Abstract
Background
CCR5-delta32 heterozygous individuals are susceptible to HIV-1. However, it is not clear if there is a relevant protective effect against transmission and a beneficial effect in terms of HIV progression which cannot be attributed to CCR5 surface density alone. Therefore we investigated HIV-1 dependency factors (HDF) which might be differently regulated in CCR5 wild type (WT) and CCR5-delta32 heterozygous individuals.
Methods
We examined CD34+ hematopoietic progenitor cells derived from bone marrow samples from 19 healthy volunteers, 12 individuals with CCR5 WT and 7 with heterozygous CCR5-delta32 deletion. Samples were analyzed using a global gene expression oligonucleotide microarray (HG-U133plus 2.0, Affymetrix Inc.).
Results
A total of 205 genes were found with altered expression (3fold difference, present call rate of 75%, p < 0.05) and 7 of these had a connection to HIV-1 pathogenesis. In 4 genes: TOP1, CXCR2, SREBF2, and TAP we found a different regulation which was consistent with a supposed beneficial effect for CCR5-delta32 heterozygotes.
Conclusion
The CCR5-delta32 deletion is associated with other HDFs in HIV-1 pathogenesis as a possible explanation for beneficial effects regarding the deletion leading to a variant expression profile in heterozygous carriers of this mutation.
Keywords: CCR5-delta32, HIV-1, Coexpression, TOP1, CXCR2, SREBF2, TAP
Background
The human immunodeficiency virus type 1 (HIV-1) genome encodes a rather small number of 15 proteins. However, during infection and interaction with the target cells, the virus exploits multiple proteins of the host for its replication. Alterations of expression or functional changes based on genetic variances of critical genes are called HIV-dependency factors (HDFs). They are required for infection or disease progression, influencing the efficacy of HIV-1 transmission, viral load, and collapse of the immune system. Understanding these HDFs might play a key role in identifying new therapeutics strategies against the infection [1].
One of the most prominent HDF is the association of HIV-1 and the chemokine receptor CCR5. In combination with CD4, CCR5 is required for internalization of the viral genome. Up to date, over 70 mutations have been described in the CCR5 gene including the intensively studied 32 base pair deletion (CCR5-delta32) that introduces a premature stop-codon into the CCR5 locus [2,3].
Interestingly, CCR5 is highly preferred from the virus and people with a homozygous CCR5-delta32 deletion gain a nearly complete protection from HIV-1 transmission. This is remarkable as CXCR4 is always expressed as an alternative coreceptor in CCR5-deficient individuals and most HIV positive patients harbor a small fraction of CXCR4 using strains (X4 type) [4]. Therefore, CCR5 deficient individuals are frequently exposed against X4 but hardly ever get infected. It is still not clear whether this observation could only be explained by a presumed selective disadvantage of X4 in the absence of CCR5 or whether there are additional effects of the CCR5 deletion causing the protection from X4 transmission.
In addition, a certain protective effect in heterozygous CCR5-delta32 carriers is also controversially discussed (Table 1). It is well known that CCR5 receptor density on the target cells determines the susceptibility against transmission and that CCR5-delta32 heterozygotes have a reduced receptor expression on the surface [5]. However, CCR5 is a dynamic receptor and several studies have shown that CCR5 density in delta32 heterozygotes could be similar to wild type individuals and therefore the CCR5 surface expression might not be the single protective factor [6,7].
Table 1.
A selection of published surveys concerning possible clinical benefits (transmission or progression) of the CCR5-delta32 heterozygous genotype
| N= | Study population | Benefit tested | Significance | Reference |
|---|---|---|---|---|
| 1427 |
HIV + vs. sero-negative control group |
Transmission |
P = 0.012 |
[8] |
| 30 |
ESN |
Transmission |
P = 0.04 |
[9] |
| 54 |
ESN |
Transmission |
P = 0.05* |
[10] |
| 2605 |
Woman |
Transmission |
OR: 0.63 (95% CI 0.44-0.90) |
[11] |
| 2996 |
High risk groups |
Transmission |
Risk = 0.30 (95% CI: 0.08-0.97) |
[12] |
| 1252 |
MSM |
Transmission |
NS |
[13] |
| 1200 |
HIV + vs control |
Transmission, progression to AIDS |
Both significant |
[14] |
| 108 |
IDU |
Progression to AIDS |
NS |
[15] |
| 127 |
Children |
Progression to AIDS |
Significant (p = 0.008) Effect is abrogated in case of a SDF1-3′A genotype |
[16] |
| 1955 |
Hemophilia patients |
Progression to AIDS |
Significant |
[17] |
| 512 | Mother-child | Transmission, progression to AIDS | NS, significant | [18] |
These controversial data suggest that there might be additional factors beside the CCR5-delta32 genotype like SDF 1-3′ which contribute to the clinical outcome of transmission and infection. (ESN = exposed seronegative, MSM = men having sex with men, IDU = injecting drug users, CI = confidence interval, OR = odds ratio, NS = not significant) *only significant in heterosexuals.
Taken together, the mechanism of a near-perfect protection of complete CCR5 deficiency as well as the observation that HIV-infected CCR5-delta32 heterozygotes may have a slower progression to AIDS has to be more elucidated and might help identifying other key players contributing to HIV pathogenesis [13,17].
Therefore, in this work we analyzed the gene expression patterns of CCR5-delta32 deletion in comparison to wild type carriers, further, how the alteration change in expression might contribute to the reduced clinical susceptibility and progression of HIV disease observed in carriers of this mutation. This might improve the understanding of the connection between HIV pathogenesis and immune system interaction of the underlying host.
Results and discussion
We screened CD34+ cells of 19 specimen contained within a blood donor bank and identified 12 healthy donors carrying wild type (WT) CCR5 while the remaining seven carried the CCR5-delta32 deletion allele heterozygously (Table 2). Gene expression analysis revealed 205 genes differentially expressed between WT and heterozygous carriers of CCR5-delta32 mutation. 96 were found to be overexpressed in heterozygotes (Additional file 1: Table S1) whereas the remaining 109 genes were found to be overexpressed in WT samples, respectively.
Table 2.
Results and characteristics of analyzed specimen
| Specimen | Gender | Age | CCR5 WT | CCR5-delta32 heterozygous | |
|---|---|---|---|---|---|
| 1 |
N0014/05 |
m |
19 |
+ |
|
| 2 |
N0015/05 |
f |
53 |
+ |
|
| 3 |
N0023/05 |
m |
24 |
+ |
|
| 4 |
N0024/05 |
f |
21 |
+ |
|
| 5 |
N0031/05 |
m |
25 |
+ |
|
| 6 |
N0035/05 |
m |
21 |
|
+ |
| 7 |
N0091/06 |
m |
30 |
+ |
|
| 8 |
N0094/06 |
m |
20 |
+ |
|
| 9 |
N0103/06 |
m |
32 |
+ |
|
| 10 |
N0106/06 |
m |
23 |
+ |
|
| 11 |
N0118/07 |
f |
28 |
|
+ |
| 12 |
N0136/07 |
f |
29 |
|
+ |
| 13 |
N0153/07 |
f |
29 |
|
+ |
| 14 |
N0158/07 |
m |
25 |
|
+ |
| 15 |
N0163/07 |
f |
23 |
|
+ |
| 16 |
N0200/08 |
f |
74 |
+ |
|
| 17 |
N0204/08 |
f |
85 |
+ |
|
| 18 |
N0305/09 |
f |
28 |
|
+ |
| 19 | N0312/09 | f | 34 | + |
CD34+ hematopoeitic progenitor cells for genotyping were obtained by bone marrow aspiration.
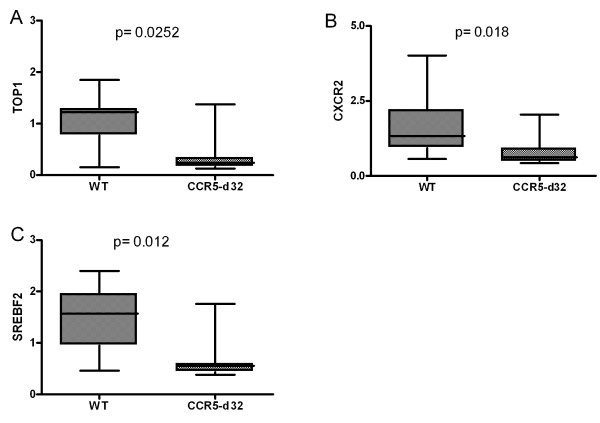
Further review of gene databases concerning the known or proposed function of these genes regarding HIV-1 infection revealed three genes in the WT group and four genes in the CCR5-delta32 group with significantly different overexpression profiles (Figures 1 and 2).
Figure 1.
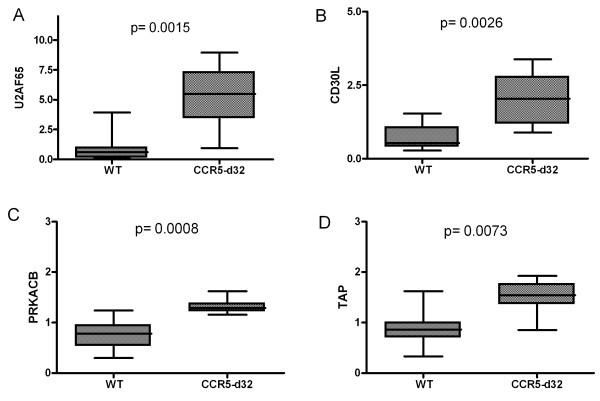
Results of the micro array gene expression analysis of 19 specimens. In the group of heterozygous CCR5-delta32 carriers 4 genes associated with HIV-1 pathogenesis have been found to be overexpressed: A. U2AF65, B. CD30L, C. PRKACB, and D. TAP. Normalized arbitrary expression levels.
Figure 2.
Results of the micro array gene expression analysis of 19 specimens. In wild type sample there were 3 genes associated with HIV-1 significantly overexpressed compared to heterozygous CCR5-delta32 carriers: A. TOP1, B. CXCR2, and C. SREBF2. Normalized arbitrary expression levels.
The theory that human host genes or polymorphic variants of genes may influence transmission or the outcome of HIV-1 infection goes back to the early 1980’s [19]. Improved techniques using high throughput assays provide insight into a detailed view in the pathogenesis of HIV-1 and possible host or HIV-dependency factors (HDF) [1]. Besides the well-characterized chemokine entry mechanism there are several other allelic mutations described in HDF genes such as IL10, IFNG, or KIR3DS1 (together with HLA-Bw4). In addition, several HLA subtypes have been found to be associated either with delayed or accelerated progression to AIDS [20].
However, some phenomena remain unsolved. Especially the role of the CCR5-delta32 deletion in transmission and maintaining the infection is under investigation and there are currently more open questions than answers. Recent progress in HIV gene therapy revealed new insights of the CCR5-delta32 deletion [21,22]. In the SB-728-T Sangamo trial with anti CCR5 zinc-finger nuclease (ZFN) treated lymphocytes in HIV patients, only one candidate with a CCR5-delta32 heterozygous genotype in this group achieved spontaneous virus control during HAART discontinuation. Taken into account that the CCR5 density in heterozygotes does not substantially differ from the density in wild types, it is astonishing that an additional knock down of less than a few percent of CCR5 expression by ZFNs has subsequently such a deep impact on viral replication. Consequently it was not surprising that Sangamo initiated a follow up trial especially for carriers of the CCR5-delta32 genotype (SB-728-902-Cohort 5) [23].
Most recently, the report of a long term control of HIV in two patients receiving an allogeneic stem cell transplantation (SCT) with a CCR5 wild type allo-graft (“Boston patients”) has brought up new questions. These patients showed a profound reduction of the viral reservoir, and declining HIV antibody levels with ongoing engraftment during the follow-up period of several years similar to the “Berlin patient” [24]. Finally, in 2013 antiretroviral medication was discontinued in these patients and none of them developed a rebound of HIV infection. Transplantation has led to a replacement of the old immune system, antiretroviral therapy has protected the new engrafting cells from re-infection, and probably GvHD has cleared residual viral reservoirs in the host. Interestingly, both patients were heterozygous for the CCR5-delta32 deletion [25].
Genes with higher expression in the wild type group
From 109 genes significantly overexpressed in the wild type group, three were recognized to have an association with HIV-1 (Table 3). Of these three, CXCR2 is expressed in higher quantities in the wild type group. Like CXCR1, CXCR2 acts as a receptor for interleukin 8 (IL-8) and is differentially expressed in subsets of lymphocytes like CD56+/CD16+ NK-cells [26]. Both receptors share intracellular cross-talk and in cells expressing either CXCR1 or CXCR2, exposure to IL-8 leads to an internalization of these receptors. This effect is significant lower in cells expressing both of the receptors like the CD56+/CD16+ NK-cells [27]. Additionally, binding of IL-8 on the CXCR1 receptors causes an additional cross-phosphorylation of both, CCR5 and CXCR4, resulting in a decreased susceptibility against HIV-1. Only the association of IL-8/CXCR1 and not IL-8/CXCR2 inherits this protective effect [28]. Most surprisingly in this context is that the HIV-1 matrix protein p17 binds to both, CXCR1 and CXCR2 in an IL-8-like manner [29]. Taken together, the higher expression of CXCR2 in the wild type group might have implications for the p17HIV-1/IL-8 pathway of CXCR1 induced CCR5 internalization resulting in an altered susceptibility.
Table 3.
Three genes with a link to HIV-1 pathogenesis were found to be significant higher expressed in the CCR5 wild type group
| Significant higher Expression in CCR5 wild type samples | ||||
|---|---|---|---|---|
| Gene |
Function (link to HIV-1) |
Beneficial effect for CCR5-delta32 carriers |
Reference |
|
| Transm. |
Repli. |
|||
| TOP1 |
Synthesis of cDNA (in combination with RT transcription of viral RNA to DNA) |
|
Possible |
[30] |
| CXCR2 |
Receptor for IL-8. Mediates neutrophil migration to sites of inflammation. (binding side of p17
HIV-1
, cross-linked to CXCR1 and indirectly cross-linked to CXCR1 mediated CCR5 internalisation) |
Possible |
|
[29] |
| SREBF2 | Cholesterol metabolism (enhancing viral replication) | Possible | [31] | |
For TOP1 and SREBF2 there are reports that overexpression of these genes might have a beneficial effect on HIV-1 infection. For CXCR2 overexpression this beneficial effect was described in case of HIV-1 transmission. (transm. = transmission, repli. = replication).
Topoisomerase 1 (TOP1) is a 91 kDa enzyme that critically regulates the topological state of DNA during transcription. TOP1 in association with reverse transcriptase is most relevant for HIV cDNA synthesis [32]. Moreover, TOP1 is essential to directly interact with HIV-1 nucleocapsid protein [33] In vitro assays with overexpression of TOP1 led to an 5fold increase of infective virions derived from these manipulated cells [34]. Our finding that TOP1 was significantly underexpressed in CCR5-delta32 samples than in wild type samples could contribute to the clinical observation of decreased rate of progression to AIDS in HIV-1-infected heterozygous carriers.
Finally, cholesterol is known to play an essential role in the life cycle of several enveloped viruses. Many of these viruses manipulate host cholesterol metabolism to facilitate their replication. The target gene of the sterol regulatory element-binding protein 2 (SREBP2) which was found to be downregulated in the heterozygous CCR5-delta32 group is TFII-I, a gene critical for HIV-1 transcription in activated T cells [31]. Experimental knockdown of SREBP2 expression by small interfering RNA (siRNA) resulted in a significant reduced viral replication. Consecutively, decreased expression of SREBP2 in CCR5-delta32 heterozygotes may contribute to benefits of viral replication and progression to AIDS.
Genes with higher expression in the CCR5-delta32 group
In the CCR5-del32 group, 96 genes have been found to be overexpressed and four of them had a link to HIV-1 infection (Table 4).
Table 4.
Four genes were found with link to HIV-1 in the CCR5-delta32 heterozygous group
| Significant higher Expression in CCR5-delta32 samples | ||||
|---|---|---|---|---|
| Gene |
Function (link to HIV-1) |
Beneficial effect for CCR5-delta32 carriers |
Reference |
|
| Transm. |
Repli. |
|||
| U2AF65 |
Splicing factor (enhances with the polypyrimidine tract of the splicosome of several HIV-1 genes) |
Unknown |
[35] |
|
| CD30L |
Regulation immune response (elevated CD30 levels during acute HIV-1 infection) |
Unknown |
[36] |
|
| PRKACB |
Catalytic subunit of camp-dependent protein kinase (coexpression with the HDF GliPR) |
Unknown |
[37] |
|
| TAP | Antigen presentation (HIV-1 abrogates TAP mediated peptide transport to MHC I presentation) | Possible | [38] | |
Only for TAP there was a beneficial effect pre-described in terms of viral transmission. For the other three genes there are currently no data concerning a beneficial effect available. (transm. = transmission, repli. = replication).
PRKACB was significantly higher expressed in the CCR5-delta32 group. PRKACB is one of five human C subunits of the PKA group which acts as a serine/threonine kinase but only PRKACB together with PRKACA and PRKX do act as functional protein kinases [39]. Concerning HIV-1 pathogenesis this kinase has been described to be involved in infection such as activating viral transcription of monocytes/macrophages in vitro, or activation of T-cells in HIV-patients with ongoing HAART [40,41]. Most interestingly is the transcriptional regulation of CCR5 expression through a cAMP/PKA/CREB dependent pathway. [42] The CCR5 promoter region contains an cyclic AMP responsive element which could be critical for the regulation of gene expression [43]. Although authors could show that a temporal increase of CCR5 expression could be mediated by PKA, the role of differentially regulated PRKACB in HIV-1 pathogenesis still needs to be further determined.
One possible mechanism of HIV-1 escape from immune recognition and consecutive cytolysis is downregulation of the MHC I expression. TAP as a transporter for antigen presentation was overexpressed in the CCR5-delta32 carrier group. It is known that HIV-1 infection abolishes the ability of cells to translocate antigenic peptides via TAP to the endoplasmatic reticulum and thus indirectly abrogates peptides from MHC I presentation [38]. Overexpression of TAP in heterozygotes might partially compensate this HIV-induced inhibition leading to an enhanced clearance of infected cells and indirectly to a decreased susceptibility in this group.
U2AF65 is a 65 kDa auxiliary factor of pre-mRNA splicing and contains a sequence-specific RNA-binding region with 3′ RNA recognition motifs and an Arg/Ser-rich domain necessary for splicing. The spliced mRNA species in HIV-1 infected cells are env/vpu, nef, rev, vpr, and vif. The binding of a 35 kDa subunit stabilizes the binding of U2AF65 with the polypyrimidine tract (PPT) of introns during spliceosome assembly [35,44]. However, there are no reports that altered expression of U2AF65 contributes to HIV-1 viral progression or susceptibility of transmission.
CD30L is mainly expressed in high levels on activated T cells interacting either with membrane bound CD30 from B or T lymphocytes or soluble CD30. Soluble CD30 has been described to be expressed in high amounts during acute HIV-1 infection [36]. Recently, it has been reported that specified CD4 and CD8 T cell clones are capable of producing T helper (Th)2-type cytokines, which release soluble CD30 into the circulation [45]. The general function of the CD30/CD30L interaction is still unsolved. It has been suggested that this interaction promotes secondary humoral immune responses. However, an in vivo test system with CD30-/- knockout mice or anti CD30L antibodies have led to conflicting results [46]. Taken together, there is no convincing explanation whether the CD30L overexpression in CCR5-delta32 heterozygotes may contribute to a potential benefit in these patients or not.
Conclusions
We identified 7 genes previously found to be associated with HIV-1 pathogenesis and furthermore were differentially expressed in CCR5 wild type and CCR5-delta32 heterozygous individuals. Out of these two, CXCR2 and TAP may have a beneficial effect concerning HIV-1 transmission, and two genes, TOP1 and SREBF2, may be favorable in terms of replication and disease progression in heterozygous CCR5-delta32 carriers. Concerning the other three genes, there was no evidence of a beneficial effect described so far. The clinical relevance of these findings should be further tested in prospective studies.
Methods
Specimen
Input material for this analysis were immunomagnetically purified CD34+ cells from bone marrow aspiration of 19 healthy volunteers, 9 male 10 female, aged 19–85 (median 25) years. All donors gave written informed consent before investigation. Prior to CD34+ selection, mononuclear cells were isolated by density gradient centrifugation through Ficoll-Hypaque (Biochrom, Berlin, Germany). The study protocol was approved by the Ethical committee II, Medical Faculty Mannheim, Heidelberg University Institutional Review Board.
CCR5 genotyping analysis
Genomic DNA was extracted from heparinised peripheral blood monocytes (PBMC) of the donors with the QIAGEN-Blood-Midi-Kit (Qiagen, Germany). Screening of the donors for the CCR5-delta32-allele was performed with a genomic PCR using primers flanking the site of the deletion (forward: 5′-CTCCCAGGAATCATCTTTACC-3′, reverse: 5′-TCATTTCGACACCGAAGCAG-3′) leading to a PCR fragment of 200 base pairs (bp) for the CCR5-allele and of 168 bp in case of a delta32 deletion. Results were confirmed by allele specific PCR and by direct sequencing using the BigDye®-Terminator-1.1.-Cycle-Sequencing-Kit (Applied Biosystems, Germany). Sequences were analyzed using the Vector-NTI-Contig-Express-software (Invitrogen, Germany).
RNA preparation and array based gene expression analysis
Total RNA was extracted from purified CD34+ hematopoietic progenitor cells using TRIzol (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s protocol. The quality of RNA was determined by the 2100 Bioanalyzer system (Agilent Technologies, Waldbronn, Germany) and only samples showing no RNA-degradation were included into the analysis. Oligonucleotide microarrays (HG-U133plus 2.0, Affymetrix Inc., Santa Clara, CA) were hybridized as described previously [47]. Data analysis was performed by the Microarray Suite 5.0 (Affymetrix), and the Genespring software 4.2 (Silicon Genetics, Redwood City, CA). The quality control parameters were in accordance to the MIAME consensus criteria for micro array data with a present call rate of at least 25% [48]. Previously, our group has shown a close correlation of micro-array data compared to quantitative PCR methods in the same sample [49].
Statistics
All samples were normalized with expression values raised to an arbitrary value of 1. Only expression values which reach a present call rate of 75% have been used. Only genes with a significant three-fold difference of expression (p < 0.05) between the CCR5 wild type and the CCR5-delta32 group were eligible for further statistical analysis. Expression analysis of the different groups was performed by using the Mann Whitney test.
Competing interests
The authors confirm that this article content has no conflicts of interest.
Authors’ contributions
GH designed to study and wrote the manuscript, CB performed CCR5 genotyping, MN analyzed micro array data, DN performed the micro array, MR and HK Contributed to conception and design of the study and interpretation of the data. Revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Supplementary Material
Results of the Affymetrix Micro Array Assay. Only expressions with 3fold difference, present call rate of 75%, and p < 0.05 have been noted.
Contributor Information
Gero Hütter, Email: gero.huetter@medma.uni-heidelberg.de.
Christian Blüthgen, Email: chris.bluethgen@gmail.com.
Martin Neumann, Email: m.neumann@charite.de.
Mark Reinwald, Email: mark.reinwald@medma.uni-heidelberg.de.
Daniel Nowak, Email: daniel.nowak@medma.uni-heidelberg.de.
Harald Klüter, Email: h.klueter@blutspende.de.
Acknowledgements
This work was supported by the Bill & Melinda Gates Foundation (Grand Challenges Explorations Grant Number: OPP1033364 ). We thank Susanne Ganepola for proofreading the manuscript.
References
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bream JH, Young HA, Rice N, Martin MP, Smith MW, Carrington M, O’Brien SJ. CCR5 promoter alleles and specific DNA binding factors. Science. 1999;284:223. doi: 10.1126/science.284.5412.223a. [DOI] [PubMed] [Google Scholar]
- Carrington M, Dean M, Martin MP, O’Brien SJ. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8:1939–1945. doi: 10.1093/hmg/8.10.1939. [DOI] [PubMed] [Google Scholar]
- Delobel P, Sandres-Saune K, Cazabat M, Pasquier C, Marchou B, Massip P, Izopet J. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2005;38:382–392. doi: 10.1097/01.qai.0000152835.17747.47. [DOI] [PubMed] [Google Scholar]
- Lin YL, Mettling C, Portales P, Reynes J, Clot J, Corbeau P. Cell surface CCR5 density determines the postentry efficiency of R5 HIV-1 infection. Proc Natl Acad Sci USA. 2002;99:15590–15595. doi: 10.1073/pnas.242134499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J, Portales P, Segondy M, Baillat V, Andre P, Avinens O, Picot MC, Clot J, Eliaou JF, Corbeau P. CD4 T cell surface CCR5 density as a host factor in HIV-1 disease progression. Aids. 2001;15:1627–1634. doi: 10.1097/00002030-200109070-00004. [DOI] [PubMed] [Google Scholar]
- Venkatesan S, Petrovic A, Van Ryk DI, Locati M, Weissman D, Murphy PM. Reduced cell surface expression of CCR5 in CCR5Delta 32 heterozygotes is mediated by gene dosage, rather than by receptor sequestration. J Biol Chem. 2002;277:2287–2301. doi: 10.1074/jbc.M108321200. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth RJ, Collman RG, Doms RW, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Trecarichi EM, Tumbarello M, De Gaetano DK, Tamburrini E, Cauda R, Brahe C, Tiziano FD. Partial protective effect of CCR5-Delta 32 heterozygosity in a cohort of heterosexual Italian HIV-1 exposed uninfected individuals. AIDS Res Ther. 2006;3:22. doi: 10.1186/1742-6405-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, MacGregor RR, Burger H, Mick R, Doms RW, Collman RG. CCR5 genotypes in sexually active couples discordant for human immunodeficiency virus type 1 infection status. J Infect Dis. 1997;176:1093–1096. doi: 10.1086/516519. [DOI] [PubMed] [Google Scholar]
- Philpott S, Weiser B, Tarwater P, Vermund SH, Kleeberger CA, Gange SJ, Anastos K, Cohen M, Greenblatt RM, Kovacs A, Minkoff H, Young MA, Miotti P, Dupuis M, Chen CH, Burger H. CC chemokine receptor 5 genotype and susceptibility to transmission of human immunodeficiency virus type 1 in women. J Infect Dis. 2003;187:569–575. doi: 10.1086/367995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, Buchbinder S, Koblin B, Seage GR III. Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. J Acquir Immune Defic Syndr. 2001;27:472–481. doi: 10.1097/00126334-200108150-00009. [DOI] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- Oh DY, Jessen H, Kucherer C, Neumann K, Oh N, Poggensee G, Bartmeyer B, Jessen A, Pruss A, Schumann RR, Hamouda O. CCR5Delta32 genotypes in a German HIV-1 seroconverter cohort and report of HIV-1 infection in a CCR5Delta32 homozygous individual. PLoS One. 2008;3:e2747. doi: 10.1371/journal.pone.0002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel J, Langendam MW, Coutinho RA, Krol A, Brouwer M, Schuitemaker H. No evidence for an effect of the CCR5 delta32/+ and CCR2b 64I/+ mutations on human immunodeficiency virus (HIV)-1 disease progression among HIV-1-infected injecting drug users. J Infect Dis. 1999;179:825–831. doi: 10.1086/314658. [DOI] [PubMed] [Google Scholar]
- Sei S, Boler AM, Nguyen GT, Stewart SK, Yang QE, Edgerly M, Wood LV, Brouwers P, Venzon DJ. Protective effect of CCR5 delta 32 heterozygosity is restricted by SDF-1 genotype in children with HIV-1 infection. Aids. 2001;15:1343–1352. doi: 10.1097/00002030-200107270-00003. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien SJ. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene: hemophilia growth and development study, multicenter AIDS cohort study, multicenter hemophilia cohort study, San Francisco City cohort, ALIVE study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Misrahi M, Teglas JP, N’Go N, Burgard M, Mayaux MJ, Rouzioux C, Delfraissy JF, Blanche S. CCR5 chemokine receptor variant in HIV-1 mother-to-child transmission and disease progression in children: French pediatric HIV infection study group. Jama. 1998;279:277–280. doi: 10.1001/jama.279.4.277. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Dean M. In search of AIDS-resistance genes. Sci Am. 1997;277:44–51. doi: 10.1038/scientificamerican0997-44. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- Hütter G, Schneider T, Thiel E. Transplantation of selected or transgenic blood stem cells - a future treatment for HIV/AIDS? J Int AIDS Soc. 2009;12:10. doi: 10.1186/1758-2652-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hütter G, Symond G. In: Treatment strategies: AIDS. 2. Hajba L, editor. London: The Cambridge Research Centre; 2011. Progress in HIV gene therapy; pp. 44–46. [Google Scholar]
- Ando D, Tang WW, Stein D, Wang S, Lee G, Holmes M, Giedlin M, Gregory P, Tabas P, June C. HAART Treatment Interruption following Adoptive Transfer of Zinc Finger Nuclease (ZFN) Modified Autologous CD4 T-cells (SB-728-T) to HIV-infected Subjects Demonstrates Durable Engraftment and Suppression of Viral Load [abstract] Chicago: Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC); 2011. 17–20 September. [Google Scholar]
- Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, Gallien S, Lin NH, Giguel FF, Lavoie L, Ho VT, Armand P, Soiffer RJ, Sagar M, Lacasce AS, Kuritzkes DR. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207:1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich T, Hanhauser E, Sirignano M, Davis B, Lee TH, Keating S, Busch M, Marty F, LaCasce A, Armand P, Soiffer R, Altfeld M, Kuritzkes D. In depth investigation of peripheral and gut HIV-1 reservoirs, HIV-specific cellular immunity, and host microchimerism following allogeneic hematopoetic stem cell transplantation. Kuala Lumpur: 7th International AIDS Society Conference on HIV Pathogenesis, Treatment and Prevention; 2013. [Google Scholar]
- Robertson MJ. Role of chemokines in the biology of natural killer cells. J Leukoc Biol. 2002;71:173–183. [PubMed] [Google Scholar]
- Attal H, Cohen-Hillel E, Meshel T, Wang JM, Gong W, Ben-Baruch A. Intracellular cross-talk between the GPCR CXCR1 and CXCR2: role of carboxyl terminus phosphorylation sites. Exp Cell Res. 2008;314:352–365. doi: 10.1016/j.yexcr.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Richardson RM, Tokunaga K, Marjoram R, Sata T, Snyderman R. Interleukin-8-mediated heterologous receptor internalization provides resistance to HIV-1 infectivity: role of signal strength and receptor desensitization. J Biol Chem. 2003;278:15867–15873. doi: 10.1074/jbc.M211745200. [DOI] [PubMed] [Google Scholar]
- Caccuri F, Giagulli C, Bugatti A, Benetti A, Alessandri G, Ribatti D, Marsico S, Apostoli P, Slevin MA, Rusnati M, Guzman CA, Fiorentini S, Caruso A. HIV-1 matrix protein p17 promotes angiogenesis via chemokine receptors CXCR1 and CXCR2. Proc Natl Acad Sci USA. 2012;109:14580–14585. doi: 10.1073/pnas.1206605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana B, Zhuang L, Moitra PK, Stantchev TS, Broder CC, Cutler ML, D’Arpa P. Human TOP1 residues implicated in species specificity of HIV-1 infection are required for interaction with BTBD2, and RNAi of BTBD2 in old world monkey and human cells increases permissiveness to HIV-1 infection. Virol J. 2010;7:332. doi: 10.1186/1743-422X-7-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HE, Linde ME, Khatua AK, Popik W, Hildreth JE. Sterol regulatory element-binding protein 2 couples HIV-1 transcription to cholesterol homeostasis and T cell activation. J Virol. 2011;85:7699–7709. doi: 10.1128/JVI.00337-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoya Y, Tokunaga K, Sawa H, Maeda M, Ueno T, Yoshikawa T, Hasegawa H, Sata T, Kurata T, Hall WW, Cullen BR, Takahashi H. Human topoisomerase I promotes HIV-1 proviral DNA synthesis: implications for the species specificity and cellular tropism of HIV-1 infection. Proc Natl Acad Sci USA. 2003;100:8442–8447. doi: 10.1073/pnas.1430827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Matsuda M, Kojima A, Sata T, Andoh T, Kurata T, Nagashima K, Hall WW. Human immunodeficiency virus type 1 reverse transcriptase: enhancement of activity by interaction with cellular topoisomerase I. Proc Natl Acad Sci USA. 1995;92:5694–5698. doi: 10.1073/pnas.92.12.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks T, Kelly J, Benn S, Kinter A, Justement J, Gold J, Redfield R, Sell KW, Fauci AS. Susceptibility of normal human lymphocytes to infection with HTLV-III/LAV. J Immunol. 1986;136:4049–4053. [PubMed] [Google Scholar]
- Domsic JK, Wang Y, Mayeda A, Krainer AR, Stoltzfus CM. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol. 2003;23:8762–8772. doi: 10.1128/MCB.23.23.8762-8772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzolo G, Vinante F, Nadali G, Krampera M, Morosato L, Chilosi M, Raiteri R, Sinicco A. High serum level of soluble CD30 in acute primary HIV-1 infection. Clin ExpImmunol. 1997;108:251–253. doi: 10.1046/j.1365-2249.1997.d01-1005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo G, Muller-Kuller T, Dietrich U, Hoelzer D, Ottmann OG, Scheuring UJ. Inhibition of HIV-1 replication by small interfering RNAs directed against glioma pathogenesis related protein (GliPR) expression. Retrovirology. 2010;7:26. doi: 10.1186/1742-4690-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O, Vey T, Kerkau T, Hunig T, Schimpl A. HIV type 1 abrogates TAP-mediated transport of antigenic peptides presented by MHC class I: transporter associated with antigen presentation. AIDS Res Hum Retroviruses. 2002;18:1319–1325. doi: 10.1089/088922202320886361. [DOI] [PubMed] [Google Scholar]
- Soberg K, Jahnsen T, Rognes T, Skalhegg BS, Laerdahl JK. Evolutionary paths of the cAMP-dependent protein kinase (PKA) catalytic subunits. PLoS One. 2013;8:e60935. doi: 10.1371/journal.pone.0060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson CC, Bryn T, Yndestad A, Eiken HG, Bjerkeli V, Froland SS, Aukrust P, Tasken K. Cytokine networks are pre-activated in T cells from HIV-infected patients on HAART and are under the control of cAMP. Aids. 2004;18:171–179. doi: 10.1097/00002030-200401230-00005. [DOI] [PubMed] [Google Scholar]
- Rabbi MF, Al-Harthi L, Saifuddin M, Roebuck KA. The cAMP-dependent protein kinase A and protein kinase C-beta pathways synergistically interact to activate HIV-1 transcription in latently infected cells of monocyte/macrophage lineage. Virology. 1998;245:257–269. doi: 10.1006/viro.1998.9158. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Pirrone V, Wigdahl B, Nonnemacher MR. Transcriptional regulation of the chemokine co-receptor CCR5 by the cAMP/PKA/CREB pathway. Biomed Pharmacother. 2011;65:293–297. doi: 10.1016/j.biopha.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers HF, Biesta PJ, Montagne LJ, Van Haastert ES, Van Der Valk P, Van Den Elsen PJ. CC chemokine receptor 5 gene promoter activation by the cyclic AMP response element binding transcription factor. Blood. 2008;112:1610–1619. doi: 10.1182/blood-2008-01-135111. [DOI] [PubMed] [Google Scholar]
- Jenkins JL, Agrawal AA, Gupta A, Green MR, Kielkopf CL. U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res. 2013;41:3859–3873. doi: 10.1093/nar/gkt046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romagnani S, Del Prete G, Maggi E, Chilosi M, Caligaris-Cappio F, Pizzolo G. CD30 and type 2 T helper (Th2) responses. J Leukoc Biol. 1995;57:726–730. doi: 10.1002/jlb.57.5.726. [DOI] [PubMed] [Google Scholar]
- Kennedy MK, Willis CR, Armitage RJ. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology. 2006;118:143–152. doi: 10.1111/j.1365-2567.2006.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WK, De Vos S, Komor M, Hoelzer D, Wachsman W, Koeffler HP. Characterization of gene expression of CD34+ cells from normal and myelodysplastic bone marrow. Blood. 2002;100:3553–3560. doi: 10.1182/blood.V100.10.3553. [DOI] [PubMed] [Google Scholar]
- Brazma A. Minimum information about a microarray experiment (MIAME)–successes, failures, challenges. Scientific World J. 2009;9:420–423. doi: 10.1100/tsw.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueller S, Komor M, Nowak D, Baldus CD, De Vos S, Hoelzer D, Ottmann OG, Hofmann WK. Identification of defects in the transcriptional program during lineage-specific in vitro differentiation of CD34(+) cells selected from patients with both low- and high-risk myelodysplastic syndrome. Exp Hematol. 2010;38:718–732. doi: 10.1016/j.exphem.2010.04.018. 732 e711-716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of the Affymetrix Micro Array Assay. Only expressions with 3fold difference, present call rate of 75%, and p < 0.05 have been noted.