Abstract
Purpose
The fibroblast growth factor 10 (FGF10) gene polymorphism rs339501 was previously reported to be associated with high myopia in a Chinese population. In the present study, we investigated whether FGF10 polymorphisms are associated with extreme myopia in a Japanese population as well.
Methods
A total of 433 Japanese patients with extreme myopia (≤ −10.00 diopters) and 542 Japanese healthy controls (+1.50 to −1.50 diopters) were recruited. We genotyped seven tagging single-nucleotide polymorphisms (SNPs), including rs339501, in FGF10. We also performed an imputation analysis to evaluate the potential association of ungenotyped FGF10 SNPs, and 34 SNPs were imputed.
Results
It was found that rs339501 and rs12517396 exhibited the strongest association with extreme myopia (p=3.9 × 10−4, corrected p [Pc]=0.0030). A significant association was also observed for rs10462070 (p=6.5 × 10−4, Pc=0.0059). These three SNPs were in strong linkage disequilibrium (D’ ≥0.99, r2 ≥0.96). However, the frequency of the A allele of rs339501 was increased in cases compared to controls, which differs from the increased frequency of the G allele in cases in the previous Chinese population.
Conclusions
Three FGF10 SNPs in complete linkage disequilibrium—rs339501, rs12517396, and rs10462070—were associated with extreme myopia in the Japanese population, and the risk allele of rs339501 differed from the previous Chinese population. Therefore, these three SNPs may not be an important risk factor for susceptibility to extreme myopia. Further studies are needed to elucidate the possible contribution of the FGF10 region in the development of extreme myopia.
Introduction
Myopia is a very common refractive error that has a significant impact on public health and economics around the world. High myopia, which is a refractive error ≤ –6 diopters (D), is a major cause of blindness associated with an increased risk of various ocular and systemic diseases, including retinal detachment, glaucoma, and cataracts [1]. The prevalence of high myopia has been reported to range from 1.0% to 9.6% in the general population, but it exhibits variable incidence in different countries, with a preponderance in Asia [2-5].
Although the cause of high myopia is unclear, family correlation studies and twin studies have shown that genetic factors play a significant role in its development [6-11], with a relationship between the genetic basis of eye growth and the development of myopia. Twin studies revealed a correlation between axial length and refractive error that was much higher in monozygotic twins compared to dizygotic twins [12,13]. The pattern of inheritance in high myopia appears to be heterogeneous, with an autosomal dominant to autosomal recessive pattern [9]. Therefore, risk factors that contribute to the development of high myopia include genetic heterogeneity and axial length [14,15]. Familial linkage studies have attempted to identify candidate genes that might contribute to myopia, and significant linkages have been reported at 18 loci, specifically MYP1 to MYP18 [16]. Many recent genome-wide association studies (GWASs) have been conducted to identify genes involved in myopia or high myopia, and many candidate loci/genes have been reported [17-27].
The fibroblast growth factor (FGF) family of proteins plays important roles in the proliferation and differentiation of a wide variety of cells and tissues. A defect in FGF10 leads to the development and differentiation of several ocular tissues [28-30]. Sclera remodeling, which is one of the important mechanisms in the development of myopia, involves alterations in both the degradation and synthesis of extracellular matrix components [31], and FGF10 can modulate extracellular matrix–associated genes [32-35]. Recently, His et al. [36] reported that the sclera of myopic mouse eyes have higher levels of FGF10 mRNA. The G allele of FGF10 polymorphism rs339501 was also found to be associated with higher FGF10 expression and the risk of extreme myopia (≤-10 D) in a Chinese population residing in Taiwan. Therefore, higher expression of FGF10 caused by the G allele of rs339501 could represent a risk for myopia. The aim of the present study was to investigate whether genetic polymorphisms in FGF10 are associated with extreme myopia in Japanese patients.
Methods
Subjects
We recruited 433 unrelated Japanese individuals with extreme myopia (refractive error ≤ –10.00 D in at least one eye) and 542 unrelated healthy Japanese controls (+1.50 to −1.50 D in both eyes) at Yokohama City University and Okada Eye Clinic in Japan. All participants were diagnosed by comprehensive ophthalmologic tests, including axial length, fundus examination, spherical power, and corneal curvature (Autorefractor; NIDEK [Gamagori, Japan] ARK-730A, ARK-700A TOPCON [Tokyo, Japan] KP-8100P, BIO and PACHY Meter AL-2000; Tomey Corporation, Nagoya, Japan). The individuals with extreme myopia had no known genetic diseases associated with myopia and/or high myopia, including glaucoma, keratoconus, or Marfan syndrome. Patient age ranged from 12 to 76 years (mean 38.1±12.0 years), and 44.4% of patients were male. The average spherical refractive errors were −11.9±2.20 D (range −6.75 to −22.75 D) in the right eye (OD) and −11.9±2.29 D (range −8.50 to −23.0 D) in the left eye (OS). The average axial length was 28.0±1.18 mm (range 26.0 to 33.1 mm) for OD and 28.0±1.23 mm (range 26.0 to 34.7 mm) for OS. The average corneal refraction was 43.8±1.46 D (range 39.5 to 47.8 D) for OD and 43.8±1.52 D (range 39.8 to 53.0 D) for OS. Control individuals were healthy volunteers and not related to each other or the patients. The controls were sex-matched (47.2% male) to the patients with an age range of 24 to 75 years (mean 40.6±12.0 years). All participants had similar social backgrounds and resided in the same urban area. Informed consent was obtained from all participants. The study methodology adhered to the tenets of the Declaration of Helsinki and was approved by the relevant ethics committees in each participating institute.
Single-nucleotide polymorphism genotyping of the FGF10 gene region
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Procedures were performed under standardized conditions to prevent variation in DNA quality. Seven tagging single nucleotide polymorphisms (SNPs) covering the FGF10 region including 10 kb upstream and downstream from the gene were selected from HapMap Japanese data (minor allele frequency ≥5%, pairwise r2 ≥0.8; Table 1; NCBI). Genotyping was performed using the TaqMan 5′ exonuclease assay with validated TaqMan primer-probe sets supplied by Applied Biosystems (Foster City, CA). PCR was performed using a reaction mixture with a total volume of 10 μl containing 1X TaqMan Universal PCR Master Mix (Applied Biosystems), 24 nm of each primer-probe set, and 3 ng genomic DNA. The PCR conditions were as follows: 95 °C for 10 min, followed by 40 cycles of denaturation at 92 °C for 15 s and annealing/ extension at 60 °C for 1 min. The probe’s fluorescence signal was detected using the StepOnePlus Real-Time PCR System (Applied Biosystems).
Table 1. The 41 FGF10 SNPs in the present study.
| SNP | Position on chromosome five (Build 37.1) | Gene location |
|---|---|---|
|
rs1448044 |
44,296,986 |
3′-UTR |
|
rs10072476 |
44,299,400 |
3′-UTR |
|
rs9292903 |
44,299,998 |
3′-UTR |
|
rs1979079 |
44,300,650 |
3′-UTR |
|
rs1374961 |
44,303,760 |
3′-UTR |
|
rs6451758 |
44,305,515 |
Intron 2 |
|
rs10462070 |
44,305,749 |
Intron 2 |
|
rs10473352 |
44,308,252 |
Intron 2 |
|
rs1374962 |
44,311,070 |
Intron 1 |
|
rs16873956 |
44,312,489 |
Intron 1 |
|
rs10060796 |
44,313,151 |
Intron 1 |
|
rs1839090 |
44,313,282 |
Intron 1 |
|
rs980510 |
44,318,532 |
Intron 1 |
|
rs13436788 |
44,318,624 |
Intron 1 |
|
rs10057630 |
44,327,864 |
Intron 1 |
|
rs4866891 |
44,328,270 |
Intron 1 |
|
rs987642 |
44,331,905 |
Intron 1 |
|
rs1011814 |
44,335,820 |
Intron 1 |
|
rs10512844 |
44,338,759 |
Intron 1 |
|
rs2330544 |
44,339,764 |
Intron 1 |
|
rs2330545 |
44,339,810 |
Intron 1 |
|
rs7708529 |
44,347,131 |
Intron 1 |
|
rs1482689 |
44,359,428 |
Intron 1 |
|
rs12517396 |
44,359,526 |
Intron 1 |
|
rs339509 |
44,360,892 |
Intron 1 |
|
rs17234079 |
44,362,204 |
Intron 1 |
|
rs1482672 |
44,362,769 |
Intron 1 |
|
rs339502 |
44,364,007 |
Intron 1 |
|
rs2121875 |
44,365,545 |
Intron 1 |
|
rs339501 |
44,365,633 |
Intron 1 |
|
rs11750845 |
44,373,060 |
Intron 1 |
|
rs1384449 |
44,377,060 |
Intron 1 |
|
rs16901816 |
44,381,698 |
Intron 1 |
|
rs2973644 |
44,384,183 |
Intron 1 |
|
rs1482679 |
44,385,415 |
Intron 1 |
|
rs2973646 |
44,387,537 |
Intron 1 |
|
rs2973649 |
44,391,161 |
5′-UTR |
|
rs1482680 |
44,392,142 |
5′-UTR |
|
rs723166 |
44,396,015 |
5′-UTR |
|
rs10473354 |
44,396,353 |
5′-UTR |
| rs10941665 | 44,398,696 | 5′-UTR |
Genotyped SNPs are indicated in bold.
Imputation analysis of the FGF10 gene region
We performed an imputation analysis to evaluate the potential association of ungenotyped SNPs in the FGF10 region, including 10 kb upstream and downstream from the gene. The genotypes of 433 cases and 542 controls were imputed using MACH v1.0 [37,38]. For the reference panel, we used Japanese data from HapMap phase 3. For quality control, we excluded SNPs from the reference panel if they had a call rate <95%, leaving 34 SNPs for the imputation. As none of the SNPs had a squared correlation between imputed and true genotypes <0.3, the 34 imputed SNPs were included in the association analysis (Table 1).
Statistical analysis
Allele frequencies, Hardy–Weinberg equilibrium, and linkage disequilibrium (LD) were assessed using Haploview 4.1 software [39]. Differences in allele haplotype frequencies between cases and controls were assessed by χ2. The obtained p values were corrected for multiple testing using a permutation test (10,000 iterations) in Haploview. A corrected p (Pc) value <0.05 was considered significant. Conditional logistic regression analysis was performed to assess the effect of each SNP on disease susceptibility using PLINK [40].
Results
The genotype frequencies of all seven tagging and 34 imputed SNPs were in Hardy–Weinberg equilibrium among both cases and controls. Figure 1 and Table 2 show the results of the association analysis of 41 SNPs in FGF10. Of the seven tagging SNPs, rs339501 exhibited a strong association with extreme myopia (p=3.9 × 10−4, Pc=0.0030), and the frequency of the A allele of rs339501 was increased in cases compared to controls (90.0% versus 84.5%, odds ratio [OR]=1.64), which is the opposite of that reported in the previous Chinese population. In other tagging SNPs, the frequencies of the A allele of rs2330545 and A allele of rs1384449 were also increased in cases compared to controls (p=0.047 and p=0.019, respectively), although this increase did not reach significance after correcting for multiple testing (Pc >0.05).
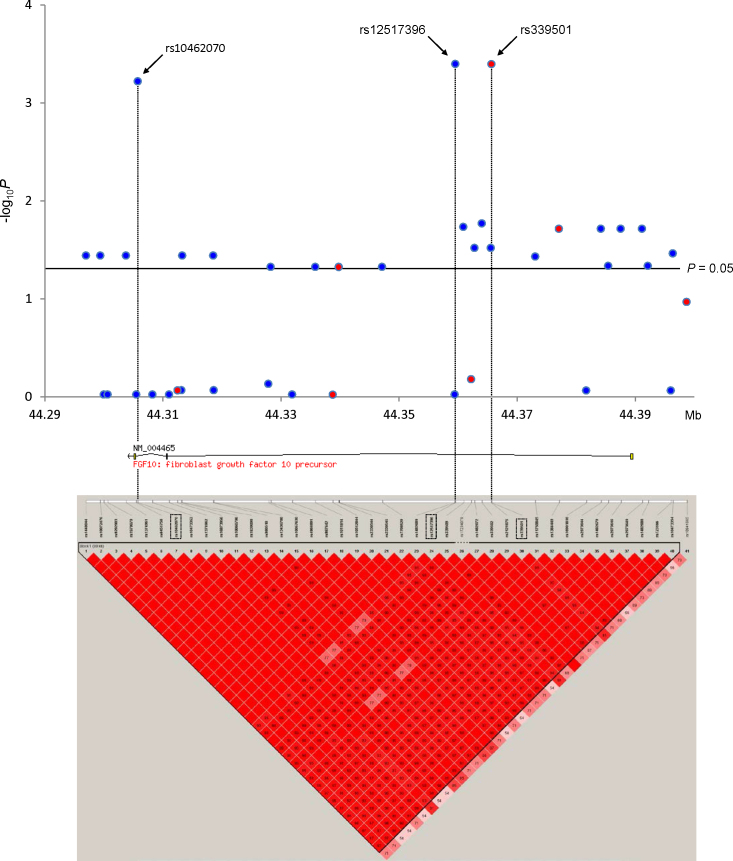
Figure 1.
In-depth single-nucleotide polymorphism analysis of the FGF10 region. The upper panel shows the distribution of association results of single-nucleotide polymorphisms (SNPs) across FGF10. Genotyped SNPs are indicated by a red circle, and imputed SNPs are indicated by a blue circle. The lower panel shows the linkage disequilibrium structure in FGF10. Higher D’ values are indicated by a brighter red.
Table 2. Allelic association results for SNPs rs10462070, rs12517396 and rs339501.
| SNP | Position on chromosome five (Build 37.1) | Gene location | Alleles | Risk allele | Risk allele frequency |
P | Pc | OR (95%CI) | |
|---|---|---|---|---|---|---|---|---|---|
| Cases (n=433) | Controls (n=542) | ||||||||
|
rs10462070 |
44,305,749 |
Intron 2 |
A/G |
A |
0.901 |
0.849 |
6.5×10−4 |
0.0059 |
1.62 (1.22–2.13) |
|
rs12517396 |
44,359,526 |
Intron 1 |
A/C |
C |
0.900 |
0.845 |
3.9×10−4 |
0.0030 |
1.64 (1.25–2.16) |
| rs339501 | 44,365,633 | Intron 1 | A/G | A | 0.900 | 0.845 | 3.9×10−4 | 0.0030 | 1.64 (1.25–2.16) |
Of 34 imputed SNPs, rs12517396 showed the strongest significance, equivalent to rs339501, and the C allele was associated with a risk of extreme myopia (p=3.9 × 10−4, Pc=0.0030, OR=1.64). The A allele of rs10462070 was also strongly associated with a risk of extreme myopia (p=6.5 × 10−4, Pc=0.0059, OR=1.62). Another 20 imputed SNPs showed moderate association (p<0.05) with the disease but this did not reach significance after correction (Pc >0.05).
Figure 1 shows the overall LD patterns for the 41 SNPs in 975 individuals. Strong LD was observed throughout the FGF10 gene region and 40 SNPs from rs1448044 to rs10473354 were located in one haplotype block (Block 1). The three SNPs with the strongest signal, rs339501—rs12517396, and rs10462070—were in complete LD in Block 1 (D’ ≥0.99, r2 ≥0.96). Twenty-two SNPs with moderate association were also in Block 1. To elucidate the effect of rs339501, rs12517396, and rs10462070 on disease susceptibility, we performed conditional logistic regression analysis. However, we could not determine which variant was the causal SNP for the observed associations in this study because of the complete LD among the three SNPs.
Discussion
Myopia is a complex disease that involves both environmental factors and multiple interacting genetic factors. In particular, determination of the role of genetic factors in high myopia has been influenced by its high prevalence, genetic heterogeneity, and potentially modulating environmental factors. In the past few years, previous GWASs have reported many genomic loci/genes that confer susceptibility to myopia [17-27]. Although Hsi et al. recently reported that FGF10 rs339501 is associated with extreme myopia (refractive error ≤ –10.00 D) but not high myopia (≤ –6.00 D) in a Chinese population using a candidate gene approach [36], the GWASs have not identified FGF10 as a myopia susceptibility gene. At least two possible explanations exist for this difference. First, the GWAS platforms may not have included the significant SNP rs339501 and other SNPs in strong LD with rs339501 that would lead to the detection of an association between the FGF10 region and myopia. Second, none of the GWASs focused on extreme myopia; they used high myopia, pathological myopia (axial length ≥28 mm), axial length, or refraction error.
The A allele frequency of rs339501 was found to have a role in the risk of extreme myopia in our Japanese population. This finding differs from the previous study of a Chinese population [36] in which extreme myopia cases had a significantly higher frequency of the G allele compared to controls. We also found that two other SNPs, rs12517396 and rs10462070, in complete LD with rs339501 were strongly associated with extreme myopia, but these SNPs were not included in the previous study. These three SNPs are intronic variants that can significantly affect gene expression levels and contribute to the development of human diseases [41-43]. Hsi et al. reported that the G allele of rs339501 significantly increases the expression of FGF10, suggesting that the increased FGF10 expression caused by the G allele increases the risk for myopia. However, because our results showed that the A allele of rs339501 is associated with a risk of extreme myopia in our Japanese population, it suggests that the G allele is not a risk factor for the susceptibility of extreme myopia in all populations.
Drastic differences in the allelic distribution of disease risk–associated SNPs among different ethnic populations have been reported in exfoliation syndrome (XFS). XFS is strongly associated with certain SNPs, including rs1048661, rs2165241, and rs3825942 of the lysyl oxidase-like 1 (LOXL1) gene, in many different ethnic groups [44-46], suggesting that LOXL1 is the major susceptibility gene for the development of XFS. However, the allelic distributions of rs1048661 and rs2165241 were different between East Asian populations, including Japanese, Chinese, and Korean, and other ethnic populations such as Caucasian, Middle Eastern, and black South African; the risk alleles of rs1048661 and rs2165241 for XFS in East Asians were the opposite of those reported for other ethnic populations [44-46]. On the other hand, the risk allele of rs3825942 for XFS was different between black South Africans and all other reported ethnicities, including East Asians and Caucasians [44-46]. Although the reasons for discrepancies in the allelic distributions of the LOXL1 SNPs among XFS patients with different ethnicities are unclear, it has been suggested that these SNPs are not the true causal variants of XFS, and that unidentified genetic variants in strong LD with these SNPs may play important roles in the development of XFS.
In this study, we found that the risk allele of FGF10 rs339501 for extreme myopia in the Japanese population is different from that reported in the Chinese population residing in Taiwan. The disparity between our results and those of the original report can be explained based on the association between XFS and LOXL1 SNPs; another FGF10 variant may be the true genetic factor and the associations observed in the present and previous studies may have resulted from strong LD with the true FGF10 variant. Variable LD patterns among different ethnic groups could explain the conflicting results; the true risk-associated allele in FGF10 may be linked to the G allele of rs339501 in the Chinese population and the A allele of rs339501 in the Japanese population. This explanation does not seem to be unreasonable because a close similarity exists in the genetic backgrounds of the Japanese and Chinese populations [47]. In addition, our study and the original study used limited sample sizes of extreme myopia (433 from Japan, 125 from Taiwan). Limited sample sizes can sometimes lead to false positive or negative results in an association study. Therefore, further association studies of FGF10 variants with larger sample sizes of Japanese, Chinese, and other ethnic populations are needed. We also need to consider the disparity in gender between the present and the original study. Men comprised 44.4% of patients with extreme myopia in the present study, whereas 65.4% of patients were men in the original study. In recent genetic studies of extreme myopia, 30–40% of the patients were men [48-50], suggesting that extreme myopia is more common in women, although the association of gender with extreme myopia still needs to be elucidated. Therefore, sampling bias may have existed in the original study.
In conclusion, we found that the FGF10 variants, including rs339501 reported in the previous study, are associated with extreme myopia in our Japanese population, whereas the disease risk–associated allele differed between the present and the previous study. Our findings suggest that the FGF10 variants studied in the present study are not an important risk factor for susceptibility to extreme myopia. However, because FGF10 variants may still affect the risk of extreme myopia, further genetic studies are needed to clarify the contribution of the FGF10 region in the development of extreme myopia.
Acknowledgments
This study was supported by JSPS KAKENHI Grant Number 23590382. We sincerely thank all of the participants for their participation in this study and all of the medical staff involved in sample collection and diagnosis.
References
- 1.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25:381–91. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 2.Fledelius HC. Myopia prevalence in Scandinavia. A survey, with emphasis on factors of relevance for epidemiological refraction studies in general. Acta Ophthalmol Suppl. 1988;185:44–50. doi: 10.1111/j.1755-3768.1988.tb02661.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilson A, Woo G. A review of the prevalence and causes of myopia. Singapore Med J. 1989;30:479–84. [PubMed] [Google Scholar]
- 4.Saw SM, Gazzard G, Koh D, Farook M, Widjaja D, Lee J, Tan DT. Prevalence rates of refractive errors in Sumatra, Indonesia. Invest Ophthalmol Vis Sci. 2002;43:3174–80. [PubMed] [Google Scholar]
- 5.Kleinstein RN, Jones LA, Hullett S, Kwon S, Lee RJ, Friedman NE, Manny RE, Mutti DO, Yu JA, Zadnik K, Collaborative Longitudinal Evaluation of Ethnicity and Refractive Error Study Group Refractive error and ethnicity in children. Arch Ophthalmol. 2003;121:1141–7. doi: 10.1001/archopht.121.8.1141. [DOI] [PubMed] [Google Scholar]
- 6.Wu MM, Edwards MH. The effect of having myopic parents: an analysis of myopia in three generations. Optom Vis Sci. 1999;76:387–92. doi: 10.1097/00006324-199906000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Guggenheim JA, Kirov G, Hodson SA. The heritability of high myopia: a reanalysis of Goldschmidt’s data. J Med Genet. 2000;37:227–31. doi: 10.1136/jmg.37.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutti DO, Mitchell GL, Moeschberger ML, Jones LA, Zadnik K. Parental myopia, near work, school achievement, and children's refractive error. Invest Ophthalmol Vis Sci. 2002;43:3633–40. [PubMed] [Google Scholar]
- 9.Farbrother JE, Kirov G, Owen MJ, Guggenheim JA. Family aggregation of high myopia: estimation of the sibling recurrence risk ratio. Invest Ophthalmol Vis Sci. 2004;45:2873–8. doi: 10.1167/iovs.03-1155. [DOI] [PubMed] [Google Scholar]
- 10.Liang CL, Yen E, Su JY, Liu C, Chang TY, Park N, Wu MJ, Lee S, Flynn JT, Juo SH. Impact of family history of high myopia on level and onset of myopia. Invest Ophthalmol Vis Sci. 2004;45:3446–52. doi: 10.1167/iovs.03-1058. [DOI] [PubMed] [Google Scholar]
- 11.Klein AP, Duggal P, Lee KE, Klein R, Bailey-Wilson JE, Klein BE. Support for polygenic influences on ocular refractive error. Invest Ophthalmol Vis Sci. 2005;46:442–6. doi: 10.1167/iovs.04-0794. [DOI] [PubMed] [Google Scholar]
- 12.Sorsby A, Fraser GR. Statistical note on the components of ocular refraction in twins. J Med Genet. 1964;1:47–9. doi: 10.1136/jmg.1.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyhne N, Sjolie AK, Kyvik KO, Green A. The importance of genes and environment for ocular refraction and its determiners: a population based study among 20–45 year old twins. Br J Ophthalmol. 2001;85:1470–6. doi: 10.1136/bjo.85.12.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saw SM, Chua WH, Gazzard G, Koh D, Tan DT, Stone RA. Eye growth changes in myopic children in Singapore. Br J Ophthalmol. 2005;89:1489–94. doi: 10.1136/bjo.2005.071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guggenheim JA, Pong-Wong R, Haley CS, Gazzard G, Saw SM. Correlations in refractive errors between siblings in the Singapore Cohort Study of risk factors for myopia. Br J Ophthalmol. 2007;91:781–4. doi: 10.1136/bjo.2006.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu L, Li ZK, Gao JR, Liu JR, Xu CT. Epidemiology, genetics and treatments for myopia. Int J Ophthalmol. 2011;4:658–69. doi: 10.3980/j.issn.2222-3959.2011.06.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hysi PG, Young TL, Mackey DA, Andrew T, Fernández-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YJ, Goh L, Khor CC, Fan Q, Yu M, Han S, Sim X, Ong RT, Wong TY, Vithana EN, Yap E, Nakanishi H, Matsuda F, Ohno-Matsui K, Yoshimura N, Seielstad M, Tai ES, Young TL, Saw SM. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118:368–75. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Qu J, Xu X, Zhou X, Zou H, Wang N, Li T, Hu X, Zhao Q, Chen P, Li W, Huang K, Yang J, He Z, Ji J, Wang T, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum Mol Genet. 2011;20:2861–8. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, Qu J, Zhang D, Zhao P, Zhang Q, Tam PO, Sun L, Zuo X, Zhou X, Xiao X, Hu J, Li Y, Cai L, Liu X, Lu F, Liao S, Chen B, He F, Gong B, Lin H, Ma S, Cheng J, Zhang J, Chen Y, Zhao F, Yang X, Chen Y, Yang C, Lam DS, Li X, Shi F, Wu Z, Lin Y, Yang J, Li S, Ren Y, Xue A, Fan Y, Li D, Pang CP, Zhang X, Yang Z. Genetic variants at 13q12.12 are associated with high myopia in the Han Chinese population. Am J Hum Genet. 2011;88:805–13. doi: 10.1016/j.ajhg.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verhoeven VJ, Hysi PG, Saw SM, Vitart V, Mirshahi A, Guggenheim JA, Cotch MF, Yamashiro K, Baird PN, Mackey DA, Wojciechowski R, Ikram MK, Hewitt AW, Duggal P, Janmahasatian S, Khor CC, Fan Q, Zhou X, Young TL, Tai ES, Goh LK, Li YJ, Aung T, Vithana E, Teo YY, Tay W, Sim X, Rudan I, Hayward C, Wright AF, Polasek O, Campbell H, Wilson JF, Fleck BW, Nakata I, Yoshimura N, Yamada R, Matsuda F, Ohno-Matsui K, Nag A, McMahon G, St Pourcain B, Lu Y, Rahi JS, Cumberland PM, Bhattacharya S, Simpson CL, Atwood LD, Li X, Raffel LJ, Murgia F, Portas L, Despriet DD, van Koolwijk LM, Wolfram C, Lackner KJ, Tönjes A, Mägi R, Lehtimäki T, Kähönen M, Esko T, Metspalu A, Rantanen T, Pärssinen O, Klein BE, Meitinger T, Spector TD, Oostra BA, Smith AV, de Jong PT, Hofman A, Amin N, Karssen LC, Rivadeneira F, Vingerling JR, Eiríksdóttir G, Gudnason V, Döring A, Bettecken T, Uitterlinden AG, Williams C, Zeller T, Castagné R, Oexle K, van Duijn CM, Iyengar SK, Mitchell P, Wang JJ, Höhn R, Pfeiffer N, Bailey-Wilson JE, Stambolian D, Wong TY, Hammond CJ, Klaver CC. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131:1467–80. doi: 10.1007/s00439-012-1176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Q, Barathi VA, Cheng CY, Zhou X, Meguro A, Nakata I, Khor CC, Goh LK, Li YJ, Lim W, Ho CE, Hawthorne F, Zheng Y, Chua D, Inoko H, Yamashiro K, Ohno-Matsui K, Matsuo K, Matsuda F, Vithana E, Seielstad M, Mizuki N, Beuerman RW, Tai ES, Yoshimura N, Aung T, Young TL, Wong TY, Teo YY, Saw SM. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8:e1002753. doi: 10.1371/journal.pgen.1002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng W, Butterworth J, Bradley DT, Hughes AE, Soler V, Calvas P, Malecaze F. A genome-wide association study provides evidence for association of chromosome 8p23 (MYP10) and 10q21.1 (MYP15) with high myopia in the French Population. Invest Ophthalmol Vis Sci. 2012;53:7983–8. doi: 10.1167/iovs.12-10409. [DOI] [PubMed] [Google Scholar]
- 26.Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, Höhn R, MacGregor S, Hewitt AW, Nag A, Cheng CY, Yonova-Doing E, Zhou X, Ikram MK, Buitendijk GH, McMahon G, Kemp JP, Pourcain BS, Simpson CL, Mäkelä KM, Lehtimäki T, Kähönen M, Paterson AD, Hosseini SM, Wong HS, Xu L, Jonas JB, Pärssinen O, Wedenoja J, Yip SP, Ho DW, Pang CP, Chen LJ, Burdon KP, Craig JE, Klein BE, Klein R, Haller T, Metspalu A, Khor CC, Tai ES, Aung T, Vithana E, Tay WT, Barathi VA. Consortium for Refractive Error and Myopia (CREAM), Chen P, Li R, Liao J, Zheng Y, Ong RT, Döring A; Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Research Group, Evans DM, Timpson NJ, Verkerk AJ, Meitinger T, Raitakari O, Hawthorne F, Spector TD, Karssen LC, Pirastu M, Murgia F, Ang W; Wellcome Trust Case Control Consortium 2 (WTCCC2), Mishra A, Montgomery GW, Pennell CE, Cumberland PM, Cotlarciuc I, Mitchell P, Wang JJ, Schache M, Janmahasathian S, Igo RP Jr, Lass JH, Chew E, Iyengar SK; Fuchs' Genetics Multi-Center Study Group, Gorgels TG, Rudan I, Hayward C, Wright AF, Polasek O, Vatavuk Z, Wilson JF, Fleck B, Zeller T, Mirshahi A, Müller C, Uitterlinden AG, Rivadeneira F, Vingerling JR, Hofman A, Oostra BA, Amin N, Bergen AA, Teo YY, Rahi JS, Vitart V, Williams C, Baird PN, Wong TY, Oexle K, Pfeiffer N, Mackey DA, Young TL, van Duijn CM, Saw SM, Bailey-Wilson JE, Stambolian D, Klaver CC, Hammond CJ. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45:314–8. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Gong B, Chen L, Zuo X, Liu X, Tam PO, Zhou X, Zhao P, Lu F, Qu J, Sun L, Zhao F, Chen H, Zhang Y, Zhang D, Lin Y, Lin H, Ma S, Cheng J, Yang J, Huang L, Zhang M, Zhang X, Pang CP, Yang Z. A genome-wide meta-analysis identifies two novel loci associated with high myopia in the Han Chinese population. Hum Mol Genet. 2013;22:2325–33. doi: 10.1093/hmg/ddt066. [DOI] [PubMed] [Google Scholar]
- 28.Govindarajan V, Ito M, Makarenkova HP, Lang RA, Overbeek PA. Endogenous and ectopic gland induction by FGF-10. Dev Biol. 2000;225:188–200. doi: 10.1006/dbio.2000.9812. [DOI] [PubMed] [Google Scholar]
- 29.Makarenkova HP, Ito M, Govindarajan V, Faber SC, Sun L, McMahon G, Overbeek PA, Lang RA. FGF10 is an inducer and Pax6 a competence factor for lacrimal gland development. Development. 2000;127:2563–72. doi: 10.1242/dev.127.12.2563. [DOI] [PubMed] [Google Scholar]
- 30.Puk O, Esposito I, Söker T, Löster J, Budde B, Nürnberg P, Michel-Soewarto D, Fuchs H, Wolf E, Hrabé de Angelis M, Graw J. A new Fgf10 mutation in the mouse leads to atrophy of the harderian gland and slit-eye phenotype in heterozygotes: a novel model for dry-eye disease? Invest Ophthalmol Vis Sci. 2009;50:4311–8. doi: 10.1167/iovs.09-3451. [DOI] [PubMed] [Google Scholar]
- 31.Hausman RE. Ocular extracellular matrices in development. Prog Retin Eye Res. 2007;26:162–88. doi: 10.1016/j.preteyeres.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 32.McBrien NA, Lawlor P, Gentle A. Scleral remodeling during the development of and recovery from axial myopia in the tree shrew. Invest Ophthalmol Vis Sci. 2000;41:3713–9. [PubMed] [Google Scholar]
- 33.Siegwart JT, Jr, Norton TT. Selective regulation of MMP and TIMP mRNA levels in tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol Vis Sci. 2005;46:3484–92. doi: 10.1167/iovs.05-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X, Pan Y, Carbe C, Powers A, Grobe K, Zhang X. Glycosaminoglycan-dependent restriction of FGF diffusion is necessary for lacrimal gland development. Development. 2012;139:2730–9. doi: 10.1242/dev.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsau C, Ito M, Gromova A, Hoffman MP, Meech R, Makarenkova HP. Barx2 and Fgf10 regulate ocular glands branching morphogenesis by controlling extracellular matrix remodeling. Development. 2011;138:3307–17. doi: 10.1242/dev.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsi E, Chen KC, Chang WS, Yu ML, Liang CL, Juo SH. A functional polymorphism at the FGF10 gene is associated with extreme myopia. Invest Ophthalmol Vis Sci. 2013;54:3265–71. doi: 10.1167/iovs.13-11814. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Willer CJ, Sanna S, Abecasis GR. Genotype Imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong L, Catoire H, Dion P, Gaspar C, Lafrenière RG, Girard SL, Levchenko A, Rivière JB, Fiori L, St-Onge J, Bachand I, Thibodeau P, Allen R, Earley C, Turecki G, Montplaisir J, Rouleau GA. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–74. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju H, Lim B, Kim M, Noh SM, Kim WH, Ihm C, Choi BY, Kim YS, Kang C. SERPINE1 intron polymorphisms affecting gene expression are associated with diffuse-type gastric cancer susceptibility. Cancer. 2010;116:4248–55. doi: 10.1002/cncr.25213. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274–86. doi: 10.1038/tpj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams SE, Whigham BT, Liu Y, Carmichael TR, Qin X, Schmidt S, Ramsay M, Hauser MA, Allingham RR. Major LOXL1 risk allele is reversed in exfoliation glaucoma in a black South African population. Mol Vis. 2010;16:705–12. [PMC free article] [PubMed] [Google Scholar]
- 45.Rautenbach RM, Bardien S, Harvey J, Ziskind A. An investigation into LOXL1 variants in black South African individuals with exfoliation syndrome. Arch Ophthalmol. 2011;129:206–10. doi: 10.1001/archophthalmol.2010.349. [DOI] [PubMed] [Google Scholar]
- 46.Kasım B, İrkeç M, Alikaşifoğlu M, Orhan M, Mocan MC, Aktaş D. Association of LOXL1 gene polymorphisms with exfoliation syndrome/glaucoma and primary open angle glaucoma in a Turkish population. Mol Vis. 2013;19:114–20. [PMC free article] [PubMed] [Google Scholar]
- 47.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu MM, Yap MK, Ho DW, Fung WY, Ng PW, Gu YS, Yip SP. Investigating the relationship between UMODL1 gene polymorphisms and high myopia: a case–control study in Chinese. BMC Med Genet. 2012;13:64. doi: 10.1186/1471-2350-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhuang W, Yang P, Li Z, Sheng X, Zhao J, Li S, Yang X, Xiang W, Rong W, Liu Y, Zhang F. Association of insulin-like growth factor-1 polymorphisms with high myopia in the Chinese population. Mol Vis. 2012;18:634–44. [PMC free article] [PubMed] [Google Scholar]
- 50.Miyake M, Yamashiro K, Nakanishi H, Nakata I, Akagi-Kurashige Y, Tsujikawa A, Moriyama M, Ohno-Matsui K, Mochizuki M, Yamada R, Matsuda F, Yoshimura N. Insulin-like growth factor 1 is not associated with high myopia in a large Japanese cohort. Mol Vis. 2013;19:1074–81. [PMC free article] [PubMed] [Google Scholar]