Abstract
During their life cycle, trypanosomes must overcome conflicting demands to ensure their survival and transmission. First, they must evade immunity without overwhelming the host. Second, they must generate and maintain transmission stages at sufficient levels to allow passage into their tsetse vector. Finally, they must rapidly commit to onward development when they enter the tsetse fly. On the basis of recent quantification and modelling of Trypanosoma brucei infection dynamics, we propose that the interplay between immune evasion and development achieves both infection chronicity and transmissibility. Moreover, we suggest that a novel form of bistable regulation ensures developmental commitment on entry into the tsetse fly midgut.
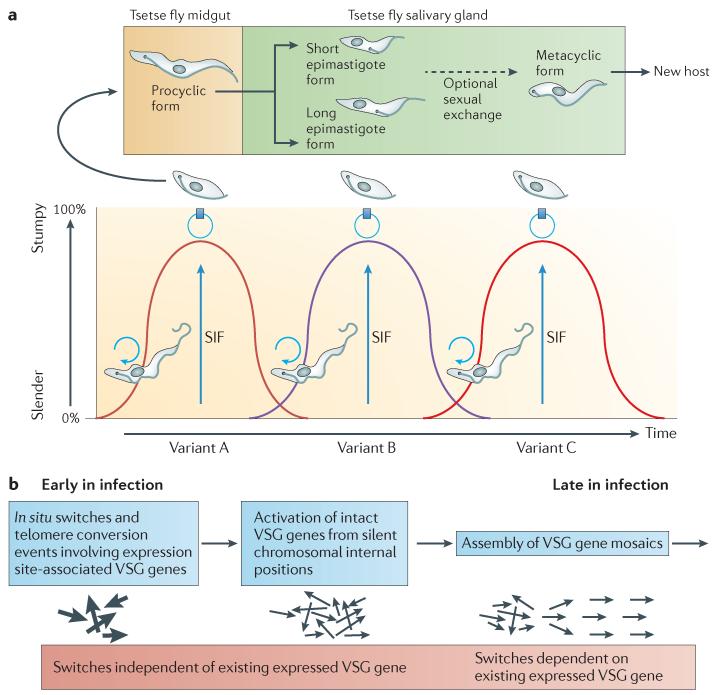
African trypanosomes, the causative agents of human African trypanosomiasis (HAT), remain a scourge in sub-Saharan Africa. Although HAT has been receding in recent years1, livestock infections remain prevalent and impose major restrictions on economic development in afflicted regions2. The distribution of the disease is governed by the distribution of the tsetse fly, the blood-feeding dipteran vector for the parasites. In the mammalian host, the parasites exist as proliferative slender forms, which establish parasitaemia, and transmissible stumpy forms, which are irreversibly arrested in their cell cycle and arise as parasite numbers increase. The transition from slender to stumpy forms seems to be the result of a density-sensing mechanism, whereby the parasites release a factor (or factors) — termed stumpy induction factor (SIF) — that accumulates as parasite numbers increase during the parasitaemia3,4. When SIF triggers the parasites to differentiate into stumpy forms, growth of the trypanosomal population is restricted (and thus host survival is prolonged) and transmission stages become predominant. Following transmission to tsetse flies, the parasites undergo cyclic development over 20–30 days5; this involves proliferation in the midgut as procyclic forms and then migration to the salivary glands, where epimastigotes attach to the salivary gland wall and multiply. At this stage the parasite can undergo meiosis and engage in sexual exchange6 before developing to detached, non-dividing metacyclic forms. These are adapted for survival on entering a new mammalian host, which occurs when they are ejected in the tsetse fly anticoagulant saliva during a blood meal by the fly (FIG. 1a).
Figure 1. The life cycle and immune evasion of African trypanosomes.
a | African trypanosomes are extracellular parasites that survive through their capacity for antigenic variation. During an infection, a procession of distinct antigen types emerges, each controlled by the host immune response. Simplistically, this generates an undulating parasitaemia. In each wave of parasitaemia, proliferative slender forms are responsible for the increase in parasite numbers. This is accompanied by the accumulation of a parasite-derived factor, stumpy induction factor (SIF), that stimulates slender cells to transform to stumpy forms. Stumpy forms are non-proliferative, being arrested in G0–G1, and are the transmission stage of the parasite, capable of developing further when ingested by the tsetse fly during a blood meal. Stumpy cells are irreversibly committed to cell cycle exit in the bloodstream and are removed from the population by a combination of immune clearance and cell ageing. When stumpy forms are taken up by a tsetse fly, they differentiate to midgut procyclic forms; slender forms that are taken up are killed. The procyclic forms proliferate before arresting and migrating to the salivary glands, where they attach as short epimastigote forms. These develop to metacyclic forms, which are unattached to the salivary gland wall and infective to mammals. b | During a trypanosomal infection in mammalian hosts, the probability of expression for variant surface glycoprotein (VSG) genes is governed by the activation mechanism of each gene. Early in the infection, in situ switches allow telomeric VSG genes that are encoded in expression sites to be activated. Subsequently, intact VSG genes that are encoded in subtelomeric regions are expressed. Later in infection, mosaic VSG genes can contribute to the repertoire. These are assembled from incomplete VSG genes, and their activation can be dependent on the previously active VSG gene (as they are created through homologous recombination), resulting in a ‘string’ of related VSG genes being expressed.
In this Opinion article we use the evidence from recent experimental and mathematical analyses to suggest how trypanosomes balance antigenic variation and developmental transitions to achieve infection chronicity, and how they ensure transmission through their irreversible commitment to development in the tsetse fly.
Antigenic variation versus development
Trypanosomal infections are sustained for months to years in mammalian hosts, despite the parasites being extracellular and exposed to the immune system in the circulation and tissues. This is possible because trypanosomes exhibit an extreme capacity for antigenic variation, whereby the proteins that constitute their surface coat (variant surface glycoproteins (VSGs)) are periodically changed7. This is achieved by changing the expressed VSG gene; the parasite genome has at least 15 telomeric VSG expression sites, each containing a VSG gene8, but only one of these sites is active at any one time9-11. By changing the active expression site, a change in VSG expression can be achieved in situ without DNA recombination. However, as well as the expression site-associated VSG genes, the trypanosome genome houses a pool of several hundred complete and incomplete VSG genes that can be activated only by recombination into an expression site10,11. Recombination is driven by homology in the gene-flanking regions or in the VSG coding regions themselves, enabling the creation of mosaic VSG genes through the assembly of related complete and incomplete silent VSG genes12-14. Together, these mechanisms combine to allow a vast range of VSG types to be expressed by the parasites, such that the population can inevitably outpace the immune system and ensure the maintenance of a chronic infection15.
The periodic control of distinct antigenic variants by antibodies creates an undulating parasitaemia profile that is characteristic of trypanosome infections (FIG. 1a). Another factor that contributes to this profile is the developmental transition between proliferative slender forms and non-proliferative stumpy forms of the parasites16-19. Stumpy cells are more resistant to antibody-dependent complement-mediated killing than slender forms and persist at the peak of each parasitaemia cycle as slender cells are killed20,21. Nonetheless, the stumpy forms are eventually removed by either immune clearance or cell senescence, generating a decrease in parasitaemia, which is recovered only after the outgrowth of a new population of slender cells that have undergone antigen switching.
This cycle of antigenic variation and the transition between slender and stumpy forms prolongs the infection and optimizes the potential for parasite transmission. However, until recently, the contributions of each parameter to the chronicity of infection could not be quantified, and the interactions between these parameters that combined to ensure transmission were unclear.
Infection chronicity and transmission
The contribution of antigenic variation to within-host dynamics
The various routes of VSG gene activation generate differential probabilities for the expression of different antigen types14,22,23 (FIG. 1b). For example, intact VSG genes in silent expression sites have a high probability of expression through in situ activation14. Similarly, intact VSG genes in subtelomeric sites can be readily activated through gene conversion events that are initiated in their extensive flanking homologous regions, which include repeat sequences such as the 70 bp repeats24, subtelomeric repeats and VSG conserved 3′ elements12. As these gene conversion events are based on gene-flanking homologies, they allow VSG gene switching independently of VSG gene sequence, such that the direction and predictability of switching events is limited22,25.
In contrast to these high-probability switches, the assembly of intact mosaic VSG genes is much less likely and is dependent on the existing expressed VSG gene, driving the expression of related VSG variants (‘strings’) that require coding-region similarity with, but exhibit distinct antigenicity from, the previously expressed VSG gene22,26.
Mathematical modelling of the impact of these different activation mechanisms predicts that a hierarchical expression of VSG types is generated25. Unpredictable high-frequency switches predominate early in the infection (when switching occurs in a ‘block’ of different antigen types, the genes of which are not related in sequence but are activated with similar probabilities) and are replaced later in the infection by switches that are determined by the previously expressed VSG gene25 (owing to the requirement for sequence homology). The relationship between the different VSG genes in given sequence similarity groups, as well as the ability of the immune system to discriminate between VSG types in or between these groups, is predicted to have a large influence on infection dynamics27. For example, a strong specific immune response could control waves of parasitaemia caused by only one or a few VSG types (such that the antigen types reach the threshold required to induce effective antibody production), whereas more diverse parasite populations (which do not reach the antibody induction threshold for any one antigen type) would be controlled by stumpy formation and senescence. Because the VSG gene expressed by the parasite population progresses through different block sizes during the course of infection, this creates the potential for considerable variation in the parasite load, the proportion of transmissible stumpy forms and the strength of host immunity generated to particular antigen types27.
The contribution of differentiation to within-host dynamics
The developmental switch from slender to stumpy forms is a key additional component of mathematical models for antigen switching and infection chronicity25,28-31. This is because the frequency of each developmental type in the population determines the overall growth of the population (as only slender cells proliferate)32, the number of parasites available for functional switching (as arrested stumpy cells are incapable of initiating new variant populations) and the kinetics of immune clearance (as slender and stumpy forms have different sensitivities to antibody-dependent killing)20,21. However, until recently, these components could not be accurately quantified because the relationship between morphology, cell cycle exit and transmission competence could not be easily determined. Fortunately, the recent identification of the PAD (proteins associated with differentiation) proteins has provided a molecular tool that can both discriminate stumpy forms and link these forms to the transmission competence of the parasite18,31.
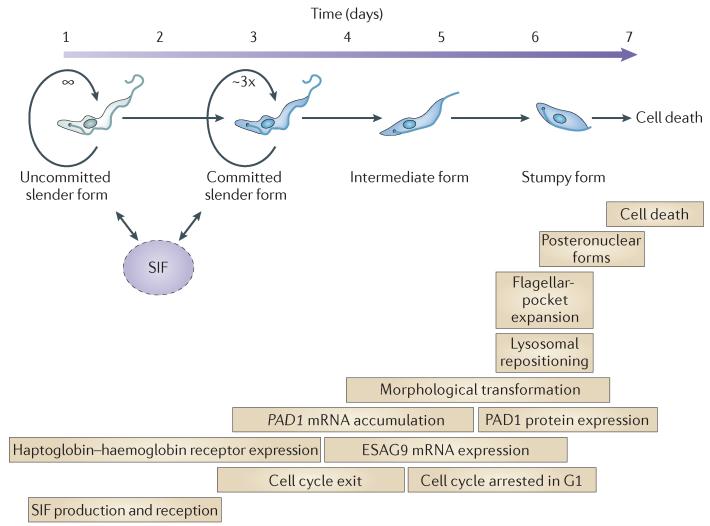
PAD proteins are carboxylate transporters that transduce the CCA (citrate and/or cis-aconitate) signal used by the parasites to detect their entry into the tsetse fly (see below). PAD1 is highly expressed on only stumpy forms of the parasite, and its mRNA is differentially expressed between slender and stumpy forms, enabling the production of stumpy forms to be monitored quantitatively by reverse transcriptase (RT)-PCR31. Combined with cytological analysis of the morphology and cell cycle status of parasites33, the detection of PAD1 mRNA has allowed the temporal order of events to be described early in a mouse infection (FIG. 2).
Figure 2. Cytological events that accompany the transition from slender to stumpy forms.
Uncommitted slender cells release and receive stumpy induction factor (SIF), stimulating them to begin the transition to stumpy forms. Mathematical modelling31 predicts that, initially, cells develop to a committed slender form that is capable of up to three rounds of cell division before cell cycle exit in G1, whereupon SIF production is lost. The resulting intermediate forms no longer express the haptoglobin-haemoglobin receptor but express mRNAs encoding PAD1 (proteins associated with differentiation 1) and ESAG9 proteins. The morphological transformation to stumpy forms involves the production of a broader cell form with a prominent, undulating membrane and the development of the mitochondrion and of diaphorase activity. As stumpy forms mature, their flagellar pocket increases in size and the lysosome is repositioned. In some cases, the nucleus moves to the cell posterior, displacing the terminal kinetoplast and generating posteronuclear stumpy forms. Stumpy cells are eventually removed from the bloodstream; mathematical modelling predicts a life expectancy of 55 hours31, in agreement with experimental observations of 48-72 hours30.
A few days after the start of the infection, the parasite population is uniformly proliferative and morphologically slender, and little PAD1 mRNA is expressed31. However, by 4 days into the infection, cells are beginning to arrest in G1, as revealed by the accumulation of cells with a single nucleus and a single kinetoplast (the specialist mitochondrial genome of the parasite; the kinetoplast is precisely replicated and segregated during the cell cycle). This arrest precedes morphological transition and represents the first detectable event in the transformation to stumpy forms, matching earlier in vitro predictions4. Between day 4 and day 5 of infection, arrested forms continue to accumulate, this being coincident with an increase in the abundance of PAD1 mRNA. Interestingly, this occurs as morphologically ‘intermediate’ forms peak, before the accumulation of morphologically stumpy forms and before the appearance of PAD1 protein. During this intermediate phase, PAD1 mRNA levels are around tenfold higher than the levels in the early slender population, but thereafter remain at this higher level and do not accumulate further as cells develop to stumpy forms31. PAD1 mRNA abundance therefore provides an early and reliable indicator of the commitment to the generation of stumpy forms, with protein expression from the transcripts being dependent on translational control. A similar mRNA profile is also shown by ESAG9 genes34,35, a diverse family of genes that were originally identified as occasional components of VSG expression sites but are also expressed from elsewhere in the genome34,36, whereas the expression of the slender-specific haptoglobin-haemoglobin receptor37 is downregulated in intermediate forms33. Following these gene expression changes, during days 6–7 cells progress morphologically to stumpy forms; characteristic changes include an expanded flagellar pocket, mitochondrial elaboration38, a reduction in flagellar length39,40, the development of a pronounced undulating membrane, and repositioning of the expanded lysosome41 to a posteronuclear position33. The nucleus itself also moves towards the posterior of the cell and in extreme cases can displace the kinetoplast at the posterior end19. These events establish a temporal description of the slender-to-stumpy transition, at least in the first wave of infection (FIG. 2).
The quantitative analysis of PAD expression data has also enabled mathematical modelling of the dominance of stumpy forms in chronic infections31, complementing the earlier models of infection dynamics that were based on antigen switching. A model based on Bayesian inference was used to infer kinetic parameters of the developmental processes, and this model, combined with the experimental dissection of the early events in the establishment of infection, generated a quantitative description of infection that fits well with published values. In the derived model (FIG. 2), slender cells commit to the production of stumpy forms before their cell cycle exit, with the committed cells replicating around three times before their arrest in G1. Although earlier mathematical models had incorporated a density-dependent differentiation component25,42, the fit of a cell density model to the experimental data was poor, whereas the prediction of a soluble density-dependent signal provided a substantially better fit, supporting the SIF concept. Slender cells and committed slender cells are predicted to produce SIF, whereas intermediate and stumpy forms are not. When parasites commit, the development to morphologically stumpy forms is completed in 70 hours, and the resultant stumpy forms are predicted to have a finite lifespan of 50 hours, a value that approximates well to the experimentally determined lifespan of stumpy cells30.
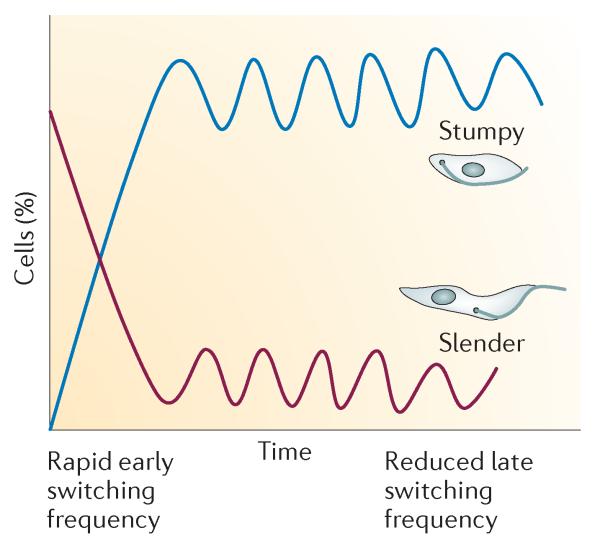
Perhaps most striking was the observation that, after the initial wave of parasitaemia, stumpy cells dominate chronic infections. An immediate implication is that the population frequency of antigenic variation will be considerably less than that if slender cells predominate. Although the mathematical models of the interaction between antigenic variation and the immune response are quite simplistic in this study, they nonetheless support there being a reduced capacity for switching in chronic infections, in which stumpy cells dominate.
How antigenic variation and differentiation ensure chronicity
These quantitative descriptions of the production and maintenance of transmission stages in trypanosomal infections have striking implications for the life history of the parasites and their epidemiology (FIG. 3). According to analysis of the first wave of parasitaemia, the early dominance of slender forms would ensure that a new host was infected by parasites that were undergoing rapid antigen switching. This is because the rate of antigen switching in natural slender trypanosomal populations is high (approximately once per 1,000 divisions43, contrasting with once per 1 million divisions for laboratory-adapted lines44,45). In the context of a host population with prior exposure to trypanosomes and the potential prevalence of herd immunity26,46, this would allow the trypanosomal population to probe the immune environment of the host using a large number of VSG types, and only cells expressing VSG types to which the host had not already been exposed would survive. This would allow the infection to get a foothold and parasite numbers to increase (FIG. 3), enabling novel mosaic VSG strings to be generated and expressed.
Figure 3. The contribution of slender and stumpy forms to a chronic infection.
Early in infection, slender cells dominate and undergo rapid switching in their variant surface glycoproteins (VSGs), assisting the establishment of the parasite population in a host that has been exposed to earlier infections or has a current infection. When the parasite population is established, it becomes predominantly stumpy, ensuring the transmissibility of the parasite (as stumpy forms are optimized for uptake by the tsetse fly vector). The small number of slender forms that are still present undergo VSG switching at a high rate and thus continue to feed new antigenic variants into the bloodstream to maintain a chronic infection.
When parasite numbers increase through the outgrowth of antigenically viable slender cells, the accumulation of SIF will stimulate these cells to become intermediate and then stumpy forms in the first wave of parasitaemia, before they are cleared by the immune response. In mice, clearance of the first wave takes 9–10 days31, which is a considerable time after an antibody response would be expected to be generated. This extended survival of the stumpy population might reflect faster hydrodynamic clearance of surface antibody by stumpy cells21 or the antibody response being insufficiently robust to rapidly clear the first wave of parasites. This scenario might be particularly relevant in field infections, in which the establishing population is antigenically heterogeneous, such that multiple antigen types in a diverse antigen block reduce the effective immune response and cause the infection profile in one peak to be dominated by stumpy formation and senescence, rather than immune responsemediated killing27. Although there is evidence for strong specific antibody responses to early variants and to the heterogeneous metacyclic VSG types that initiate infection47, in practice there is likely to be a complex balance between antibody-mediated and developmental control of early parasitaemia, and this balance might be context or infection specific.
After the first wave of parasitaemia, chronic infections show a complex profile in which the high proportion of intermediate and stumpy cells is sustained31 (FIG. 3). This ensures that transmission stages are always available should a tsetse fly take a blood meal48 — an important consideration in locations where the vectorial capacity of tsetse flies is low and effective transmission is unlikely49. Furthermore, a dominance of stumpy forms will considerably reduce the frequency of antigenic variation in the population, as only replicating slender cells are capable of generating new variants. This slows the exposure of the antigen repertoire available in the infecting population and also prevents the host becoming overwhelmed by an onslaught of new variants switching at high frequency27. The infection is sustained, however, because a small, probably fluctuating population of slender cells remains, switching at the high frequency described for natural trypanosomal populations45 (FIG. 3). These slender cells feed new variants into the population, their high switching rate ensuring that the improbable construction of mosaic VSGs that is required in chronic infections can occur and sustain the infection.
This infection profile serves to carefully balance the emergence of new variants with the transmissibility of the parasite population, and the resulting control of the parasite population also extends the life of the host.
Sensing cues for transmission
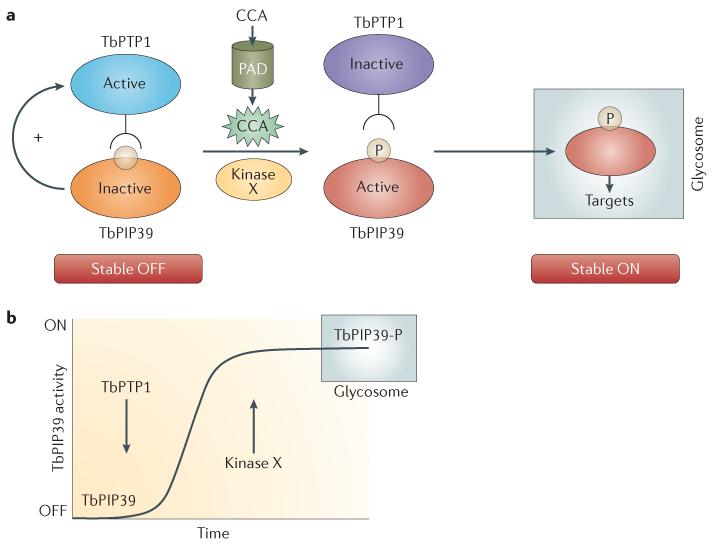
A further balancing act occurs at the molecular level when stumpy trypanosomes initiate differentiation in the tsetse fly midgut. A key trigger for this seems to be blood citrate (present at ~130 μM50) and, potentially, CCA in the tsetse fly midgut51. These metabolites are transported by PAD proteins, which are expressed on stumpy forms, but this transport occurs only on entry into the fly18. This is because trypanosomes become sensitized to CCA when the temperature is lowered to 20 °C52, upregulating the expression of PAD2 and promoting the surface access of this protein18 and potentially of other surface proteins that are important in environmental sensing. CCA seem to affect the activity of trypanosomal phosphotyrosine phosphatase 1 (TbPTP1)53 (FIG. 4). Specifically, TbPTP1 dephosphorylates and thus deactivates its substrate, the serine/threonine-specific phosphatase TbPIP39, to maintain stumpy forms in a state that is poised for differentiation but not yet committed54. Furthermore, the phosphatase activity of TbPTP1 is promoted by the association with its substrate54. TbPIP39 is a member of an unusual class of DXDXT phosphatases, for which structural modelling has predicted the existence of a citratebinding pocket55. Interestingly, the presence of CCA prevents the activation of TbPTP1 by TbPIP39 (REF. 54), allowing TbPIP39 to become (and to remain) phosphorylated at a tyrosine residue by an unidentified kinase. Phosphorylated TbPIP39 is more active than the unphosphorylated form, but both are transported to the glycosome (a peroxisome-like organelle that compart-mentalizes the glycolytic enzymes in trypanosomatids) owing to a carboxy-terminal peroxisomal-targeting signal 1 (PTS1) motif. In the glycosome, the active phosphatase can dephosphorylate targets in that organelle but is itself protected from dephosphorylation through compartmentalization away from TbPTP1. This generates an irreversible commitment to differentiation, creating a molecular bistable switch.
Figure 4. TbPTP1-TbPIP39 signalling and bistability.
a | In stumpy forms of trypanosomes, differentiation is prevented by the activity of trypanosomal phosphotyrosine phosphatase 1 (TbPTP1), which dephosphorylates (and so represses) the serine/threonine-specific phosphatase TbPIP39. The interaction between TbPIP39 and TbPTP1 stimulates the activity of TbPTP1, thus reinforcing the inactivation of TbPIP39 and generating a stable OFF state. In the presence of CCA (citrate and/or cis-aconitate), the stimulation of TbPTP1 is lost, potentially through steric interference between TbPIP39 and TbPTP1 (generated by the presence of a TbPIP39 citrate-binding pocket). This favours the phosphorylation (and so activation) of TbPIP39 by an unknown kinase (kinase X). Both active and inactive TbPIP39 are unidirectionally trafficked to the glycosome, where active TbPIP39 dephosphorylates unidentified glycosomal targets. As phosphorylated TbPIP39 is locked in the glycosomes, it cannot be dephosphorylated by TbPTP1, thus ensuring its irreversible activation. This generates a stable ON state. b | The activity of TbPIP39 in response to the actions of TbPTP1 and kinase X. TbPIP39 is activated by phosphorylation, but this is rendered irreversible by the compartmentalization of the enzyme in the glycosome. PAD, proteins associated with differentiation.
Bistable switches, in which two alternative states of activation are stable but intermediates between them are not, govern cell fate decisions in a wide range of biological systems, most notably in controlling the unidirectionality of mitotic events56-58. Similarly to cell cycle progression, developmental transitions in the trypanosomal parasite must be irreversible59 and preferably all-or-none responses, ensuring that cells can lock into the developmental response when triggered by a potentially transient environmental cue. Hence, in the stumpy form TbPIP39 reinforces its own repression by activating TbPTP1, thus stabilizing the OFF state (FIG. 4). By contrast, in the presence of CCA TbPIP39 is activated, generating an ON state. Molecular switches of this kind ordinarily operate in one cellular compartment and thereby require stringent irreversibility. In the trypanosome system, however, irreversibility is ensured by sequestration of phosphorylated TbPIP39 in the glycosome such that TbPTP1 can no longer access its target. This could assist the parasite in responding to a transient CCA signal, and indeed the parasites commit to irreversible differentiation after only 1 hour of exposure to CCA in vitro60. The compartmentalization of the transduced signal also raises further questions. For example, is TbPIP39 targeted to all glycosomes or preferentially associated with a subset that is important for differentiation? This represents an interesting question, because the composition and activity of glycosomes are developmentally regulated, and there is a rapid turnover of these organelles via autophagy during differentiation61. Active TbPIP39 may label the glycosomes in bloodstream-stage cells for degradation, may protect newly formed procyclic glycosomes, and/or may change the metabolic activity of glycosomes through its effects on the constituent enzymes and their interactions with the few other signalling molecules that can be recognized bioinformatically as being glycosome associated62. As the only characterized signalling molecule to target a peroxisome-like organelle in any eukaryotic cell, TbPIP39 has considerable potential to assist our understanding of organelle dynamics and regulation.
Although the TbPTP1-TbPIP39 signalling pathway provides a coherent and straightforward link from an external signal to an internal metabolic organelle, several questions remain unanswered. Beyond the need to identify glycosomal targets of TbPIP39, for example, an alternative potential substrate of TbPTP1 has been identified63, the role (if any) of which in differentiation is unclear. Other differentiation stimuli in addition to CCA have also been described64-68, but their links to TbPTP1–TbPIP39 signalling are unknown. Clearly the interconnections, if there are any, between these different molecules and signals remain to be uncovered.
Why stumpy forms persist
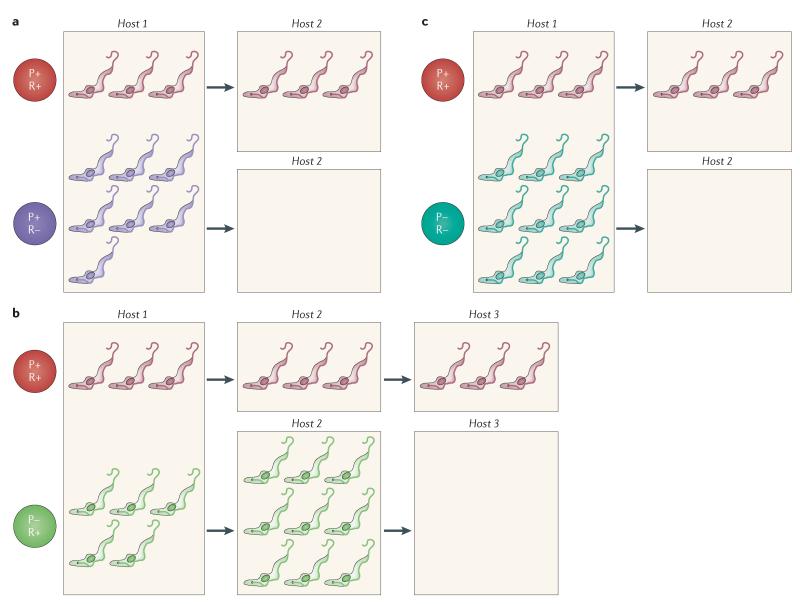
Although there is much interesting literature on the evolution of the VSG archive, the activation mechanisms of this archive and the contributions of these mechanisms to infection chronicity14,22,67, another obvious target for selection is parasite development via SIF signalling (FIG. 5). Mutants that are unable to respond to SIF would have an advantage in the context of a single host, as they would outgrow the background SIF-responsive population (FIG. 5a). Indeed, trypanosomes that are less responsive to SIF have been readily selected in the laboratory and are termed monomorphs68-71. However, the inability of such monomorphs to generate stumpy forms means that these lines would have reduced transmissibility, restricting their spread beyond the originating host and rendering them non-viable in the long term (FIG. 5a). There may also be other selection pressures that operate on SIF signalling. For example, if the production of SIF is costly, it may be advantageous to remain responsive to the signal but to stop producing it, effectively becoming a cheater72. This could provide growth advantages in the host, although these advantages would be of limited evolutionary benefit if the bottlenecks during passage through the tsetse fly73 are such that clonal populations are likely to be transmitted. In this case, the absence of SIF-producing benefactors after transmission of the cheater population would result in cells that are unable to produce stumpy forms and so have limited potential for further transmission (and also have increased virulence) (FIG. 5b). Cells that neither produce nor respond to SIF would have a further short-term advantage but, as monomorphs, would be unable to sustain transmission (FIG. 5c).
Figure 5. The impact of defects in stumpy induction factor production or detection on parasite growth and transmission.
a | Cells that produce and respond to stumpy induction factor (SIF) (P+R+ cells) generate transmissible stumpy forms and are maintained throughout multiple host–vector transactions. If cells no longer respond to SIF (P+R− cells), they gain a short-term advantage because they outgrow the P+R+ cells (which respond to SIF and therefore exit the cell cycle). However, because these P+R− cells produce less transmissible forms, their long-term viability is lower. b | Cells that can respond to the SIF produced by others in the population but do not produce SIF themselves (P−R+ cells) would have a growth advantage if the production of SIF were costly. This could promote their transmission to a new host. However, in this new host the absence of SIF benefactors would reduce the number of transmission stages and so limit the long-term viability of these cells. c | For cells that neither produce nor respond to SIF (P−R− cells), there would be a short-term advantage, as they would continue to proliferate and would out-grow other cells (including P+R−cells) because they do not incur the costs associated with SIF production. However, their transmission potential would be greatly reduced owing to the absence of stumpy forms, selecting against them in the long term.
A more complex possibility arises if cells respond to their own density-sensing signal but not to that of competitor trypanosomal populations. In this scenario, trypanosomes would be able to establish in an already-infected host or, as has been observed74, outcompete other trypanosomal strains in a co-infection. Specialized SIF-producing or SIF-sensing pathways would need to evolve for different strains to prevent interference from each other, but the likelihood of such evolution would depend on the complexity of the signalling system. Alternatively, different trypanosomal populations may exhibit different sensitivities to (or production levels of) SIF in different hosts, such that the parasites optimize their transmission depending on the host context or the probability of co-infection59,75. Although this could involve selection of mutants, epigenetic mechanisms could also be implicated. For example, the VSG expression site is believed to contribute to host optimization76 through the use of microheterogeneity in transferrin receptors that are encoded at this site (ESAG6 or ESAG7)77 or through the presence of the gene encoding serum resistance-associated protein (SRA; a protein that confers resistance to the lytic factor in human serum) in Trypanosoma brucei rhodesiense78,79. Other expression site-associated genes are of unknown specific function but nonetheless show heterogeneity. Hence, if the function of any of these genes is related to SIF production or sensitivity, then the use of the various expression sites could determine transmissibility in different contexts, providing a mechanism for optimization in hosts in which SIF shows differential turnover, for example.
Importantly, these scenarios for interparasite signalling and parasite selection are not merely theoretical concepts. As well as commonly used laboratory strains of T. brucei, several trypanosomes have been reported not to generate stumpy forms or to be of uncertain morphological type, including Trypanosoma congolense80, Trypanosoma vivax81,82 and Trypansoma evansi81. Although T. evansi are mechanically transmitted and so do not require a specialized transmission stage, all of these different trypanosomal species must exhibit growth control if they are to prevent rapid host death. Moreover, the historical literature32,80,82-84 reports that all of these species can show morphological heterogeneity, including the potential to generate growth-limited forms that resemble intermediate forms82, and this is further supported by the detection of ESAG9 transcripts (which are normally enriched in stumpy forms of the trypanosomes that produce these cells) in T. brucei equiperdum, another mechanically transmitted trypanosomatid36. Therefore, a contribution by a density-sensing mechanism is likely to be important in different trypanosomal species and subspecies, including those that do not generate the morphologically extreme stumpy forms that are obvious in T. brucei. A proper understanding of the implications of SIF production and signalling for the dynamics of trypanosomal infections, both in single infections and in co-infections, will require the identification of the chemical entity itself and its signalling pathway. Thus far, SIF is known to be <500 Da and stable in culture3, although its turnover is necessary in vivo to allow antigenically distinct slender populations to replace earlier antigen types after immune clearance of those earlier types.
Conclusion and perspectives
The components of infection chronicity represent a series of interconnected unidirectional transitions. This unidirectionality is imposed by the host immune system and also by the molecular control of parasite development both in the blood and on transmission to the tsetse fly. As well as providing an added layer of complexity to the well-rehearsed but sometimes simplistic descriptions of antigenic variation, these interdependencies emphasize the elegant balance that trypanosomes must maintain in their mammalian host to optimize their longevity and, so, their transmissibility. Interestingly, the theme that emerges is shared between many vector-borne pathogens: the investments in immune evasion, virulence and transmission are balanced to ensure survival in the long term and through multiple cycles of interaction with both the host and the vector75. Moreover, the fundamental concepts of parasite developmental transitions have broad relevance in the context of eukaryotic cell fate determination and the evolution of complex signalling mechanisms and interactions58. Understanding the complexity with which components interact at the molecular, individual and population scales will require considerable coordination between experimental and theoretical approaches. Nonetheless, this coordination is needed if therapies that target transmission are to be developed and their consequences for parasite selection (as well as their potential for therapeutic breakdown) are to be predicted.
Acknowledgements
The authors are grateful to J. D. Barry for helpful comments on the manuscript and for providing useful additional insight during the preparation of the manuscript. Work in the K.R.M. laboratory is supported by the Wellcome Trust and by a Wellcome Trust Strategic Award to the Centre for Immunity, Infection and Evolution at the University of Edinburgh, UK. Work in the N.J.S. group is supported by the Wellcome Trust (grant number 082601/Z/07/Z).
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Keith R. Matthews’s homepage: http://www.biology.ed.ac.uk/research/groups/kmatthews/index.htm
References
- 1.Barrett MP. The rise and fall of sleeping sickness. Lancet. 2006;367:1377–1378. doi: 10.1016/S0140-6736(06)68591-7. [DOI] [PubMed] [Google Scholar]
- 2.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 3.Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J. Cell Sci. 1997;110:2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- 4.Reuner B, Vassella E, Yutzy B, Boshart M. Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol. Biochem. Parasitol. 1997;90:269–280. doi: 10.1016/s0166-6851(97)00160-6. [DOI] [PubMed] [Google Scholar]
- 5.Roditi I, Lehane MJ. Interactions between trypanosomes and tsetse flies. Curr. Opin. Microbiol. 2008;11:345–351. doi: 10.1016/j.mib.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Peacock L, et al. Identification of the meiotic life cycle stage of Trypanosoma brucei in the tsetse fly. Proc. Natl Acad. Sci. USA. 2011;108:3671–3676. doi: 10.1073/pnas.1019423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudenko G. African trypanosomes: the genome and adaptations for immune evasion. Essays Biochem. 2011;51:47–62. doi: 10.1042/bse0510047. [DOI] [PubMed] [Google Scholar]
- 8.Hertz-Fowler C, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 10.Navarro M, Penate X, Landeira D. Nuclear architecture underlying gene expression in Trypanosoma brucei. Trends Microbiol. 2007;15:263–270. doi: 10.1016/j.tim.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Borst P. Antigenic variation and allelic exclusion. Cell. 2002;109:5–8. doi: 10.1016/s0092-8674(02)00711-0. [DOI] [PubMed] [Google Scholar]
- 12.Horn D, Barry JD. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- 13.Marcello L, Barry JD. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison LJ, Marcello L, McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell. Microbiol. 2009;11:1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 15.Honigberg BM, Cunningham I, Stanley HA, Su-Lin KE, Luckins AG. Trypanosoma brucei: antigenic analysis of bloodstream, vector, and culture stages by the quantitative fluorescent antibody methods. Exp. Parasitol. 1976;39:496–522. doi: 10.1016/0014-4894(76)90052-7. [DOI] [PubMed] [Google Scholar]
- 16.Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- 17.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean SD, Marchetti R, Kirk K, Matthews KA. surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson M. Notes on the polymorphism of Trypanosoma gambiense in the blood and its relation to the exogenous cycle in Glossina palpalis. Proc. R. Soc. Lond. B. 1912;85:241–539. [Google Scholar]
- 20.McLintock LM, Turner CM, Vickerman K. Comparison of the effects of immune killing mechanisms on Trypanosoma brucei parasites of slender and stumpy morphology. Parasite Immunol. 1993;15:475–480. doi: 10.1111/j.1365-3024.1993.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 21.Engstler M, et al. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 22.Morrison LJ, Majiwa P, Read AF, Barry JD. Probabilistic order in antigenic variation of Trypanosoma brucei. Int. J. Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Pays E. Pseudogenes, chimaeric genes and the timing of antigen variation in African trypanosomes. Trends Genet. 1989;5:389–391. doi: 10.1016/0168-9525(89)90181-9. [DOI] [PubMed] [Google Scholar]
- 24.Boothroyd CE, et al. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lythgoe KA, Morrison LJ, Read AF, Barry JD. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc. Natl Acad. Sci. USA. 2007;104:8095–8100. doi: 10.1073/pnas.0606206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcello L, Barry JD. From silent genes to noisy populations-dialogue between the genotype and phenotypes of antigenic variation. J. Eukaryot. Microbiol. 2007;54:14–17. doi: 10.1111/j.1550-7408.2006.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gjini E, Haydon DT, Barry JD, Cobbold CA. Critical interplay between parasite differentiation, host immunity, and antigenic variation in trypanosome infections. Am. Nat. 2010;176:424–439. doi: 10.1086/656276. [DOI] [PubMed] [Google Scholar]
- 28.Seed JR, Black SJ. A proposed densitydependent model of long slender to short stumpy transformation in the African trypanosomes. J. Parasitol. 1997;83:656–662. [PubMed] [Google Scholar]
- 29.Seed JR, Black SJ. A revised arithmetic model of long slender to short stumpy transformation in the African trypanosomes. J. Parasitol. 1999;85:850–854. [PubMed] [Google Scholar]
- 30.Turner CMR, Aslam N, Dye C. Replication, differentiation, growth and the virulence of Trypanosoma brucei infections. Parasitology. 1995;111:289–300. doi: 10.1017/s0031182000081841. [DOI] [PubMed] [Google Scholar]
- 31.Macgregor P, Savill NJ, Hall D, Matthews KR. Transmission stages dominate trypanosome within-host dynamics during chronic infections. Cell Host Microbe. 2011;9:310–318. doi: 10.1016/j.chom.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro SZ, Naessen J, Liesegang B, Moloo SK, Magondu J. Analysis by flow cytometry of DNA synthesis during the life cycle of African trypanosomes. Acta Trop. 1984;41:313–323. [PubMed] [Google Scholar]
- 33.Vanhollebeke B, Uzureau P, Monteyne D, Perez-Morga D, Pays E. Cellular and molecular remodeling of the endocytic pathway during differentiation of Trypanosoma brucei bloodstream forms. Eukaryot. Cell. 2010;9:1272–1282. doi: 10.1128/EC.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnwell EM, et al. Developmental regulation and extracellular release of a VSG expression-site-associated gene product from Trypanosoma brucei bloodstream forms. J. Cell Sci. 2010;123:3401–3411. doi: 10.1242/jcs.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen BC, Sivam D, Kifer CT, Myler PJ, Parsons M. Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics. 2009;10:482. doi: 10.1186/1471-2164-10-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florent IC, Raibaud A, Eisen H. A family of genes related to a new expression site-associated gene in Trypanosoma equiperdum. Mol. Cell. Biol. 1991;11:2180–2188. doi: 10.1128/mcb.11.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhollebeke B, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320:677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 38.Vickerman K. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature. 1965;208:762–766. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]
- 39.Tyler KM, Matthews KR, Gull K. The bloodstream differentiation-division of Trypanosoma brucei studied using mitochondrial markers. Proc. Biol. Sci. 1997;264:1481–1490. doi: 10.1098/rspb.1997.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler KM, Matthews KR, Gull K. Anisomorphic cell division by African trypanosomes. Protist. 2001;152:367–378. doi: 10.1078/1434-4610-00074. [DOI] [PubMed] [Google Scholar]
- 41.Brickman MJ, Balber AE. Trypanosoma brucei brucei and T. b. gambiense: stumpy bloodstream forms express more CB1 epitope in endosomes and lysosomes than slender forms. J. Eukaryot. Microbiol. 1994;41:533–536. doi: 10.1111/j.1550-7408.1994.tb01512.x. [DOI] [PubMed] [Google Scholar]
- 42.Savill NJ, Seed JR. Mathematical and statistical analysis of the Trypanosoma brucei slender to stumpy transition. Parasitology. 2004;128:53–67. doi: 10.1017/s0031182003004256. [DOI] [PubMed] [Google Scholar]
- 43.Turner CMR, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99:67–75. doi: 10.1017/s0031182000061035. [DOI] [PubMed] [Google Scholar]
- 44.Horn D, Cross GA. Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol. Biochem. Parasitol. 1997;84:189–201. doi: 10.1016/s0166-6851(96)02794-6. [DOI] [PubMed] [Google Scholar]
- 45.Turner CM. The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol. Lett. 1997;153:227–231. doi: 10.1111/j.1574-6968.1997.tb10486.x. [DOI] [PubMed] [Google Scholar]
- 46.Barry JD, et al. What the genome sequence is revealing about trypanosome antigenic variation. Biochem. Soc. Trans. 2005;33:986–989. doi: 10.1042/BST20050986. [DOI] [PubMed] [Google Scholar]
- 47.Barry JD, Emery DL. Parasite development and host responses during the establishment of Trypanosoma brucei infection transmitted by tsetse fly. Parasitology. 1984;88:67–84. doi: 10.1017/s0031182000054354. [DOI] [PubMed] [Google Scholar]
- 48.Van den Bossche P, et al. Transmissibility of Trypanosoma brucei during its development in cattle. Trop. Med. Int. Health. 2005;10:833–839. doi: 10.1111/j.1365-3156.2005.01467.x. [DOI] [PubMed] [Google Scholar]
- 49.Baylis M. The daily feeding rate of tsetse (Diptera: Glossinidae) on cattle at Galana Ranch, Kenya and comparison with trypanosomiasis incidence. Acta Trop. 1997;65:81–96. doi: 10.1016/s0001-706x(97)00655-4. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs SL, Lee ND. Determination of citric acid in serum and urine using Br82. J. Nucl. Med. 1964;5:297–301. [PubMed] [Google Scholar]
- 51.Brun R, Schonenberger M. Stimulating effect of citrate and cis-aconitate on the transformation of Trypanosoma brucei bloodstream forms to procyclic forms in vitro. Z. Parasitenkd. 1981;66:17–24. doi: 10.1007/BF00941941. [DOI] [PubMed] [Google Scholar]
- 52.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szoor B, Wilson J, McElhinney H, Tabernero L, Matthews KR. Protein tyrosine phosphatase TbPTP1: a molecular switch controlling life cycle differentiation in trypanosomes. J. Cell Biol. 2006;175:293–303. doi: 10.1083/jcb.200605090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szoor B, Ruberto I, Burchmore R, Matthews K. A novel phosphatase cascade regulates differentiation in trypanosomes via a glycosomal signaling pathway. Genes Dev. 2010;24:1306–1316. doi: 10.1101/gad.570310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamenski T, Heilmeier S, Meinhart A, Cramer P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell. 2004;15:399–407. doi: 10.1016/j.molcel.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 56.Ferrell JE., Jr. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- 57.Pomerening JR, Sontag ED, Ferrell JE., Jr. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature Cell Biol. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 58.Domingo-Sananes MR, Kapuy O, Hunt T, Novak B. Switches and latches: a biochemical tug-of-war between the kinases and phosphatases that control mitosis. Phil. Trans. R. Soc. 2011;366:3584–3594. doi: 10.1098/rstb.2011.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews KR. Controlling and coordinating development in vector-transmitted parasites. Science. 2011;331:1149–1153. doi: 10.1126/science.1198077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matthews KR, Gull K. Commitment to differentiation and cell cycle re-entry are coincident but separable events in the transformation of African trypanosomes from their bloodstream to their insect form. J. Cell Sci. 1997;110:2609–2618. doi: 10.1242/jcs.110.20.2609. [DOI] [PubMed] [Google Scholar]
- 61.Herman M, Perez-Morga D, Schtickzelle N, Michels PA. Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei. Autophagy. 2008;4:294–308. doi: 10.4161/auto.5443. [DOI] [PubMed] [Google Scholar]
- 62.Opperdoes FR, Szikora JP. In silico prediction of the glycosomal enzymes of Leishmania major and trypanosomes. Mol. Biochem. Parasitol. 2006;147:193–206. doi: 10.1016/j.molbiopara.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 63.Chou S, Jensen BC, Parsons M, Alber T, Grundner C. The Trypanosoma brucei life cycle switch TbPTP1 is structurally conserved and dephosphorylates the nucleolar protein NOPP44/46 . Biol. Chem. 2010;285:22075–22081. doi: 10.1074/jbc.M110.108860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yabu Y, Takayanagi T. Trypsin-stimulated transformation of Trypanosoma brucei gambiense bloodstream forms to procyclic forms in vitro. Parasitol. Res. 1988;74:501–506. doi: 10.1007/BF00531625. [DOI] [PubMed] [Google Scholar]
- 65.Rolin S, Hancocq-Quertier J, Paturiaux-Hanocq F, Nolan DP, Pays E. Mild acid stress as a differentiation trigger in Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;93:251–262. doi: 10.1016/s0166-6851(98)00046-2. [DOI] [PubMed] [Google Scholar]
- 66.Haanstra JR, et al. A domino effect in drug action: from metabolic assault towards parasite differentiation. Mol. Microbiol. 2011;79:94–108. doi: 10.1111/j.1365-2958.2010.07435.x. [DOI] [PubMed] [Google Scholar]
- 67.Frank SA. A model for the sequential dominance of antigenic variants in African trypanosome infections. Proc. Bio. Sci. 1999;266:1397–1401. doi: 10.1098/rspb.1999.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashcroft M. A comparison between syringe-passaged and a tsetse-fly transmitted line of a strain of Trypanosma rhodesiense. Ann. Trop. Med. Parasitol. 1960;54:44–70. doi: 10.1080/00034983.1960.11685955. [DOI] [PubMed] [Google Scholar]
- 69.Matthews KR, Ellis JR, Paterou A. Molecular regulation of the life cycle of African trypanosomes. Trends Parasitol. 2004;20:40–47. doi: 10.1016/j.pt.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 70.Turner CM. The use of experimental artefacts in African trypanosome research. Parasitol. Today. 1990;6:14–17. doi: 10.1016/0169-4758(90)90385-h. [DOI] [PubMed] [Google Scholar]
- 71.Fairbairn H, Culwick A. The modification of Trypanosma rhodesiense on prolonged syringe passage. Ann. Trop. Med. Parasitol. 1947;41:26. doi: 10.1080/00034983.1947.11685308. [DOI] [PubMed] [Google Scholar]
- 72.Brown SP, Taddei F. The durability of public goods changes the dynamics and nature of social dilemmas. PLoS ONE. 2007;2:e593. doi: 10.1371/journal.pone.0000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberle M, Balmer O, Brun R, Roditi I. Bottlenecks and the maintenance of minor genotypes during the life cycle of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001023. doi: 10.1371/journal.ppat.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison WI, Wells PW, Moloo SK, Paris J, Murray M. Interference in the establishment of superinfections with Trypanosoma congolense in cattle. J. Parasitol. 1982;68:755–764. [PubMed] [Google Scholar]
- 75.Pollitt LC, Macgregor P, Matthews K, Reece SE. Malaria and trypanosome transmission: different parasites, same rules? Trends Parasitol. 2011;27:197–203. doi: 10.1016/j.pt.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pays E, Lips S, Nolan D, Vanhamme L, Perez-Morga D. The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts. Mol. Biochem. Parasitol. 2001;114:1–16. doi: 10.1016/s0166-6851(01)00242-0. [DOI] [PubMed] [Google Scholar]
- 77.Bitter W, Gerrits H, Kieft R, Borst P. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature. 1998;391:499–502. doi: 10.1038/35166. [DOI] [PubMed] [Google Scholar]
- 78.Xong HV, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95:839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 79.Pays E, Vanhollebeke B. Human innate immunity against African trypanosomes. Curr. Opin. Immunol. 2009;21:493–498. doi: 10.1016/j.coi.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 80.Vickerman K. The fine structure of Trypanosoma congolense in its bloodstream phase. J. Protozool. 1969;16:54–69. doi: 10.1111/j.1550-7408.1969.tb02233.x. [DOI] [PubMed] [Google Scholar]
- 81.Stevens JR, Brisse S. In: in The Trypanosomiases. Maudlin I, Holmes P, Miles M, editors. CABI; 2004. p. 25. Ch. 1. [Google Scholar]
- 82.Gardiner PR, Wilson AJ. Trypanosoma (Duttonefla) vivax. Parasitol. Today. 1987;3:49–52. doi: 10.1016/0169-4758(87)90213-4. [DOI] [PubMed] [Google Scholar]
- 83.Nantulya VM, Doyle JJ, Jenni L. Studies on Trypanosoma (nannomonas) congolense. I. On the morphological appearance of the parasite in the mouse. Acta Trop. 1978;35:329–337. [PubMed] [Google Scholar]
- 84.Miles MA. Pleomorphism demonstrated in a clone of an akinetoplastic strain of Trypanosoma evansi. Trans. R. Soc. Trop. Med. Hyg. 1970;64:471. doi: 10.1016/0035-9203(70)90045-3. [DOI] [PubMed] [Google Scholar]