Abstract Abstract
Using the fluorescence in situ hybridization (FISH), the presence of (TTAGG)n telomeric sequence was detected in the chromosomes of Lethocerus patruelis (Stål, 1854) belonging to the family Belostomatidae (Heteroptera: Nepomorpha). This sequence was exclusively present at the ends of chromosomes in this species. This is the first evidence of the insect-type TTAGG telomeric repeats in Heteroptera.
Keywords: Chromosomes, FISH, (TTAGG)n telomeric repeat, true bugs, Nepomorpha, Belostomatidae, Lethocerus patruelis
Introduction
Telomeres are specific nucleoprotein structures at the ends of chromosomes and are responsible for their stability. Information on the telomere structure and function is presently available for many animals, plants and fungi (Fuchs et al. 1995, McKnight and Shippen 2004, Traut et al. 2007, Zakian 2012). The telomeres of insect species are predominantly composed of a pentanucleotide sequence repeat (TTAGG)n (reviewed in Frydrychová et al. 2004). On the other hand, there are some higher taxa known to have lost this telomeric motif during their evolution, and Heteroptera are repeatedly referred to as one of such groups (Sahara et al. 1999, Frydrychová et al. 2004, Vitková et al. 2005, Lukhtanov and Kuznetsova 2010, Grozeva et al. 2011, Kuznetsova et al. 2011).
In this paper we report the molecular structure of telomeres at the physical ends of chromosomesin Lethocerus patruelis (Stål, 1854) (Nepomorpha: Belostomatidae).
Material and methods
Spread chromosome preparations were made from testes of Lethocerus patruelis and stained using a Shiff-Giemsa method as described in Grozeva et al. (in press). The molecular structure of telomeres was investigated by fluorescence in situ hybridization of chromosomes (FISH) with a (TTAGG)n probe. In addition, we used an 18S rDNA probe to reveal the location of ribosomal clusters, NORs, on Lethocerus patruelis chromosomes. In these experiments we followed the protocol described in Grozeva et al. (2011). Fluorescence images were taken with a Leica DFC 345 FX camera using Leica Application Suite 3.7 software with an Image Overlay module.
Results
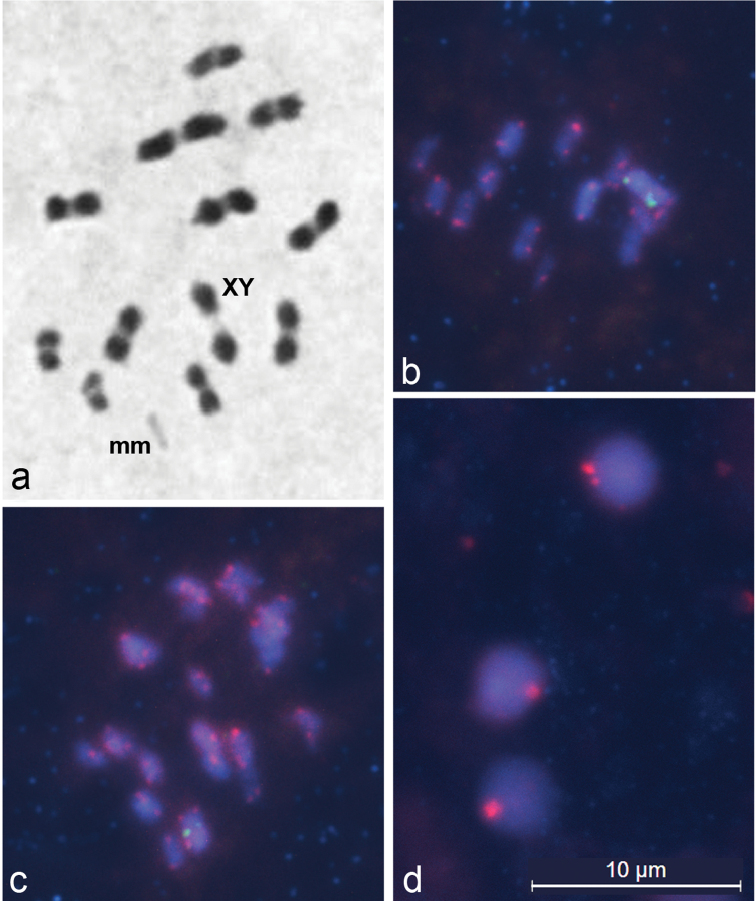
At first metaphases in Lethocerus patruelis males, 11 autosomal bivalents, each with one (sometimes two) terminal or subterminal chiasmata, a bivalent of m-chromosomes (micro-chromosomes) and a XY- pseudo-bivalent could be seen (Fig. 1a). Figures 1b-d show the results of fluorescence in situ hybridization with pentanucleotide (TTAGG)n and 18S rDNA probes to several meiotic spreads. At metaphase nuclei, TTAGG fluorescent signals (red) are clearly seen at all chromosomal ends, whereas rDNA clusters (green) are clearly evident on the X and Y chromosomes (Fig. 1b, c). Prominent telomere clustering at the periphery of spermatid nuclei (Fig. 1d) creates one large while sometimes a small number of TTAGG signals (red).
Figures 1.
Meiotic chromosomes of Lethocerus patruelis subjected to standard staining (a) and FISH (b–d). a metaphase I showing n = 11AA + mm + XY; b–d representative FISH images of metaphase I chromosomes (b, c) and spermatids (d) hybridized with probes against 18S rDNA and telomeres, showing ribosomal clusters (green) on X and Y chromosomes (b, c), and TTAGG repeats (red) located at the ends of chromosomes (b, c) and clustered at the periphery of spermatid nuclei (d).
Discussion
The standard karyotype of Lethocerus patruelis males is 2n = 22A + 2m + XY as it was recently shown by Grozeva et al. (in press). We found that Lethocerus patruelis displayed FISH rDNA sites both on X and Y chromosomes. This is as expected since CMA3-staining performed by Grozeva et al. (in press) has revealed GC rich clusters (typically pointed to NORs) on the sex chromosomes in this species. In other Belostomatidae species studied in this respect, NORs are known to be located either on sex chromosomes or on a pair of autosomes, the co-generic species sometimes differing in this pattern (reviewed in Grozeva et al. 2011).
DNA of the telomeres consists of short nucleotide motifs (combinations) repeated thousands and millions of times. Comparative analysis of these motifs in various groups of organisms has shown that they are evolutionarily stable, and, having once appeared during the evolution, mark taxa and phylogenetic lineages of high rank (Traut et al. 2007).
Quite recently, Frydrychová et al. (2004) assembled and analyzed the data available on the telomeric sequences in Insecta, and, together with some original observations, they interpreted these character data in a phylogenetic framework. Conclusions in that work are largely congruent with those previously proposed by Sahara et al. (1999). The great majority of insect species share the telomeres composed of (TTAGG)n repeat. Since the same telomere composition is characteristic of the rest of arthropods, the (TTAGG)n telomeric motif is considered an ancestral one in Insecta. Many higher-level insect groups preserved this motif; however several orders, e.g. Dermaptera, Heteroptera, Diptera and some others, are suggested to have lost this telomeric sequence during the evolution (Sahara et al. 1999, Frydrychová et al. 2004, Vitková et al. 2005, Lukhtanov and Kuznetsova 2010).
We emphasize, however, that the problem of telomere composition in different insect orders is still not adequately explored and in most cases, the available data concern one or more species only (see Fig. 6 in Frydrychová et al. 2004). On the other hand, in one of the better studied orders, Coleoptera (data are available for more than 20 species), both (TTAGG)n-positive and (TTAGG)n-negative species have been reported (Frydrychová and Marec 2002, Frydrychová et al. 2004).
In Heteroptera, the absence of the (TTAGG)n telomeric motif was firstly shown for Halyomorpha halys (Stål, 1855) (Pentatomidae) studied using Southern hybridization (Okazaki et al. 1993: as Halyomorpha mista (Uhler, 1860)) and Pyrrhocoris apterus (Linnaeus, 1758) (Pyrrhocoridae) subjected to both Southern hybridization and FISH (Sahara et al. 1999). On the other hand, this sequence was revealed in telomeres of non-heteropteran Hemiptera and some other Paraneoptera (Frydrychova et al., 2004).
Originally proposed by Sahara et al. (1999) and accepted at a later time by other authors (Frydrychova et al. 2004, Vitková et al. 2005, Lukhtanov and Kuznetsova 2010), the hypothesis for the loss of (TTAGG)n sequence in true bugs has received further support owing to the discovery of Grozeva et al. (2011) that five more species studied by FISH and Dot-blot hybridization are also (TTAGG)n- negative. Based on evidence provided by Okazaki et al. (1993), Sahara et al. (1999) and Grozeva et al. (2011), (TTAGG)n motif is known to be absent in seven species of true bugs. These species represent phylogenetically distant families, such as Pentatomidae (Halyomorpha halys, Eurydema oleracea (Linnaeus, 1758), Graphosoma lineatum (Linnaeus, 1758)) and Pyrrhocoridae (Pyrrhocoris apterus) belonging to the infraorder Pentatomomorpha and also Miridae (Deraeocoris rutilus (Herrich-Schaffer, 1838), Megaloceroea recticornis (Geoffroy, 1785)) and Cimicidae (Cimex lectularius (Linnaeus, 1758) belonging to the infraorder Cimicomorpha.
Our results of FISH with a (TTAGG)n probe strongly demonstrated that (TTAGG)n sequence was located at the telomeres of all chromosomes in Lethocerus patruelis. The finding of the insect-type (TTAGG)n telomeric motif in Lethocerus patruelis is thus clearly indicative of the heterogeneity of Heteroptera in telomere organization. The family Belostomatidae, to which this species belongs, is classified within the infraorder Nepomorpha (or true water bugs). The data on telomeres imply that true water bugs preserved the plesiomorphic telomere structure, whereas Cimicomorpha and Pentatomomorpha have the apomorphic state of this character, which can be considered a synapomorphy of these infraorders. This conclusion is consistent with the generally accepted opinion that Cimicomorpha and Pentatomomorpha represent a monophyletic lineage, and Nepomorpha has a basal position within Heteroptera (Wheeler et al. 1993; Mahner 1993, Scherbakov and Popov 2002, Xie et al. 2008, Weirauch and Schuh 2011).
Acknowledgements
The study was supported (for VK and BA) by the RFBR (grant 11-04-00734), programs of the Presidium of the RAS “Gene Pools and Genetic Diversity” and “Origin of the Biosphere and Evolution of Geo-biological Systems” and the Ministry of Education and Science of the Russian Federation, and (for SG) by DO-02-259 NSF Sofia, BAS.
References
- Frydrychová R, Marec F. (2002) Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera). Genetica 115: 179-187.10.1023/A:1020175912128 [DOI] [PubMed] [Google Scholar]
- Frydrychová R, Grossmann P, Trubač P, Vitková M, Marec F. (2004) Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 47: 163-178.10.1139/g03-100 [DOI] [PubMed] [Google Scholar]
- Fuchs J, Brandes A, Schubert I. (1995) Telomere sequence localization and karyotype evolution in higher plants. Plant Systematics and Evolution 196: 227-241.10.1007/BF00982962 [Google Scholar]
- Grozeva S, Kuznetsova V, Anokhin B. (2011) Karyotypes, male meiosis and comparative FISH mapping of 18S ribosomal DNA and telomeric (TTAGG)n repeat in eight species of true bug (Hemiptera, Heteroptera). Comparative Cytogenetics 5 (4): 355-374.10.3897/CompCytogen.v5i4.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva S, Kuznetsova V, Simov N, Langurov M, Dalakchieva S. (in press) Sex chromosome “pre-reduction” in male meiosis of Lethocerus patruelis (Stål, 1854) (Heteroptera, Belostomatidae) with some notes on the distribution of the species. ZooKeys. [DOI] [PMC free article] [PubMed]
- Kuznetsova VG, Grozeva SM, Nokkala S, Nokkala Ch. (2011) Cytogenetics of the true bug infraorder Cimicomorpha (Hemiptera, Heteroptera): a review. ZooKeys 154: 31-70.10.3897/zookeys.154.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Kuznetsova VG. (2010) What Genes and Chromosomes Say about the Origin and Evolution of Insects and Other Arthropods. Russian Journal of Genetics 46 (9): 1115-1121.10.1134/S1022795410090279 (Originally published in: Genetika, 2010, 46(9): 1258–1265). [PubMed] [Google Scholar]
- Mahner M. (1993) Systema Cryptoceratorum Phylogeneticum (Insecta, Heteroptera). Zoologica 48: 1-302 [Google Scholar]
- McKnight TD, Shippen DE. (2004) Plant telomere biology. Plant Cell 16: 794-803.10.1105/tpc.160470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara K, Marec F, Traut W. (1999) TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome research 7: 449– 460. 10.1023/A:1009297729547 [DOI] [PubMed]
- Scherbakov DE, Popov YA. (2002) Superorder Cimicidea Laicharting, 1781. Order Hemiptera Linne, 1758. The bugs, cicadas, plantlice, scale insects, etc. In: Rasnitsyn AP (Ed) History of Insects. DLJ Quicke, PP. 143–157. Dordrecht, The Netherlands: Kluwer Academic, 517.
- Traut W, Szczepanowski M, Vitková M, Opitz C, Marec F, Zrzavy J. (2007) The telomere repeat motif of basal Metazoa. Chromosome Research 15 (3): 371-382 [DOI] [PubMed] [Google Scholar]
- Vitková M, Kral J, Traut W, Zrzavy J, Marec F. (2005) The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Research 13: 145-156.10.1007/s10577-005-7721-0 [DOI] [PubMed] [Google Scholar]
- Weirauch C, Schuh RT. (2011) Systematics and Evolution of Heteroptera: 25 Years of Progress. Annual Review of Entomology 56: 487-510.10.1146/annurev-ento-120709-144833 [DOI] [PubMed] [Google Scholar]
- Wheeler WC, Schuh RT, Bang R. (1993) Cladistic relationships among higher groups of Heteroptera: congruence between morphological and molecular data sets. Entomologica Scandinavica 24: 121-137.10.1163/187631293X00235 [Google Scholar]
- Xie Q, Tian Y, Zheng L, Bu W. (2008) 18S rRNA hyper-elongation and the phylogeny of Euhemiptera (Insecta: Hemiptera). Molecular Phylogenetics and Evolution 47: 463-471.10.1016/j.ympev.2008.01.024 [DOI] [PubMed] [Google Scholar]
- Zakian VA. (2012) Telomeres: the beginnings and ends of eukaryotic chromosomes. Experimental Cell Research 318 (12): 1456-1460.10.1016/j.yexcr.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]