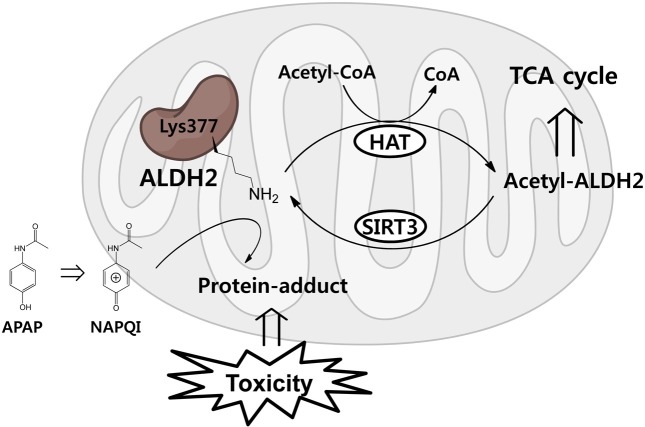
Exacerbated drug-toxicity by protein acetylation: Lysine acetylation contributes to drug-induced hepatotoxicity. In mitochondria, SIRT 3 is the primary mitochondrial deacetylase that modulates mitochondrial metabolic and oxidative stress regulatory pathways (26). Mitochondrial aldehyde dehydrogenase 2 (ALDH 2) is a direct SIRT 3 substrate with an acetylation site at Lys377. The acetaminophen reactive metabolite, NAPQI, binds to ALDH 2 at Lys377 and reduces its activity (20) (Fig. 2). In other words, the maintenance of lysine acetylation competes to bind with toxic metabolites at the same residue.
Fig. 2. Scheme for functional change of ALDH 2 by lysine acetylation or NAPQI-adduct formation at Lys377.
Protein acetylation and oxidative stress: Cumulative oxidative stress, resulting from the production of reactive oxygen species (ROS) during respiration, is believed to be a major cause of aging and numerous diseases. Increased SIRT 3 expression induced by calorie restriction (CR) deacetylates two critical lysine residues on SOD 2 and promotes its antioxidative activity (27,28). Furthermore, mitochondrial SIRT 3 was found to be down-regulated by chronic ethanol consumption or a high-fat diet (24,29). Following SIRT 3 down-regulation, the acetylations of IDH, ALDH, and SOD 2 are significantly increased, and altered redox balance in hepatic mitochondria can alter NADP+/NADPH ratio and increase fatty acid production due to TCA cycle dysregulation, aldehyde-associated ROS generation, and increased superoxide levels.
Hazardous metal toxicity and histone acetylation: Chronic exposure to arsenic in drinking water, especially in utero or perinatal exposure, can initiate neurological and cognitive dysfunctions and memory impairments (18). Arsenic exposure during embryonic life causes global hypo-acetylation at H3K9, changes in functional annotation, and the significant representation of Krüppel associated box (KRAB) transcription factors in brain samples from exposed pups. In addition, arsenic is associated with an increased risk of bladder cancer in man, and this carcinogenic effect is has been attributed to the induction of epigenetic changes at H4K16 acetylation, which leads to aberrant gene expression (30).
Lysine formylation. Lysine formylation is the shortest type of PTM, and has been reported for biological and chemical modifications in vitro. In 1999, N-terminus lysine formylation was reported in Escherichia coli (E. coli) (31). Obviously attention must be paid to the possibility of chemical modifications that may occur to protein samples during sample handling and manipulation prior to analysis by tandem mass spectrometry (32). Whereas N-terminus lysine formylation has been studied in detail, N(epsilon)-lysine formylation was discovered among chemical modifications of histones and other nuclear proteins (33). Core and linker histones are formylated at multiple lysine residues located both in the tails and globular domains of histones, in which lysine residues are known to be involved in the organization of nucleosomal particles or residues that have important roles in DNA binding. Moreover, in chromosomal proteins, formylation is relatively abundant, suggesting that it may interfere with epigenetic mechanisms governing chromatin function, which could lead to cellular deregulation and disease.
Toxic mechanisms of lysine formylation mainly involve in the formation of stable lysine adducts in proteins following exposure to toxicants. The major reactive compound generated by the metabolism of trichloroethylene (TCE) by cytochrome P450 (CYP) can form N6-formyllysine protein adducts (34). These TCE-derived protein adducts can be used as a basis for considering exposure and the risk of TCE in humans. Furthermore, the N6-formylation of lysine occurs as a protein secondary modification and appears to arise from products of DNA oxidation in cells (35). The N6-formyl modification of lysine may interfere with the signaling functions of lysine acetylation, which is recognized as an important determinant of methylation and gene expression in mammalian cells, and thus, this modification may contribute to the pathophysiology of oxidative and nitrosative stress. In addition, formylation at lysine residues might cause hapten-protein binding with 2,5-dimethyl-p-benzoquinonediimine, the oxidation of intermediates of allergenic p-amino aromatic compounds, which is the lysineinduced N-formylation to generate antigenic reaction (36).
Lysine propionylation and butyrylation. In 2007, lysine propionylation and butyrylation were discovered in histones and confirmed by in vitro labeling and by mass spectrometry based peptide mapping (17,37). Propionyl-CoA and butyryl-CoA, are structurally similar to acetyl-CoA and differ by one or two CH2 units. In addition, lysine propionylation has been identified in non-histone proteins in eukaryotic cells, such as, in p53, p300, and CREB binding-protein (38). Furthermore, propionylation at H3 lysine Lys23 was detected in the leukemia cell line U937 by mass spectrometry and Western analysis using a specific antibody (39). Propionylation levels in U937 cells reduce remarkably during monocytic differentiation, indicating that this modification is dynamically regulated. Furthermore, charge neutralization and nonpolar enhancement by propionylation of lysine residues appears to be highly effective for promoting surface hydrophobicity, and important driver of protein aggregation (40).
Lysin malonylation and succinylation. Lysine malonylation and succinylation were novel types of lysine PTMs, and were originally detected by mass spectrometry and protein sequence-database searching in 2011 (41). Lysine malonylation is a dynamic and evolutionarily conserved PTM observed in mammalian and bacterial cells, and SIRT 5, a member of the class III lysine deacetylases, can catalyze lysine demalonylation and lysine desuccinylation both in vitro and in vivo. Lysine succinylated and malonylated peptides in histone protein were verified by the MS/MS of synthetic peptides, HPLC co-elution, and isotopic labeling in HeLa cells, mouse embryonic fibroblasts, Drosophila S2, and Saccharomyces cerevisiae cells (42). Mutagenesis of succinylation sites followed by functional assays suggested that histone lysine succinylation could have unique functional consequences.
SIRT 5 exhibits demalonylase and desuccinylase activity, in addition, lysine succinylation and malonylation are abundant mitochondrial protein modifications (43). Succinyl- CoA is an intermediate in the TCA cycle and a precursor of porphyrin synthesis, whereas malonyl-CoA is the precursor of de novo fatty acid synthesis and a critical inhibitor of fatty acid oxidation. Malonyl-CoA also regulates, directly or indirectly, physiological or pathological conditions, such as, muscle contraction, cardiac ischemia, β-cell insulin secretion, and the hypothalamic control of appetite (44). Although the biological roles of lysine malonylation and succinylation are known to be related to cellular metabolism, no toxicological research has been conducted on biological roles.
Table 1.
Summarizing enzymes involved in protein lysine acylation and deacylation
| Acylations | Acyltransferases | Deacylases | |||
|---|---|---|---|---|---|
|
| |||||
| Class | Subclass | Members | Class | Members | |
|
| |||||
| Acetylation | GNAT-family | GCN5L | Class I | HDAC 1,2,3,8 | |
| PCAF | |||||
| Tip60 | |||||
| A | MYST-family | HBOI | Class II | HDAC 4,5,6,7,9,10 | |
| etc | |||||
| p300/CBP | |||||
| Others | TFIIIC complex | Class III | SIRT 1-7 | ||
| etc | |||||
| B | HAT1 | ||||
| Formylation | unknown | unknown | |||
| Propionylation | p300, CBP | unknown | |||
| Butrylation | p300, CBP | unknown | |||
| Malonylation | unknown | SIRT 5 | |||
| Succinylation | unknown | SIRT 5 | |||
| Crotonylation | unknown | unknown | |||
Lysine crotonylation. Recently, histone lysine crotonylation was found to mark X/Y-linked genes that are active in post-meiotic male germ cells (45). The unique structure and genomic localization of histone crotonylation at lysine residues suggests that it is mechanistically and functionally different from histone lysine acetylation (46). Specifically, in human somatic and mouse male germ cell genomes, histone lysine crotonylation marks either active promoters or potential enhancers. In male germinal cells immediately following meiosis, lysine crotonylation is enriched on sex chromosomes and marks testis-specific genes, including a significant proportion of X-linked genes that escape sex chromosome inactivation in haploid cells.
PERSPECTIVES
The emerging field of toxicoproteomics has been boosted by quantitative and qualitative proteomic technologies and their increasing applications in toxicology research (47). Toxicoproteomics uses the discovery potential of proteomics in toxicology research by applying global protein measurement technologies to biofluids and tissues after host exposure to injurious agents (48). During the past decade, toxicoproteomics has developed in parallel with proteomic technology in the following regards: the analysis of global protein expression, the use of high performance LC-MS/MS platforms and stable isotope labeling technology, targeted quantitative analysis in multiple-reaction monitoring mode, and PTM analysis by enrichment technology.
Specially, the exploration of novel PTMs and the identifications of their biological roles are a focus in the proteomic research field. Reversible PTMs can regulate the activities and localizations of intracellular proteins, and are crucial for understanding biological roles (1,3,49). Lysine acetylation, the best-known type of lysine acylation PTM, regulates various crucial roles in biological systems (9). In current review, we consider toxicity to originate from the abnormal regulation of lysine acetylation, such as, the dysregulation of modulating enzymes like acetyltransferase/deacetylase, the blocking of protein acetylation at specific lysine residues and the subsequent modulation of protein functions, and the dysregulation of histone acetylation as an epigenetic marker (24,26,30). However, new types of lysine acylations, such as, propionylation, butyrylation, malonylation, succinylation, and crotonylation continue to be detected, and their biological roles have yet to be determined.
In conclusion, the abnormal regulations of PTMs could be closely associated with the toxicities of many xenobiotics. However, although protein acylation regulates many pivotal biological functions, the possible roles of protein acylation at lysine residues in the mechanisms responsible for the toxic activities of most xenobiotics have not been comprehensively investigated. Those in the field of toxicology should pay attention to the toxicoproteomic approach involving PTMs, because the toxicology is changing, and the new paradigms arising from toxicoproteomics are destined to play important roles.
Acknowledgments
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (Grant No: A112026).
References
- 1.Walsh C.T., Garneau-Tsodikova S., Gatto G.J. Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. (2005);44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Mann M., Jensen O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. (2003);21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 3.Cantin G.T., Yates J.R. 3rd. Strategies for shotgun identification of post-translational modifications by mass spectrometry. J. Chromatogr. A. (2004);1053:7–14. doi: 10.1016/j.chroma.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 4.Lu Z., Cheng Z., Zhao Y., Volchenboum S.L. Bioinformatic analysis and post-translational modification crosstalk prediction of lysine acetylation. PLoS One. (2011c);6:e28228. doi: 10.1371/journal.pone.0028228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keck J.M., Jones M.H., Wong C.C., Binkley J., Chen D., Jaspersen S.L., Holinger E.P., Xu T., Niepel M., Rout M.P., Vogel J., Sidow A., Yates J.R. 3rd., Winey M. A cell cycle phosphoproteome of the yeast centrosome. Science. (2011);332:1557–1561. doi: 10.1126/science.1205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter T. Signaling--2000 and beyond. Cell. (2000);100:113–127. doi: 10.1016/S0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 7.Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., Brunak S., Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling. (2010);3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 8.Norris K.L., Lee J.Y., Yao T.P. Acetylation goes global: the emergence of acetylation biology. Sci. Signaling. (2009);2:pe76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S.C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N.V., White M., Yang X.J., Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. (2006);23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. (2009);325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 11.Lin H., Su X., He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. (2012);7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J.Y., Lin Y.Y., Sheu J.C., Wu J.T., Lee F.J., Chen Y., Lin M.I., Chiang F.T., Tai T.Y., Berger S.L., Zhao Y., Tsai K.S., Zhu H., Chuang L.M., Boeke J.D. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell. (2011a);146:969–979. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang C., Lin S.H., Huang F., Pan J., Josic D., Yu-Lee L.Y. Acetylation of RNA processing proteins and cell cycle proteins in mitosis. J. Proteome. (2010);9:4554–4564. doi: 10.1021/pr100281h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carafa V., Nebbioso A., Altucci L. Sirtuins and disease: the road ahead. Front. Pharmacol. (2012);3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z.B., Zeng R., Xiong Y., Guan K.L., Zhao S., Zhao G.P. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. (2010);327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S.M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K.L. Regulation of cellular metabolism by protein lysine acetylation. Science. (2010);327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., Falck J.R., Peng J., Gu W., Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics. (2007);6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cronican A.A., Fitz N.F., Carter A., Saleem M., Shiva S., Barchowsky A., Koldamova R., Schug J., Lefterov I. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS One. (2013);8:e53478. doi: 10.1371/journal.pone.0053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebastián C., Zwaans B.M., Silberman D.M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A.R., Toiber D., Cosentino C., Greenson J.K., MacDonald A.I., McGlynn L., Maxwell F., Edwards J., Giacosa S., Guccione E., Weissleder R., Bernstein B.E., Regev A., Shiels P.G., Lombard D.B., Mostoslavsky R. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. (2012);151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z., Bourdi M., Li J.H., Aponte A.M., Chen Y., Lombard D.B., Gucek M., Pohl L.R., Sack M.N. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. (2011b);12:840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peserico A., Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. (2011);2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X.J., Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. (2008);31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shepard B.D., Tuma P.L. Alcohol-induced protein hyperacetylation: mechanisms and consequences. World J. Gastroenterol. (2009);15:1219–1230. doi: 10.3748/wjg.15.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritz K.S., Galligan J.J., Hirschey M.D., Verdin E., Petersen D.R. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J. Proteome Res. (2012);11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You M., Liang X., Ajmo J.M., Ness G.C. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am. J. Gastrointest. Liver Physiol. (2008);294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 26.Shimazu T., Hirschey M.D., Hua L., Dittenhafer-Reed K.E., Schwer B., Lombard D.B., Li Y., Bunkenborg J., Alt F.W., Denu J.M., Jacobson M.P., Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. (2010);12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X., Brown K., Hirschey M.D., Verdin E., Chen D. Calorie restriction reduces oxidative stress by SIRT3- mediated SOD2 activation. Cell Metab. (2010);12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B., Grueter C.A., Harris C., Biddinger S., Ilkayeva O.R., Stevens R.D., Li Y., Saha A.K., Ruderman N.B., Bain J.R., Newgard C.B., Farese R.V. Jr., Alt F.W., Kahn C.R., Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. (2010);464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kendrick A.A., Choudhury M., Rahman S.M., McCurdy C.E., Friederich M., Van Hove J.L., Watson P.A., Birdsey N., Bao J., Gius D., Sack M.N., Jing E., Kahn C.R., Friedman J.E., Jonscher K.R. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem. J. (2011);433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jo W.J., Ren X., Chu F., Aleshin M., Wintz H., Burlingame A., Smith M.T., Vulpe C.D., Zhang L. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol. Appl. Pharmacol. (2009);241:294–302. doi: 10.1016/j.taap.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkins M.R., Gasteiger E., Gooley A.A., Herbert B.R., Molloy M.P., Binz P.A., Ou K., Sanchez J.C., Bairoch A., Williams K.L., Hochstrasser D.F. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. (1999);289:645–657. doi: 10.1006/jmbi.1999.2794. [DOI] [PubMed] [Google Scholar]
- 32.Osés-Prieto J.A., Zhang X., Burlingame A.L. Formation of epsilon-formyllysine on silver-stained proteins: implications for assignment of isobaric dimethylation sites by tandem mass spectrometry. Mol. Cell. Proteomics. (2007);6:181–192. doi: 10.1074/mcp.M600279-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Wisniewski J.R., Zougman A., Mann M. Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. (2008);36:570–577. doi: 10.1093/nar/gkm1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai H., Guengerich F.P. Acylation of protein lysines by trichloroethylene oxide. Chem. Res. Toxicol. (2000);13:327–335. doi: 10.1021/tx000003p. [DOI] [PubMed] [Google Scholar]
- 35.Jiang T., Zhou X., Taghizadeh K., Dong M., Dedon P.C. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. (2007);104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eilstein J., Giménez-Arnau E., Duché D., Rousset F., Lepoittevin J.P. Mechanistic studies on the lysineinduced N-formylation of 2,5-dimethyl-p-benzoquinonediimine. Chem. Res. Toxicol. (2007);20:1155–1161. doi: 10.1021/tx700040s. [DOI] [PubMed] [Google Scholar]
- 37.Zhang K., Chen Y., Zhang Z., Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. J. Proteome Res. (2009);8:900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S.S., Gu W., Zhao Y. Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteomics. (2009);8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu B., Lin Y., Darwanto A., Song X., Xu G., Zhang K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. (2009);284:32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Es-Haghi A., Shariatizi S., Ebrahim-Habibi A., Nemat-Gorgani M. Amyloid fibrillation in native and chemically-modified forms of carbonic anhydrase II: role of surface hydrophobicity. Biochim. Biophys. Acta. (2012);1824:468–477. doi: 10.1016/j.bbapap.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B.M., Tishkoff D., Ho L., Lombard D., He T.C., Dai J., Verdin E., Ye Y., Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics. (2011);10:M111.012658.. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J.D., Zhao Y. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics. (2012);11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newman J.C., He W., Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J. Biol. Chem. (2012);287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu. Rev. Nutr. (2008);28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 45.Montellier E., Rousseaux S., Zhao Y., Khochbin S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic malespecific gene expression. BioEssays. (2012);34:187–193. doi: 10.1002/bies.201100141. [DOI] [PubMed] [Google Scholar]
- 46.Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. (2011);146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George J., Singh R., Mahmood Z., Shukla Y. Toxicoproteomics: new paradigms in toxicology research. Toxicol. Mech. Methods. (2010);20:415–423. doi: 10.3109/15376511003667842. [DOI] [PubMed] [Google Scholar]
- 48.Wetmore B.A., Merrick B.A. Toxicoproteomics: proteomics applied to toxicology and pathology. Toxicol. Pathol. (2004);32:619–642. doi: 10.1080/01926230490518244. [DOI] [PubMed] [Google Scholar]
- 49.Beltrao P., Albanèse V., Kenner L.R., Swaney D.L., Burlingame A., Villèn J., Lim W.A., Fraser J.S., Frydman J., Krogan N.J. Systematic functional prioritization of protein posttranslational modifications. Cell. (2012);150:413–425. doi: 10.1016/j.cell.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]