Abstract
Purpose
Age-related macular degeneration (AMD) is a major cause of blindness in developed countries. The molecular pathogenesis of early events in AMD is poorly understood. We investigated differential gene expression in samples of human retinal pigment epithelium (RPE) and choroid from early AMD and control maculas with exon-based arrays.
Methods
Gene expression levels in nine human donor eyes with early AMD and nine control human donor eyes were assessed using Affymetrix Human Exon ST 1.0 arrays. Two controls did not pass quality control and were removed. Differentially expressed genes were annotated using the Database for Annotation, Visualization and Integrated Discovery (DAVID), and gene set enrichment analysis (GSEA) was performed on RPE-specific and endothelium-associated gene sets. The complement factor H (CFH) genotype was also assessed, and differential expression was analyzed regarding high AMD risk (YH/HH) and low AMD risk (YY) genotypes.
Results
Seventy-five genes were identified as differentially expressed (raw p value <0.01; ≥50% fold change, mean log2 expression level in AMD or control ≥ median of all average gene expression values); however, no genes were significant (adj. p value <0.01) after correction for multiple hypothesis testing. Of 52 genes with decreased expression in AMD (fold change <0.5; raw p value <0.01), 18 genes were identified by DAVID analysis as associated with vision or neurologic processes. The GSEA of the RPE-associated and endothelium-associated genes revealed a significant decrease in genes typically expressed by endothelial cells in the early AMD group compared to controls, consistent with previous histologic and proteomic studies. Analysis of the CFH genotype indicated decreased expression of ADAMTS9 in eyes with high-risk genotypes (fold change = –2.61; raw p value=0.0008).
Conclusions
GSEA results suggest that RPE transcripts are preserved or elevated in early AMD, concomitant with loss of endothelial cell marker expression. These results are consistent with the notion that choroidal endothelial cell dropout or dedifferentiation occurs early in the pathogenesis of AMD.
Introduction
Age-related macular degeneration (AMD) is the leading cause of blindness among the elderly in developed countries [1]. AMD involves the progressive loss of photoreceptor cells from the macular region of the retina, resulting in impaired vision and, in advanced stages, blindness. At least three cell layers undergo changes in AMD, including the photoreceptor cells, retinal pigment epithelium (RPE), and choriocapillaris. The RPE regulates the activities of the photoreceptor cells and choriocapillaris. For example, RPE cells actively phagocytose photoreceptor cell outer segments, recycle vitamin A, shuttle debris from the photoreceptor cells to the bloodstream, and import glucose, oxygen, and other components to accommodate the high metabolic demands of the retina [2], in addition to providing trophic support to the choriocapillaris [3,4]. The choroid serves as a high-volume transportation courier, delivering nutrients to the RPE and accepting waste products for further processing elsewhere in the body. The preclinical and early stages of AMD are recognizable by increased formation of lipid-rich sub-RPE deposits termed drusen and altered RPE pigmentation [5,6].
The photoreceptor cells, RPE, and choriocapillaris endothelial cells form an interdependent complex. Injury or dysfunction in any of these layers leads to loss of the other two in several chorioretinal diseases. A more complete understanding of the early sequelae of events in AMD is necessary to guide new therapies. Numerous interdependent biologic processes have been implicated in the pathogenesis of AMD, including increased activity of the complement cascade, infiltration of cells mediating inflammatory responses, increased oxidative stress, and altered lipid metabolism [7,8]. Although RPE cells are typically viewed as the primary cells affected in AMD, changes in the microvasculature of the choroid (choriocapillaris) have also been reported in association with drusen, including dropout of vessels [9,10] and decreased blood flow [11]. In a subset of advanced AMD cases, choroidal neovascular membranes (CNVs) form as blood vessels from the choroid breach the RPE and proliferate either beneath the RPE or in the sub-retinal space. Expression of vascular endothelial growth factor (VEGF), a marker of hypoxia, has been implicated in the formation of CNVs [12]. In current medical practice, only after CNVs have appeared and photoreceptor cell death has occurred can therapeutic measures be taken to slow further vision loss [13].
Despite considerable progress in unraveling genetic risk factors for AMD, major challenges remain. The relationships between the biologic processes remain uncertain, and the initial molecular conditions driving development of AMD are poorly understood. Evaluating gene expression in early AMD, intermediate AMD, and advanced AMD is one approach to advancing exploration of these problems. The first large-scale study of gene expression in the AMD-affected retina and RPE and choroid tissue identified changes between various stages of AMD, including apoptotic and neovascular pathways in advanced AMD [14]. As part of a study examining the relationship between AMD and gene methylation, Hunter and colleagues examined gene expression in AMD and normal samples [15]. They found that expression of glutathione S-transferase isoform mu1 (GSTM1) and mu5 (GSTM5), antioxidant isoenzymes, was reduced in the RPE and choroid of eyes affected by AMD compared to control eyes; this reduction was correlated with hypermethylation of the GSTM1 promoter [15]. However, only two of the samples were classified as early AMD, and analysis was performed without respect to AMD grade.
To further investigate the molecular events initiating AMD, we evaluated gene expression in early AMD and normal RPE and choroid tissues using exon-based microarrays (Affymetrix). Our analysis identified a statistically significant decrease in expression of endothelial genes in early AMD with preservation of RPE-specific transcripts, suggesting that vessel loss or dedifferentiation may precede advanced damage in AMD.
Methods
Tissue acquisition
Donor eyes were obtained through the Iowa Lions Eye Bank (Iowa City, IA) after receiving informed consent in accordance with the tenets of the Declaration of Helsinki. Donor age, sex, cause of death, and diagnostic status are listed in Table 1. Eyes were dissected, and 6 mm punches were taken from the macula, centered on the fovea centralis. Macular RPE and choroid were separated from the neural retina, flash frozen in liquid nitrogen, and stored at −80 °C. The neural retina was used for separate experiments. In this manner, RNA was stabilized within 4 h of death. RNA was extracted from the frozen RPE and choroids using a commercially available kit (RNeasy Mini Kit; Qiagen, Valencia, CA).
Table 1. Donor information.
| Sample | Batch | Gender | CFH genotype | Age | Cause of death | Age-related maculopathy |
|---|---|---|---|---|---|---|
| ARM1 |
A |
F |
HY |
78 |
Coronary artery disease |
RPE changes |
| ARM2 |
A |
F |
HY |
80 |
Intracerebral hemorrhage |
RPE changes |
| ARM3 |
A |
M |
HY |
90 |
Respiratory failure |
Macular drusen |
| ARM4 |
A |
F |
HY |
91 |
Pneumonia |
Macular drusen |
| ARM5 |
A |
F |
YY |
91 |
Not available |
RPE changes; neovascular membrane in contralateral eye |
| ARM6 |
A |
M |
HH |
78 |
Respiratory failure |
RPE changes |
| ARM7 |
B |
F |
YY |
81 |
Ischemic bowel |
RPE changes |
| ARM8 |
B |
M |
HH |
92 |
Pneumonia |
Numerous large drusen, no atrophy or exudate |
| ARM9 |
B |
M |
HH |
77 |
Not available |
RPE changes |
| CTRL1 |
B |
M |
HY |
93 |
Cardiac arrest |
Normal fundus exam <2 years |
| CTRL2 |
A |
M |
YY |
83 |
Pneumonia |
Normal fundus exam <2 years; large cup to disc ratio |
| CTRL3* |
B |
M |
HY |
84 |
Subdural hematoma |
Normal fundus exam >2 years |
| CTRL4 |
B |
F |
HY |
77 |
Not available |
Normal fundus exam but old records; normal gross appearance |
| CTRL5 |
A |
M |
YY |
81 |
Respiratory failure |
Normal fundus exam <2 years |
| CTRL6 |
A |
F |
HH |
87 |
Aortic stenosis |
Normal fundus exam <2 years |
| CTRL7 |
A |
M |
HY |
77 |
Renal failure |
Normal fundus exam <2 years |
| CTRL8 |
B |
F |
YY |
83 |
Not available |
Normal fundus exam <2 years |
| CTRL9* | A | M | HY | 77 | Brain tumor | Normal fundus exam <2 years |
Asterisks (*) denote samples that did not pass quality control metrics. Batch indicates the batch of array processing. RPE changes refer to regions where RPE is depigmented or hypopigmented [32].
CFH risk-allele genotyping
To characterize how the risk allele in complement factor H (CFH; single nucleotide polymorphism [SNP] rs1061170) affects gene expression in the RPE and choroid, genotyping was performed on DNA from either whole blood (collected in EDTA-coated tubes) or extraocular muscle. DNA was isolated using established methods with the Gentra system (Qiagen, Valencia, CA) or the Qiagen DNeasy Blood & Tissue Kit, for blood and muscle, respectively. Genotyping of this polymorphism was performed using TaqMan predesigned SNP genotyping assays (Applied Biosystems) in a high-throughput system (Fluidigm, San Francisco, CA).
Microarray processing
Exon expression levels of the RPE and choroid RNA were determined using Affymetrix GeneChip Human Exon 1.0 ST arrays processed in two batches at the University of Iowa DNA Facility. Samples were used essentially as previously described [16]. Briefly, single primer isothermal amplification was used to convert 25 ng total RNA to cDNA with the WT-Ovation Pico RNA Amplification System (NuGEN Technologies, San Carlos, CA). The resulting cDNA was purified using a Qiagen QIAquick PCR Purification column, converted to sense target (ST)-cDNA using the WT-Ovation Exon Module v1 (NuGEN Technologies), and purified once more. ST-cDNA was then fragmented (85 nt mean length), and the NuGEN FL-Ovation cDNA Biotin Module, v2 (NuGEN Technologies) was used to biotin-label the fragments according to the manufacturer’s guidelines. This product was combined with Affymetrix eukaryotic hybridization buffer (Affymetrix, Santa Clara, CA) and hybridized to Affymetrix Human Exon 1.0 ST arrays (Affymetrix). An Affymetrix Model 3000 scanner with 7G upgrade was used to scan the arrays. Data were collected with GeneChip operating software (ver. 1.4; Affymetrix) and saved as CEL files.
Microarray analysis
Preprocessing, expression analysis, and quality control
To assess differential expression and alternative splicing, CEL files were normalized by robust multiarray average (RMA) [17] to facilitate comparisons across arrays using AltAnalyze (ver. 2.0.7 beta) with gene and transcript data from the Ensembl 65 database and human genome build Hg19 [18]. RMA, a common algorithm for processing microarray data, corrects for background fluorescence, normalizes data for comparison between arrays, and produces log2-transformed estimates of probeset expression. Technical variation among microarrays is well known and often arises when arrays are processed in separate batches (i.e., prepared by different people, on different days, etc.) [19]. To address this issue in our data set, initial probeset-level expression estimates were corrected for batch effects while preserving AMD status, CFH genotype, and sex covariates using the ComBat algorithm [20]. ComBat removes batch effects more effectively than several other procedures [21]. The batch-corrected data were reimported into AltAnalyze and processed as follows: masking of cross-hybridizing probesets; filtering probesets expressed below non-log level (i.e., expression intensity before log2 transformation) of 70; gene-level differential expression using a moderated t test; and computation of false discovery rate (FDR) adjusted p values to account for multiple hypothesis testing correction [22]. Heatmap generation and hierarchical clustering were performed using the R statistical software (ver. 3.0.0) [23]. To cluster genes based on the similarity of the expression pattern across samples, we used a Pearson-based distance metric (1 minus Pearson’s correlation coefficient) [24]. To assess alternative splicing, the splicing index and MIDAS [25] procedures in AltAnalyze were used. Putatively alternatively spliced transcripts were visualized using DomainGraph (ver. 3.0) [18], a plugin for Cytoscape (ver. 2.8.1) [26]. Quality control metrics, including distance between arrays and comparison of array intensity distributions, were calculated using the arrayQualityMetrics package (ver. 3.16.0) [27] for R. After outlying arrays were removed, the final AltAnalyze results were recomputed.
Gene set analysis
The Ensembl BioMart tool was used to map identifiers from previous gene expression studies to Ensembl IDs [28]. Gene set enrichment analysis (GSEA; ver. 14) [29] implemented with GenePattern (ver. 3.6.0) [30] was used to evaluate the overrepresentation of custom gene sets with AMD or control samples. For GSEA, phenotype permutation was performed using 1,000 permutations and two gene sets, an RPE-specific set and an endothelium-associated set, compiled based on literature search. Annotation of differentially expressed genes was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Tools with default parameters [31].
Results
Quality control
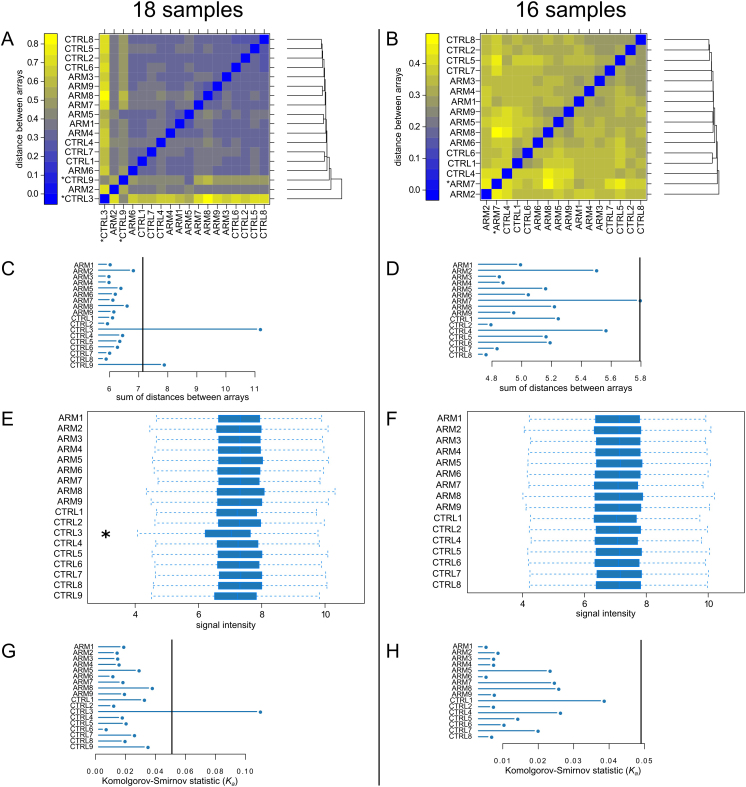
We defined early AMD as RPE changes (depigmentation or hypopigmentation) and/or macular drusen without geographic atrophy (GA) or CNV [32] as determined by chart review. Eyes with CNV or geographic atrophy were excluded. Since early AMD was classified in the donors by ophthalmoscopy, completely ruling out a rare event such as occult CNV is impossible in the early AMD samples. With the exception of one donor who later did not pass quality control, all unaffected control donors were required to have had a normal fundus exam within two years of death. Based on these criteria, nine samples were classified as early AMD (designated ARM1-9) and nine samples were classified as controls (designated CTRL1-9; Table 1). To assess the statistical equivalence of donor ages in each group, we performed Welch’s two-sample t test, which gave a p value of 0.54. The mean RNA integrity numbers for samples processed in batch A was 6.76 (standard deviation [SD] 0.34) and for batch B was 6.22 (SD 0.59; Table 1). Following normalization in AltAnalyze (Figure 1), samples CTRL3 and CTRL9 were flagged as potential outliers by arrayQualityMetrics analysis [27] based on their sum of distances to other arrays (Figure 2A,C). Additionally, the gene-level signal intensity distribution of CTRL3 was lower than that of other arrays and was flagged by arrayQualityMetrics’s Kolmogorov–Smirnov-based outlier detection module (Figure 2E,G). Given the magnitude of the difference between these two arrays and the other arrays, we removed these two arrays from the data set and reprocessed the remaining 16 samples (Figure 2B,D,F,H). With this reduced data set, ARM7 was flagged as an outlier based on its sum of distances to other arrays (Figure 2B,D). However, as the magnitude of this difference was marginal, we retained this sample in the data set. After CTRL3 and CTRL9 were removed, the p value for age between the two groups was 0.70.
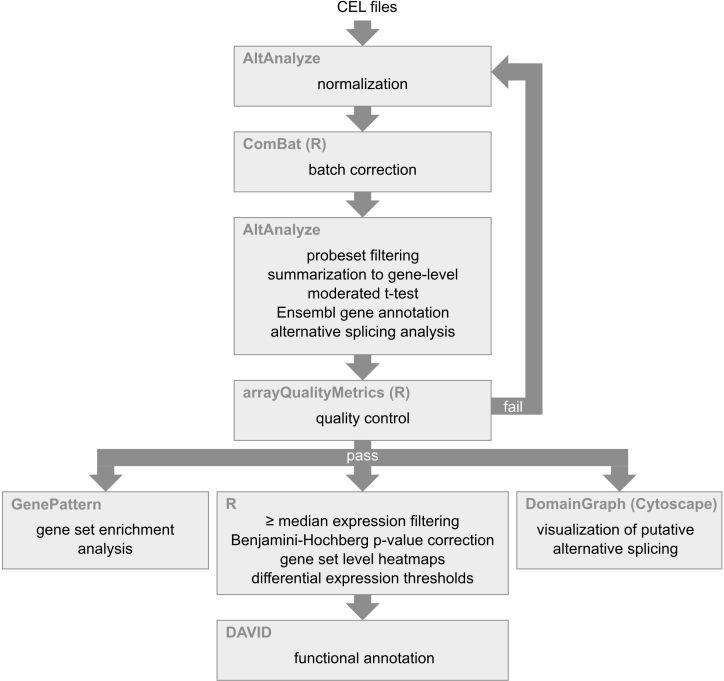
Figure 1.
Overview of bioinformatics pipeline. Software is shown in the upper left corner of the boxes while the analytic process is indicated in the center of the boxes.
Figure 2.
Quality control plots generated by arrayQualityMetrics before and after removal of CTRL3 and CTRL9. A and B are a false color heatmaps indicating the distances between arrays, computed as the absolute mean distance between the data. C and D indicate the sum of distances computed for A and B. For C the outlier threshold was 7.14 (vertical bar), and for D the outlier threshold was 5.79 (vertical bar). When analyzing all 18 samples, both CTRL3 and CTRL9 were flagged as exceeding the outlier threshold (A and C). When CTRL3 and CTRL9 were omitted, only ARM7 exceeded the threshold (D). E and F are boxplots of the gene-level signal intensity distribution across the arrays. For E and F, the Kolmogorov-Smirnov statistic Ka was calculated based on the distanced between each individual array and the pooled distribution of all arrays. G and H show Ka for each array with outlier thresholds (vertical bars) of 0.0509 and 0.049, respectively. Only CTRL3 was flagged by this metric (G).
Differentially expressed genes
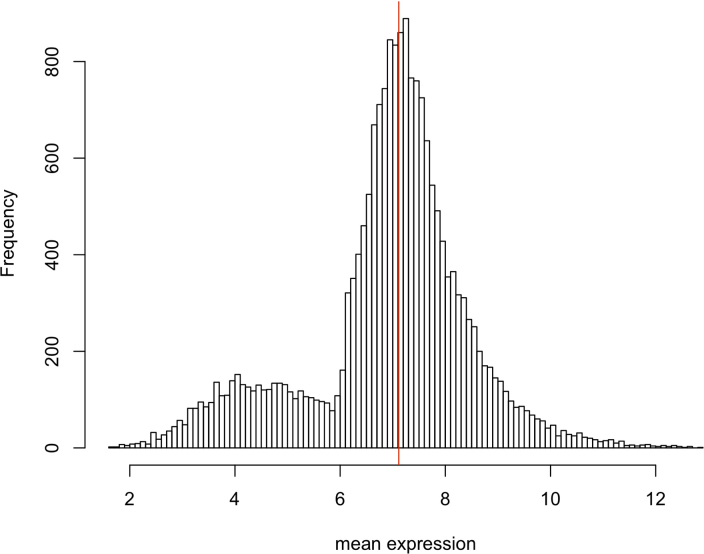
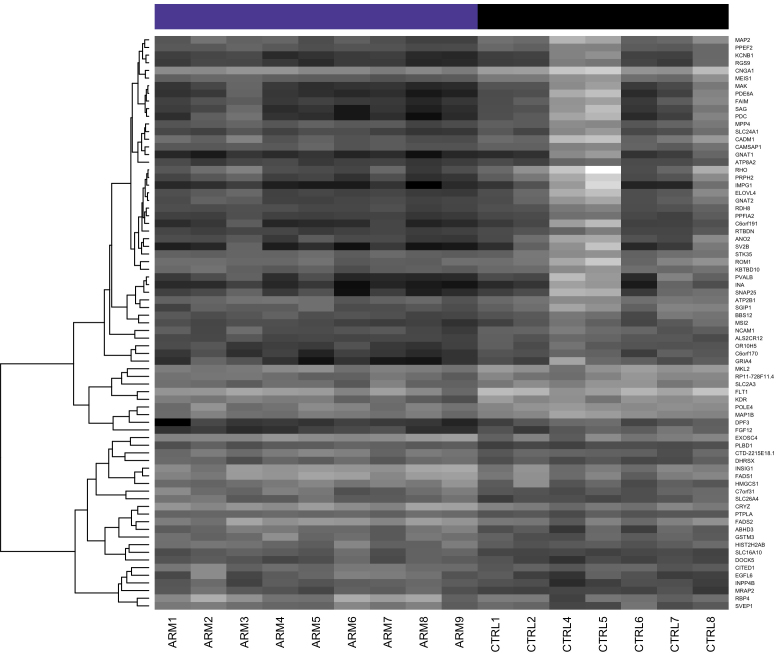
AltAnalyze mapped the exon array probesets to 42,187 unique Ensembl identifiers. To reduce false positive observations, we filtered this set by selecting only protein coding genes with mean log2 expression greater than or equal to the median expression of all protein-coding genes (7.11; Figure 3), which resulted in 10,286 genes. We found 75 genes differentially expressed in AMD compared to controls (at least 50% difference in expression; moderated t test raw p value <0.01; Table 2; Figure 4). None of the genes were significantly differentially expressed after FDR correction (adj. p value <0.1). We submitted the 52 downregulated genes and the 23 upregulated genes to DAVID for annotation. Of genes upregulated in early AMD, three genes (FADS1, FADS2, PTPLA) were identified as implicated in biosynthesis of unsaturated fatty acids (fold enrichment = 86.68; Benjamini corrected p value = 0.0052). Of genes downregulated in early AMD, 28 terms were significantly enriched (fold enrichment from 2 to 128; Benjamini-corrected p value <0.01). These terms were associated with vision, sensory perception, and the plasma membrane. No transcripts flagged by AltAnalyze as putative splicing events appeared to be alternatively spliced upon close visual inspection of the probeset data in DomainGraph.
Figure 3.
Distribution of mean expression before median thresholding. Mean expression is log2 of gene-level signal intensity. The median indicated by the red line.
Table 2. Differentially expressed genes in early AMD versus control samples.
| Ensembl ID | Symbol | Mean AMD | Mean Control | Fold change | Raw p value | FDR adj. P value | Name |
|---|---|---|---|---|---|---|---|
| ENSG00000138207 |
RBP4 |
9.01 |
7.92 |
2.12 |
0.0089 |
0.4149 |
retinol binding protein 4, plasma |
| ENSG00000198759 |
EGFL6 |
7.78 |
6.77 |
2.01 |
0.0089 |
0.4149 |
EGF-like-domain, multiple 6 |
| ENSG00000158201 |
ABHD3 |
8.01 |
7.13 |
1.84 |
0.0044 |
0.3607 |
abhydrolase domain containing 3 |
| ENSG00000186480 |
INSIG1 |
9.25 |
8.44 |
1.76 |
0.0024 |
0.3582 |
insulin induced gene 1 |
| ENSG00000112972 |
HMGCS1 |
8.39 |
7.58 |
1.76 |
0.0095 |
0.4149 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| ENSG00000116791 |
CRYZ |
9.20 |
8.39 |
1.75 |
0.0002 |
0.2057 |
crystallin, zeta (quinone reductase) |
| ENSG00000135324 |
MRAP2 |
7.41 |
6.61 |
1.75 |
0.0006 |
0.2515 |
melanocortin 2 receptor accessory protein 2 |
| ENSG00000165124 |
SVEP1 |
8.16 |
7.37 |
1.74 |
0.0052 |
0.3792 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| ENSG00000149485 |
FADS1 |
9.22 |
8.43 |
1.73 |
0.0035 |
0.3582 |
fatty acid desaturase 1 |
| ENSG00000112394 |
SLC16A10 |
7.43 |
6.67 |
1.70 |
0.0001 |
0.1976 |
solute carrier family 16, member 10 (aromatic amino acid transporter) |
| ENSG00000109452 |
INPP4B |
7.41 |
6.66 |
1.69 |
0.0031 |
0.3582 |
inositol polyphosphate-4-phosphatase, type II, 105 kDa |
| ENSG00000091137 |
SLC26A4 |
7.80 |
7.04 |
1.69 |
0.0030 |
0.3582 |
solute carrier family 26, member 4 |
| ENSG00000134824 |
FADS2 |
9.08 |
8.34 |
1.67 |
0.0089 |
0.4149 |
fatty acid desaturase 2 |
| ENSG00000153790 |
C7orf31 |
8.06 |
7.32 |
1.67 |
0.0089 |
0.4149 |
chromosome 7 open reading frame 31 |
| ENSG00000251606 |
CTD-2215E18.1 |
8.28 |
7.55 |
1.66 |
0.0006 |
0.2515 |
Uncharacterized protein |
| ENSG00000178896 |
EXOSC4 |
9.03 |
8.32 |
1.63 |
0.0039 |
0.3582 |
exosome component 4 |
| ENSG00000125931 |
CITED1 |
8.14 |
7.47 |
1.60 |
0.0013 |
0.3582 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich C-terminal domain, 1 |
| ENSG00000184270 |
HIST2H2AB |
8.04 |
7.41 |
1.55 |
0.0034 |
0.3582 |
histone cluster 2, H2ab |
| ENSG00000121316 |
PLBD1 |
7.40 |
6.77 |
1.54 |
0.0001 |
0.1976 |
phospholipase B domain containing 1 |
| ENSG00000134202 |
GSTM3 |
8.04 |
7.42 |
1.53 |
0.0095 |
0.4149 |
glutathione S-transferase mu 3 (brain) |
| ENSG00000165996 |
PTPLA |
7.81 |
7.21 |
1.51 |
0.0000 |
0.1157 |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| ENSG00000147459 |
DOCK5 |
7.25 |
6.65 |
1.51 |
0.0062 |
0.4088 |
dedicator of cytokinesis 5 |
| ENSG00000169084 |
DHRSX |
7.68 |
7.09 |
1.51 |
0.0035 |
0.3582 |
dehydrogenase/reductase (SDR family) X-linked |
| ENSG00000070961 |
ATP2B1 |
7.75 |
8.37 |
−1.53 |
0.0041 |
0.3582 |
ATPase, Ca2+ transporting, plasma membrane 1 |
| ENSG00000186260 |
MKL2 |
8.50 |
9.12 |
−1.54 |
0.0038 |
0.3582 |
MKL/myocardin-like 2 |
| ENSG00000125834 |
STK35 |
7.66 |
8.29 |
−1.54 |
0.0057 |
0.4062 |
serine/threonine kinase 35 |
| ENSG00000155749 |
ALS2CR12 |
6.77 |
7.39 |
−1.54 |
0.0004 |
0.2281 |
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 |
| ENSG00000115350 |
POLE4 |
8.41 |
9.04 |
−1.55 |
0.0028 |
0.3582 |
polymerase (DNA-directed), epsilon 4 (p12 subunit) |
| ENSG00000130559 |
CAMSAP1 |
7.34 |
7.98 |
−1.56 |
0.0028 |
0.3582 |
calmodulin regulated spectrin-associated protein 1 |
| ENSG00000132932 |
ATP8A2 |
6.46 |
7.13 |
−1.59 |
0.0030 |
0.3582 |
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| ENSG00000181004 |
BBS12 |
7.07 |
7.75 |
−1.60 |
0.0098 |
0.4191 |
Bardet-Biedl syndrome 12 |
| ENSG00000149294 |
NCAM1 |
7.06 |
7.74 |
−1.60 |
0.0068 |
0.4149 |
neural cell adhesion molecule 1 |
| ENSG00000254528 |
RP11–728F11.4 |
7.98 |
8.67 |
−1.62 |
0.0018 |
0.3582 |
|
| ENSG00000156194 |
PPEF2 |
7.19 |
7.89 |
−1.63 |
0.0024 |
0.3582 |
protein phosphatase, EF-hand calcium binding domain 2 |
| ENSG00000132026 |
RTBDN |
6.52 |
7.25 |
−1.66 |
0.0032 |
0.3582 |
retbindin |
| ENSG00000080511 |
RDH8 |
6.82 |
7.56 |
−1.66 |
0.0046 |
0.3607 |
retinol dehydrogenase 8 (all-trans) |
| ENSG00000139220 |
PPFIA2 |
6.46 |
7.21 |
−1.68 |
0.0043 |
0.3582 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| ENSG00000082126 |
MPP4 |
7.27 |
8.02 |
−1.68 |
0.0088 |
0.4149 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| ENSG00000153944 |
MSI2 |
6.61 |
7.36 |
−1.68 |
0.0064 |
0.4141 |
musashi homolog 2 (Drosophila) |
| ENSG00000239474 |
KBTBD10 |
7.54 |
8.30 |
−1.69 |
0.0025 |
0.3582 |
kelch repeat and BTB (POZ) domain containing 10 |
| ENSG00000131711 |
MAP1B |
8.24 |
9.04 |
−1.74 |
0.0002 |
0.2057 |
microtubule-associated protein 1B |
| ENSG00000102755 |
FLT1 |
9.15 |
9.97 |
−1.76 |
0.0059 |
0.4088 |
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) |
| ENSG00000172519 |
OR10H5 |
6.51 |
7.33 |
−1.77 |
0.0011 |
0.3582 |
olfactory receptor, family 10, subfamily H, member 5 |
| ENSG00000059804 |
SLC2A3 |
7.88 |
8.77 |
−1.86 |
0.0003 |
0.2094 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| ENSG00000128052 |
KDR |
8.08 |
8.99 |
−1.89 |
0.0015 |
0.3582 |
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| ENSG00000198515 |
CNGA1 |
8.87 |
9.90 |
−2.05 |
0.0005 |
0.2515 |
cyclic nucleotide gated channel alpha 1 |
| ENSG00000108370 |
RGS9 |
6.12 |
7.19 |
−2.10 |
0.0059 |
0.4088 |
regulator of G-protein signaling 9 |
| ENSG00000074621 |
SLC24A1 |
6.62 |
7.71 |
−2.13 |
0.0062 |
0.4088 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| ENSG00000143995 |
MEIS1 |
7.49 |
8.59 |
−2.14 |
0.0000 |
0.1172 |
Meis homeobox 1 |
| ENSG00000149489 |
ROM1 |
7.80 |
8.94 |
−2.20 |
0.0036 |
0.3582 |
retinal outer segment membrane protein 1 |
| ENSG00000146350 |
C6orf170 |
6.09 |
7.26 |
−2.25 |
0.0001 |
0.2057 |
chromosome 6 open reading frame 170 |
| ENSG00000114279 |
FGF12 |
6.20 |
7.38 |
−2.27 |
0.0004 |
0.2281 |
fibroblast growth factor 12 |
| ENSG00000134183 |
GNAT2 |
6.85 |
8.08 |
−2.35 |
0.0036 |
0.3582 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| ENSG00000158445 |
KCNB1 |
6.13 |
7.37 |
−2.37 |
0.0047 |
0.3608 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| ENSG00000118473 |
SGIP1 |
7.24 |
8.50 |
−2.38 |
0.0004 |
0.2281 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| ENSG00000182985 |
CADM1 |
7.50 |
8.86 |
−2.58 |
0.0047 |
0.3608 |
cell adhesion molecule 1 |
| ENSG00000078018 |
MAP2 |
7.25 |
8.63 |
−2.60 |
0.0016 |
0.3582 |
microtubule-associated protein 2 |
| ENSG00000205683 |
DPF3 |
5.87 |
7.26 |
−2.62 |
0.0012 |
0.3582 |
D4, zinc and double PHD fingers, family 3 |
| ENSG00000047617 |
ANO2 |
6.73 |
8.16 |
−2.70 |
0.0043 |
0.3582 |
anoctamin 2 |
| ENSG00000158234 |
FAIM |
6.41 |
7.96 |
−2.92 |
0.0020 |
0.3582 |
Fas apoptotic inhibitory molecule |
| ENSG00000118402 |
ELOVL4 |
6.78 |
8.35 |
−2.97 |
0.0039 |
0.3582 |
ELOVL fatty acid elongase 4 |
| ENSG00000111837 |
MAK |
6.18 |
7.76 |
−3.00 |
0.0020 |
0.3582 |
male germ cell-associated kinase |
| ENSG00000203756 |
C6orf191 |
5.94 |
7.56 |
−3.07 |
0.0092 |
0.4149 |
chromosome 6 open reading frame 191 |
| ENSG00000114349 |
GNAT1 |
5.42 |
7.12 |
−3.24 |
0.0034 |
0.3582 |
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| ENSG00000100362 |
PVALB |
6.18 |
7.88 |
−3.26 |
0.0084 |
0.4149 |
parvalbumin |
| ENSG00000152578 |
GRIA4 |
5.84 |
7.55 |
−3.28 |
0.0031 |
0.3582 |
glutamate receptor, ionotrophic, AMPA 4 |
| ENSG00000132639 |
SNAP25 |
6.20 |
7.96 |
−3.38 |
0.0087 |
0.4149 |
synaptosomal-associated protein, 25 kDa |
| ENSG00000148798 |
INA |
5.36 |
7.17 |
−3.49 |
0.0074 |
0.4149 |
internexin neuronal intermediate filament protein, alpha |
| ENSG00000130561 |
SAG |
6.03 |
7.84 |
−3.51 |
0.0049 |
0.3688 |
S-antigen; retina and pineal gland (arrestin) |
| ENSG00000112619 |
PRPH2 |
6.61 |
8.50 |
−3.73 |
0.0014 |
0.3582 |
peripherin 2 (retinal degeneration, slow) |
| ENSG00000116703 |
PDC |
5.93 |
7.86 |
−3.83 |
0.0096 |
0.4149 |
phosducin |
| ENSG00000163914 |
RHO |
7.26 |
9.23 |
−3.93 |
0.0051 |
0.3754 |
rhodopsin |
| ENSG00000132915 |
PDE6A |
5.87 |
7.88 |
−4.02 |
0.0045 |
0.3607 |
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| ENSG00000185518 |
SV2B |
5.43 |
7.60 |
−4.50 |
0.0045 |
0.3607 |
synaptic vesicle glycoprotein 2B |
| ENSG00000112706 | IMPG1 | 5.29 | 7.50 | −4.62 | 0.0060 | 0.4088 | interphotoreceptor matrix proteoglycan 1 |
Genes were filtered based on at least 50% fold change between AMD and control and raw p value <0.01.
Figure 4.
Heatmap of differentially expressed genes shown in Table 2. Dark shading indicates low expression; light shading indicates high expression.
Comparison with previous studies
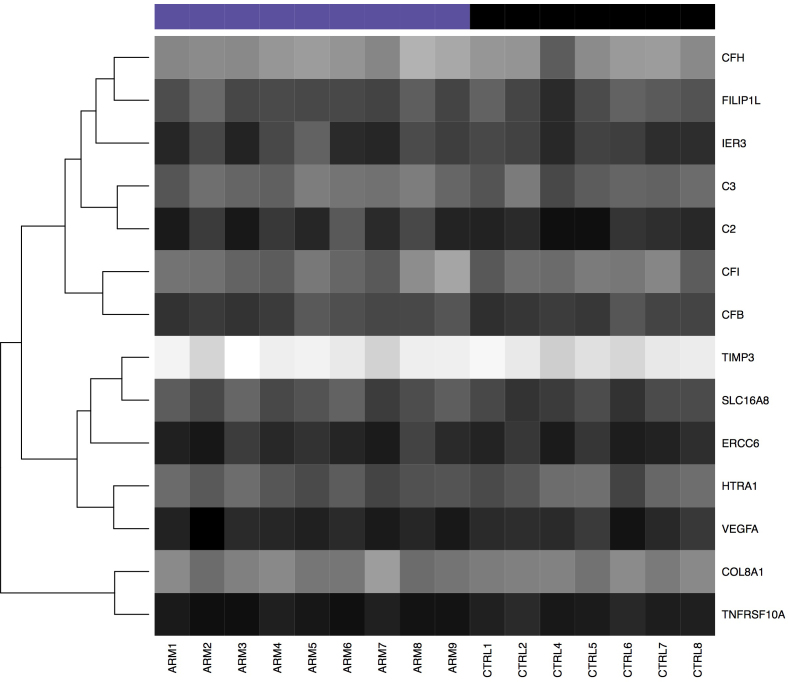
Genome-wide association studies have identified several genes harboring polymorphisms associated with AMD. We compiled a list of these genes and examined their expression levels in our data set (Table 3; Figure 5). Some genes (e.g., ADAMTS9, APOE, ARMS2, B3GALT1, CCR3, CETP, CFHR2, CFHR3, COL10A1, DDR1, LIPC, RAD51B, TGFBR1, TLR3) did not meet our filtering criteria (i.e., mean expression in AMD or control groups > median expression of all protein coding genes). Of the genes previously associated with AMD, none were differentially expressed (raw p value <0.01).
Table 3. Genes previously associated with AMD by genetic studies.
| Ensembl ID | Symbol | Mean AMD | Mean Control | Fold change | Raw p value | FDR adj. P value | Name |
|---|---|---|---|---|---|---|---|
| ENSG00000100156 |
SLC16A8 |
8.26 |
7.88 |
1.31 |
0.0129 |
0.4358 |
solute carrier family 16, member 8 (monocarboxylic acid transporter 3) [43] |
| ENSG00000166278 |
C2 |
7.49 |
7.19 |
1.24 |
0.1434 |
0.6643 |
complement component 2 [43,45,46] |
| ENSG00000125730 |
C3 |
8.80 |
8.55 |
1.19 |
0.1226 |
0.6470 |
complement component 3 [40,47-49] |
| ENSG00000000971 |
CFH |
9.64 |
9.43 |
1.16 |
0.2662 |
0.7440 |
complement factor H [41,50-52] |
| ENSG00000100234 |
TIMP3 |
11.37 |
11.22 |
1.11 |
0.3122 |
0.7823 |
TIMP metallopeptidase inhibitor 3 [40,49,53,54] |
| ENSG00000243649 |
CFB |
7.93 |
7.81 |
1.09 |
0.4199 |
0.8396 |
complement factor B [43,45,46] |
| ENSG00000205403 |
CFI |
8.93 |
8.84 |
1.07 |
0.6688 |
0.9296 |
complement factor I [43,55] |
| ENSG00000225830 |
ERCC6 |
7.34 |
7.29 |
1.03 |
0.7487 |
0.9492 |
excision repair cross-complementing rodent repair deficiency, complementation group 6 [56] |
| ENSG00000137331 |
IER3 |
7.72 |
7.67 |
1.03 |
0.8162 |
0.9636 |
immediate early response 3 [43] |
| ENSG00000168386 |
FILIP1L |
8.14 |
8.17 |
−1.02 |
0.8496 |
0.9718 |
filamin A interacting protein 1-like [43] |
| ENSG00000144810 |
COL8A1 |
9.15 |
9.20 |
−1.04 |
0.7052 |
0.9370 |
collagen, type VIII, alpha 1 [43,57] |
| ENSG00000166033 |
HTRA1 |
8.36 |
8.50 |
−1.10 |
0.4118 |
0.8344 |
HtrA serine peptidase 1 [58-60] |
| ENSG00000104689 |
TNFRSF10A |
6.88 |
7.12 |
−1.18 |
0.0136 |
0.4398 |
tumor necrosis factor receptor superfamily, member 10a [43,61] |
| ENSG00000112715 | VEGFA | 7.11 | 7.36 | −1.19 | 0.0735 | 0.5904 | vascular endothelial growth factor A [12,62] |
Figure 5.
Heatmap of genes previously associated with or possibly implicated in AMD. Dark shading indicates low expression; light shading indicates high expression.
Analysis of genes associated with either RPE or vascular endothelium
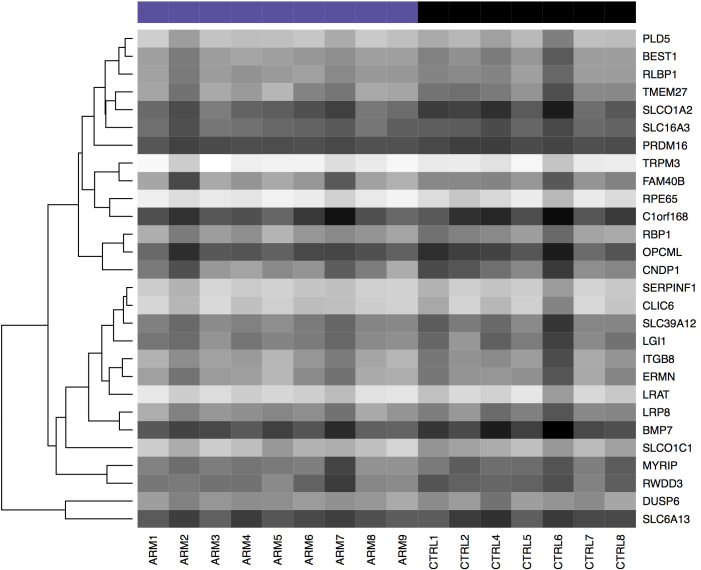
We next evaluated genes with cell-type-specific expression in either the RPE or endothelium. For RPE genes, we took the top 35 genes identified as highly expressed in the RPE versus the retina and choroid in the recent manuscript by Booij and colleagues [33]. We added SERPINF1 (PEDF), another marker of RPE, to this set [34]. Of these, 28 mapped to Ensembl IDs in our filtered set and were included for analysis. Thirteen RPE-expressed genes showed between 12% and 72% increased expression (raw p value <0.1; Table 4; Figure 6).
Table 4. Genes highly expressed in the RPE.
| Ensembl ID | Symbol | Mean AMD | Mean Control | Fold change | Raw p value | FDR adj. P value | Name |
|---|---|---|---|---|---|---|---|
| ENSG00000147003 |
TMEM27 |
9.25 |
8.47 |
1.72 |
0.0130 |
0.4358 |
transmembrane protein 27 [33] |
| ENSG00000084453 |
SLCO1A2 |
7.82 |
7.06 |
1.69 |
0.0232 |
0.4779 |
solute carrier organic anion transporter family, member 1A2 [33] |
| ENSG00000150656 |
CNDP1 |
8.76 |
8.04 |
1.64 |
0.0813 |
0.5984 |
carnosine dipeptidase 1 (metallopeptidase M20 family) [33] |
| ENSG00000101144 |
BMP7 |
7.29 |
6.64 |
1.57 |
0.0361 |
0.5074 |
bone morphogenetic protein 7 [33] |
| ENSG00000105855 |
ITGB8 |
9.46 |
8.86 |
1.52 |
0.0627 |
0.5757 |
integrin, beta 8 [33] |
| ENSG00000157193 |
LRP8 |
9.09 |
8.52 |
1.48 |
0.0333 |
0.4942 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor [33] |
| ENSG00000139155 |
SLCO1C1 |
10.15 |
9.68 |
1.39 |
0.0425 |
0.5262 |
solute carrier organic anion transporter family, member 1C1 [33] |
| ENSG00000122481 |
RWDD3 |
8.31 |
7.84 |
1.38 |
0.0942 |
0.6178 |
RWD domain containing 3 [33] |
| ENSG00000136541 |
ERMN |
9.28 |
8.83 |
1.36 |
0.1648 |
0.6759 |
ermin, ERM-like protein [33] |
| ENSG00000167995 |
BEST1 |
9.29 |
8.84 |
1.36 |
0.0621 |
0.5736 |
bestrophin 1 [33] |
| ENSG00000180287 |
PLD5 |
10.21 |
9.77 |
1.35 |
0.0695 |
0.5816 |
phospholipase D family, member 5 [33] |
| ENSG00000170011 |
MYRIP |
8.46 |
8.03 |
1.35 |
0.1063 |
0.6341 |
myosin VIIA and Rab interacting protein [33] |
| ENSG00000187889 |
C1orf168 |
7.15 |
6.75 |
1.33 |
0.2653 |
0.7427 |
chromosome 1 open reading frame 168 [33] |
| ENSG00000148482 |
SLC39A12 |
8.60 |
8.20 |
1.32 |
0.1679 |
0.6783 |
solute carrier family 39 (zinc transporter), member 12 [33] |
| ENSG00000116745 |
RPE65 |
11.29 |
10.90 |
1.32 |
0.0376 |
0.5105 |
retinal pigment epithelium-specific protein 65 kDa [33] |
| ENSG00000183715 |
OPCML |
7.40 |
7.04 |
1.28 |
0.2173 |
0.7119 |
opioid binding protein/cell adhesion molecule-like [33] |
| ENSG00000114115 |
RBP1 |
9.29 |
8.93 |
1.28 |
0.1930 |
0.6977 |
retinol binding protein 1, cellular [33] |
| ENSG00000108231 |
LGI1 |
8.58 |
8.23 |
1.27 |
0.2161 |
0.7105 |
leucine-rich, glioma inactivated 1 [33] |
| ENSG00000128578 |
FAM40B |
9.07 |
8.73 |
1.26 |
0.3842 |
0.8192 |
family with sequence similarity 40, member B [33] |
| ENSG00000159212 |
CLIC6 |
10.51 |
10.19 |
1.25 |
0.2284 |
0.7221 |
chloride intracellular channel 6 [33] |
| ENSG00000141526 |
SLC16A3 |
7.97 |
7.67 |
1.23 |
0.0985 |
0.6231 |
solute carrier family 16, member 3 (monocarboxylic acid transporter 4) [33] |
| ENSG00000121207 |
LRAT |
10.84 |
10.57 |
1.21 |
0.2578 |
0.7374 |
lecithin retinol acyltransferase (phosphatidylcholine–retinol O-acyltransferase) [33] |
| ENSG00000140522 |
RLBP1 |
9.18 |
8.94 |
1.18 |
0.2469 |
0.7340 |
retinaldehyde binding protein 1 [33] |
| ENSG00000132386 |
SERPINF1 |
10.54 |
10.31 |
1.17 |
0.2352 |
0.7252 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 [34] |
| ENSG00000010379 |
SLC6A13 |
7.31 |
7.11 |
1.14 |
0.3584 |
0.8067 |
solute carrier family 6 (neurotransmitter transporter, GABA), member 13 [33] |
| ENSG00000083067 |
TRPM3 |
11.43 |
11.25 |
1.13 |
0.3586 |
0.8067 |
transient receptor potential cation channel, subfamily M, member 3 [33] |
| ENSG00000142611 |
PRDM16 |
7.28 |
7.12 |
1.12 |
0.0982 |
0.6231 |
PR domain containing 16 [33] |
| ENSG00000139318 | DUSP6 | 9.18 | 9.10 | 1.06 | 0.6565 | 0.9264 | dual specificity phosphatase 6 [33] |
Figure 6.
Heatmap of genes highly expressed in the RPE. Dark shading indicates low expression; light shading indicates high expression.
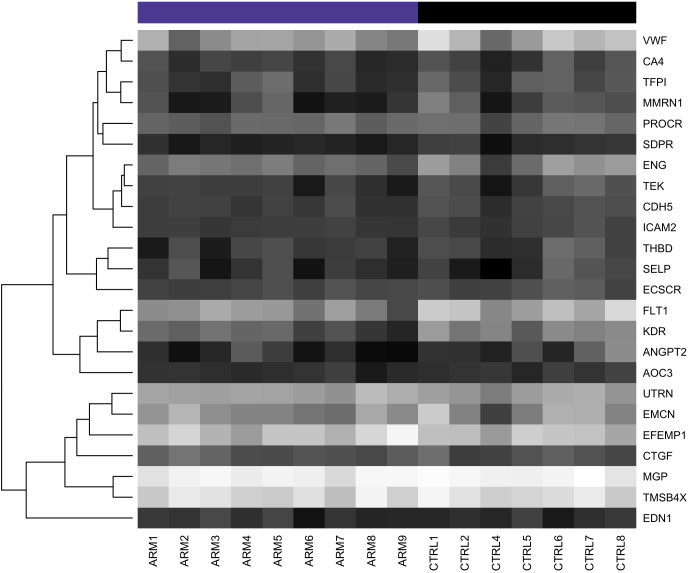
As a proxy for choroidal endothelial cells, we compiled a list of 24 genes with known expression in the endothelium and compared the fold change between the AMD and control samples. Endothelium-associated genes were based on gene lists identified as specific to the endothelium [35] or from a literature search. Seven of these genes showed decreased expression in early AMD between 20% and 189% (raw p value <0.1; Table 5; Figure 7), which suggested decreased numbers of vascular endothelial cells in the choroid and/or dedifferentiation of extant endothelial cells. Thus, there appeared to be a trend toward increased expression of RPE-expressed genes and decreased expression of endothelium-expressed genes in eyes with early AMD.
Table 5. Genes expressed in endothelial cells.
| Ensembl ID | Symbol | Mean AMD | Mean Control | Fold change | Raw p value | FDR adj. P value | Name |
|---|---|---|---|---|---|---|---|
| ENSG00000115380 |
EFEMP1 |
10.25 |
10.05 |
1.15 |
0.3126 |
0.7823 |
EGF containing fibulin-like extracellular matrix protein 1 [35] |
| ENSG00000152818 |
UTRN |
9.66 |
9.50 |
1.12 |
0.2251 |
0.7199 |
utrophin [35] |
| ENSG00000118523 |
CTGF |
8.17 |
8.01 |
1.11 |
0.3471 |
0.8008 |
connective tissue growth factor [35] |
| ENSG00000078401 |
EDN1 |
7.28 |
7.18 |
1.07 |
0.5917 |
0.9029 |
endothelin 1 [35] |
| ENSG00000164035 |
EMCN |
9.22 |
9.26 |
−1.02 |
0.9159 |
0.9858 |
endomucin [35] |
| ENSG00000101000 |
PROCR |
8.44 |
8.50 |
−1.04 |
0.6952 |
0.9347 |
protein C receptor, endothelial [35] |
| ENSG00000111341 |
MGP |
10.96 |
11.05 |
−1.06 |
0.3872 |
0.8200 |
matrix Gla protein [35] |
| ENSG00000205542 |
TMSB4X |
10.57 |
10.67 |
−1.07 |
0.5172 |
0.8821 |
thymosin beta 4, X-linked [35] |
| ENSG00000174175 |
SELP |
7.27 |
7.41 |
−1.10 |
0.6747 |
0.9312 |
selectin P (granule membrane protein 140 kDa, antigen CD62) [63,64] |
| ENSG00000108622 |
ICAM2 |
7.53 |
7.68 |
−1.11 |
0.1335 |
0.6557 |
intercellular adhesion molecule 2 [35] |
| ENSG00000131471 |
AOC3 |
7.21 |
7.38 |
−1.12 |
0.2033 |
0.7008 |
amine oxidase, copper containing 3 (vascular adhesion protein 1) [65] |
| ENSG00000249751 |
ECSCR |
7.77 |
7.94 |
−1.12 |
0.1685 |
0.6783 |
endothelial cell-specific chemotaxis regulator [35] |
| ENSG00000167434 |
CA4 |
7.52 |
7.72 |
−1.15 |
0.3435 |
0.7992 |
carbonic anhydrase IV [66] |
| ENSG00000179776 |
CDH5 |
7.55 |
7.81 |
−1.20 |
0.0621 |
0.5736 |
cadherin 5, type 2 (vascular endothelium) [35] |
| ENSG00000168497 |
SDPR |
6.92 |
7.25 |
−1.26 |
0.0529 |
0.5567 |
serum deprivation response [35] |
| ENSG00000003436 |
TFPI |
7.65 |
8.01 |
−1.28 |
0.1613 |
0.6758 |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) [35] |
| ENSG00000178726 |
THBD |
7.41 |
7.82 |
−1.34 |
0.1061 |
0.6340 |
thrombomodulin [35] |
| ENSG00000120156 |
TEK |
7.40 |
7.83 |
−1.35 |
0.1085 |
0.6363 |
TEK tyrosine kinase, endothelial [67] |
| ENSG00000106991 |
ENG |
8.57 |
9.01 |
−1.35 |
0.1025 |
0.6275 |
endoglin [35] |
| ENSG00000110799 |
VWF |
9.34 |
9.87 |
−1.45 |
0.0680 |
0.5797 |
von Willebrand factor [35] |
| ENSG00000138722 |
MMRN1 |
7.25 |
7.96 |
−1.64 |
0.0551 |
0.5579 |
multimerin 1 [35] |
| ENSG00000091879 |
ANGPT2 |
6.94 |
7.72 |
−1.72 |
0.0420 |
0.5244 |
angiopoietin 2 [67] |
| ENSG00000102755 |
FLT1 |
9.15 |
9.97 |
−1.76 |
0.0059 |
0.4088 |
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) [35] |
| ENSG00000128052 | KDR | 8.08 | 8.99 | −1.89 | 0.0015 | 0.3582 | kinase insert domain receptor (a type III receptor tyrosine kinase) [35] |
Genes in this list were previously reported as associated with endothelial cells.
Figure 7.
Heatmap of genes associated with endothelial cells. Note the trend toward decreased expression in the AMD samples (ARM 1–9). Dark shading indicates low expression; light shading indicates high expression.
To calculate the enrichment of the RPE and endothelium gene sets, we performed GSEA [29]. For each gene set, GSEA calculates (a) an enrichment score (ES) based on the rank distribution of individual genes within the set from among all unique genes symbols in our filtered set (n=10,286), (b) a normalized enrichment score (NES) that accounts for the size of the set, (c) a nominal p value based on phenotype permutation, e.g., AMD or control status of samples, and (d) an FDR q value to control for the gene set size and multiple hypothesis testing. GSEA suggests a greater trend for the overall enrichment of the endothelium-associated gene set in AMD samples (ES = –0.78; NES = –1.39; nominal p value=0.133; FDR q value = 0.08) than the enrichment of RPE-specific genes in the control samples (ES = 0.81; NES = 1.3; nom. P value = 0.167; FDR q value = 0.191). Since an FDR q value of less than 0.25 is considered significant for GSEA [29], these data suggest a significant decrease in endothelial cell transcripts.
Analysis of CFH risk genotypes
Last, we reanalyzed the data set by stratifying based on genotypes at the rs1061170 SNP in CFH (11 high-risk, YH/HH samples; five low-risk, YY samples), independent of AMD affection status. The same filtering criteria were applied as in the previous analysis, retaining 10,288 protein-coding genes. Thirty-five genes were identified as differentially expressed (at least 50% difference in expression; moderated t test raw p value <0.01; Table 6). None of the genes were significantly differentially expressed after FDR correction (p value <0.1).
Table 6. Differentially expressed genes high-risk CFH genotypes (YH/HH) versus low-risk CFH genotype (YY).
| Ensembl ID | Symbol | Mean AMD | Mean Control | Fold change | Raw p value | FDR adj. P value | Name |
|---|---|---|---|---|---|---|---|
| ENSG00000127083 |
OMD |
7.19 |
5.97 |
2.33 |
0.0047 |
0.8828 |
osteomodulin |
| ENSG00000146374 |
RSPO3 |
8.03 |
6.88 |
2.21 |
0.0058 |
0.8828 |
R-spondin 3 |
| ENSG00000011465 |
DCN |
8.86 |
7.72 |
2.19 |
0.0008 |
0.6616 |
decorin |
| ENSG00000090104 |
RGS1 |
8.10 |
7.10 |
2.00 |
0.0076 |
0.8828 |
regulator of G-protein signaling 1 |
| ENSG00000186439 |
TRDN |
8.04 |
7.08 |
1.95 |
0.0068 |
0.8828 |
triadin |
| ENSG00000176971 |
FIBIN |
8.87 |
7.98 |
1.86 |
0.0004 |
0.6215 |
fin bud initiation factor homolog (zebrafish) |
| ENSG00000146233 |
CYP39A1 |
7.31 |
6.46 |
1.80 |
0.0065 |
0.8828 |
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| ENSG00000196344 |
ADH7 |
7.66 |
6.94 |
1.65 |
0.0017 |
0.7811 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| ENSG00000182230 |
FAM153B |
7.23 |
6.54 |
1.61 |
0.0056 |
0.8828 |
family with sequence similarity 153, member B |
| ENSG00000116667 |
C1orf21 |
10.29 |
9.65 |
1.56 |
0.0000 |
0.1533 |
chromosome 1 open reading frame 21 |
| ENSG00000121769 |
FABP3 |
7.48 |
6.85 |
1.55 |
0.0088 |
0.8828 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| ENSG00000071189 |
SNX13 |
8.46 |
7.82 |
1.55 |
0.0020 |
0.7811 |
sorting nexin 13 |
| ENSG00000115607 |
IL18RAP |
7.46 |
8.05 |
−1.50 |
0.0083 |
0.8828 |
interleukin 18 receptor accessory protein |
| ENSG00000089157 |
RPLP0 |
7.00 |
7.59 |
−1.51 |
0.0029 |
0.7833 |
ribosomal protein, large, P0 |
| ENSG00000177076 |
ACER2 |
6.82 |
7.41 |
−1.51 |
0.0027 |
0.7833 |
alkaline ceramidase 2 |
| ENSG00000146592 |
CREB5 |
6.66 |
7.26 |
−1.51 |
0.0070 |
0.8828 |
cAMP responsive element binding protein 5 |
| ENSG00000182853 |
VMO1 |
6.58 |
7.18 |
−1.52 |
0.0051 |
0.8828 |
vitelline membrane outer layer 1 homolog (chicken) |
| ENSG00000141753 |
IGFBP4 |
9.07 |
9.71 |
−1.56 |
0.0082 |
0.8828 |
insulin-like growth factor binding protein 4 |
| ENSG00000167772 |
ANGPTL4 |
7.66 |
8.32 |
−1.58 |
0.0003 |
0.6215 |
angiopoietin-like 4 |
| ENSG00000198959 |
TGM2 |
8.87 |
9.61 |
−1.68 |
0.0006 |
0.6616 |
transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) |
| ENSG00000104332 |
SFRP1 |
6.96 |
7.77 |
−1.76 |
0.0067 |
0.8828 |
secreted frizzled-related protein 1 |
| ENSG00000113083 |
LOX |
6.29 |
7.14 |
−1.80 |
0.0055 |
0.8828 |
lysyl oxidase |
| ENSG00000167236 |
CCL23 |
7.10 |
8.00 |
−1.86 |
0.0066 |
0.8828 |
chemokine (C-C motif) ligand 23 |
| ENSG00000068366 |
ACSL4 |
7.33 |
8.33 |
−2.00 |
0.0028 |
0.7833 |
acyl-CoA synthetase long-chain family member 4 |
| ENSG00000138135 |
CH25H |
6.20 |
7.21 |
−2.01 |
0.0057 |
0.8828 |
cholesterol 25-hydroxylase |
| ENSG00000102265 |
TIMP1 |
10.09 |
11.11 |
−2.03 |
0.0035 |
0.8828 |
TIMP metallopeptidase inhibitor 1 |
| ENSG00000156804 |
FBXO32 |
6.83 |
7.96 |
−2.19 |
0.0000 |
0.1533 |
F-box protein 32 |
| ENSG00000179431 |
FJX1 |
8.10 |
9.28 |
−2.27 |
0.0008 |
0.6616 |
four jointed box 1 (Drosophila) |
| ENSG00000163638 |
ADAMTS9 |
5.74 |
7.12 |
−2.61 |
0.0008 |
0.6616 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| ENSG00000124102 |
PI3 |
6.40 |
7.79 |
−2.63 |
0.0006 |
0.6616 |
peptidase inhibitor 3, skin-derived |
| ENSG00000205362 |
MT1A |
8.34 |
9.75 |
−2.66 |
0.0082 |
0.8828 |
metallothionein 1A |
| ENSG00000159167 |
STC1 |
9.09 |
10.62 |
−2.90 |
0.0027 |
0.7833 |
stanniocalcin 1 |
| ENSG00000064886 |
CHI3L2 |
6.20 |
7.83 |
−3.10 |
0.0070 |
0.8828 |
chitinase 3-like 2 |
| ENSG00000162992 |
NEUROD1 |
5.41 |
7.30 |
−3.69 |
0.0092 |
0.8828 |
neurogenic differentiation 1 |
| ENSG00000115602 | IL1RL1 | 5.35 | 7.33 | −3.93 | 0.0001 | 0.3640 | interleukin 1 receptor-like 1 |
DAVID analysis of the 12 genes with increased expression in the high-risk genotype revealed no significantly enriched terms (with Benjamini corrected p value <0.01). In the DAVID analysis of the 23 genes with lower expression in the high-risk genotype, DAVID identified eight terms with significant enrichment (fold enrichment from 3 to 8; Benjamini corrected p value <0.01). These terms were associated with extracellular secretion, signal peptides, and disulfide bonds.
Discussion
AMD is a complex disease that shows altered function and viability of photoreceptor cells, RPE, and choriocapillaris endothelial cells. Although our understanding of the genetics of AMD has progressed in the last decade, many basic questions about the pathogenesis of this disease remain. In the current study, we evaluated gene expression at the exon level in AMD and control eyes and found a loss of transcripts expressed by choroidal endothelial cells.
Limitations of the current study include small sample size (due to stringent inclusion criteria) and the lack of strong signatures of differential expression between the case and control samples, obviating validation of single genes. Increased sample size may reveal some genes with a small difference between the cases and controls, which remain significant after multiple hypothesis testing. However, this is unlikely. Previous analysis of AMD-affected tissues with microarray technologies required the use of non-standard methods to identify altered gene expression (expression correlation between two data sets with p<0.1 and 25% fold change) [15] or the use of sensitive clustering algorithms to identify patterns across many AMD grades simultaneously [14]. At the tissue level in our data set, the apparent trends among the RPE and endothelial genes (overall direction of fold change and p<0.1) are supported by the GSEA results. These gene set-level changes are small in our samples.
Cross-contamination by photoreceptor-specific transcripts
We observed an elevation of select photoreceptor-specific genes (i.e., RHO, PDC) across all control samples compared to the AMD-affected samples, particularly noticeable in CTRL4 and CTRL5. High expression of retinal transcripts is commonly reported in gene expression studies of the RPE and choroid [14,36,37], and various strategies have been taken to deal with cross-contamination (e.g., laser capture microdissection [33], flagging genes with high expression in one tissue that appear in another tissue [14]). Cross-contamination is not surprising, as photoreceptor cell outer segments are partially interdigitated and ensheathed by the apical microvilli of the RPE and supported by the interphotoreceptor matrix [38]. Although most photoreceptor cell transcripts are expected to be within the outer nuclear layer and inner segments, Van Soest and colleagues suggest that these transcripts may be present at the interface between the photoreceptor outer segments and apical RPE [37]. The photoreceptor cell-specific gene expression we observed could be an artifact of mechanical separation or a stochastic individual variation in adhesion, and this random event might have occurred to a higher degree in the control eyes. However, we hypothesize that the pattern of elevated retinal contamination in the controls, which was absent from AMD cases, may be due to decreased adhesion of the retina to the RPE in the AMD samples compared to the controls. Although adhesion between the neural retina and the RPE has been examined in primates [39], to our knowledge this has not been investigated in the context of early AMD. Alternatively, if neural retina components are uniformly present in all samples, then the decreased expression of retina-specific genes in the AMD samples could indicate that expression within photoreceptors is decreased even at the earliest stages of AMD.
Intriguingly, genes with lower expression in AMD than in controls included KDR and FLT1, known VEGF receptors. Hierarchical clustering reveals that these genes do not segregate with the neural retina-specific genes and that expression of this sub-cluster of genes is consistent across controls and is not affected by CTRL4 and CTRL5 (Figure 1).
Loss of endothelial-specific gene expression
Our group has previously shown that the density of microvasculature decreases as the volume of sub-RPE drusen increases [9]. Similarly, studies of blood flow in eyes with drusen show susceptibility of the choroidal vasculature to degenerative changes in AMD [11]. In elegant whole mount studies of eyes with advanced AMD, McLeod and colleagues found a linear relationship between the RPE and choriocapillaris, with the loss of either layer affecting the integrity of the other [10]. From these studies, with a sample size similar to that in the current report, the authors concluded that RPE loss precedes endothelium loss in GA, whereas endothelium loss precedes RPE loss in CNV [10]. Our data indicate that gene expression of endothelium-related genes decreases in early AMD before any atrophy or neovascular change develops.
Our results are also consistent with state-of-the-art proteomics studies of Bruch’s membrane–choroid preparations, in which the authors found the endothelial cell proteins von Willebrand factor (VWF) and carbonic anhydrase 4 (CA4) are reduced in early and mid-stage AMD and advanced dry AMD, respectively [40]. The same authors showed no loss of RPE proteins RPE65 and CRALBP (RLBP1). In fact, the levels of these proteins were elevated in Bruch’s membrane, consistent with our findings at the mRNA level. Taken together, these studies suggest loss or dedifferentiation of choroidal endothelial cells before loss of the RPE in eyes with AMD. In addition, expression of the choroidal endothelial genes ICAM1, SELE, and PLVAP was decreased in some AMD classes in the report by Newman et al. (supplemental data) [14].
Decreased expression of ADAMTS9 in samples with high AMD risk CFH genotype
Eyes with high-risk CFH genotypes were also assessed for gene expression changes. We previously found increased membrane attack complex formation in eyes homozygous for the high-risk allele [41], and complement activation may affect gene expression in nearby cells. Although not significant after multiple hypothesis testing correction, expression of ADAMTS9 transcripts was lower in samples with high-risk CFH genotypes than in samples with the low-risk genotype. This gene is a member of the ADAMTS (a disintegrin-like and metalloprotease domain with thrombospondin type I motifs) protein family with the ability to cleave aggrecan and versican [42]. Recently a SNP (rs6795735) located 32.5 kb upstream of the ADAMTS9 transcription start site was associated with increased risk of AMD in a meta-analysis of genome-wide association (GWA) data [43]. ADAMTS9 is expressed in ARPE-19 cells [44] and several types of microvascular endothelial cells [42], although to our knowledge localization of ADAMTS9 has not been examined in human RPE and choroid samples. In microvascular endothelial cells, ADAMTS9 suppresses angiogenesis, albeit not via sequestration of VEGF165 [42]. Further experimentation is necessary to elucidate the relationship between CFH activity and ADAMTS9 function in early AMD.
In conclusion, we performed microarray analysis of human donor maculas and found early loss of choriocapillaris endothelial cell markers in early AMD, with preservation of RPE cell transcripts. These results have potential importance for therapy. Recently, the replacement of RPE cells in AMD has been contemplated as a treatment for AMD. These studies, as well as proteomic [40], anatomic [9,10], and clinical [11] studies, strongly suggest caution in this type of approach, since transplanting healthy RPE into a macula with a degenerated choriocapillaris may be fruitless. In addition to replacing photoreceptor cells and RPE, strategies for replacing lost choriocapillaris are necessary to fully restore function in AMD.
Acknowledgments
The National Institutes of Health R01 grants R01EY017451 and R01EY016822; the authors thank the eye donors and their families, and the Iowa Lions Eye Bank for their key role in providing human donor eyes for research. Supported in part by: Alcon Research, Ltd.; the Hansjoerg E.J.W Kolder, MD, PhD. Professorship for Best Disease Research; and the Howard Hughes Medical Institute.
References
- 1.Coleman HR, Chan C-C, Ferris FL, Chew EY. Age-related macular degeneration. Lancet. 2008;372:1835–45. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 3.Saint-Geniez M, Kurihara T, Sekiyama E, Maldonado AE, D’Amore PA. An essential role for RPE-derived soluble VEGF in the maintenance of the choriocapillaris. Proc Natl Acad Sci USA. 2009;106:18751–6. doi: 10.1073/pnas.0905010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurihara T, Westenskow PD, Bravo S, Aguilar E, Friedlander M. Targeted deletion of Vegfa in adult mice induces vision loss. J Clin Invest. 2012;122:4213–7. doi: 10.1172/JCI65157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curcio CA, Millican CL. Basal linear deposit and large drusen are specific for early age-related maculopathy. Arch Ophthalmol. 1999;117:329–39. doi: 10.1001/archopht.117.3.329. [DOI] [PubMed] [Google Scholar]
- 6.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–36. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- 8.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullins RF, Johnson MN, Faidley EA, Skeie JM, Huang J. Choriocapillaris vascular dropout related to density of drusen in human eyes with early age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:1606–12. doi: 10.1167/iovs.10-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, Lutty GA. Relationship between RPE and choriocapillaris in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:4982–91. doi: 10.1167/iovs.09-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunwald JE, Metelitsina TI, Dupont JC, Ying G-S, Maguire MG. Reduced foveolar choroidal blood flow in eyes with increasing AMD severity. Invest Ophthalmol Vis Sci. 2005;46:1033–8. doi: 10.1167/iovs.04-1050. [DOI] [PubMed] [Google Scholar]
- 12.Kvanta A, Algvere PV, Berglin L, Seregard S. Subfoveal fibrovascular membranes in age-related macular degeneration express vascular endothelial growth factor. Invest Ophthalmol Vis Sci. 1996;37:1929–34. [PubMed] [Google Scholar]
- 13.Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, Kim RY, the MARINA Study Group. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–31. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 14.Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA, Cooper JB, Hageman GS, Anderson DH, Johnson LV, Radeke MJ. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Medicine. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter A, Spechler PA, Cwanger A, Song Y, Zhang Z, Ying G-S, Hunter AK, Dezoeten E, Dunaief JL. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53:2089–105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swiderski RE, Nishimura DY, Mullins RF, Olvera MA, Ross JL, Huang J, Stone EM, Sheffield VC. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest Ophthalmol Vis Sci. 2007;48:3329–40. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–62. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker WL, Liao IH, Gilbert DL, Wong B, Pollard KS, McCulloch CE, Lit L, Sharp FR. Empirical Bayes accomodation of batch-effects in microarray data using identical replicate reference samples: application to RNA expression profiling of blood from Duchenne muscular dystrophy patients. BMC Genomics. 2008;9:494. doi: 10.1186/1471-2164-9-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PLoS ONE. 2011;6:e17238. doi: 10.1371/journal.pone.0017238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 23.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2011. [Google Scholar]
- 24.Gentleman R, Ding B, Dut S, Ibrahim J. Distance Measures in DNA Microarray Data Analysis. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY Springer Science+Business Media; 2005. p. 189–208. [Google Scholar]
- 25.Affymetrix. Alternative transcript analysis methods for exon arrays. Affymetrix whitepapers. 2005
- 26.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25:415–6. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 31.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, de Jong PT, Klaver CC, Klein BE, Klein R. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39:367–74. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 33.Booij JC, ten Brink JB, Swagemakers SMA, Verkerk AJMH, Essing AHW, van der Spek PJ, Bergen AAB. A new strategy to identify and annotate human RPE-specific gene expression. PLoS ONE. 2010;5:e9341. doi: 10.1371/journal.pone.0009341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele FR, Chader GJ, Johnson LV, Tombran-Tink J. Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc Natl Acad Sci USA. 1993;90:1526–30. doi: 10.1073/pnas.90.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernat JA, Crawford GE, Ogurtsov AY, Collins FS, Ginsburg D, Kondrashov AS. Distant conserved sequences flanking endothelial-specific promoters contain tissue-specific DNase-hypersensitive sites and over-represented motifs. Hum Mol Genet. 2006;15:2098–105. doi: 10.1093/hmg/ddl133. [DOI] [PubMed] [Google Scholar]
- 36.Bowes Rickman C, Ebright JN, Zavodni ZJ, Yu L, Wang T, Daiger SP, Wistow G, Boon K, Hauser MA. Defining the human macula transcriptome and candidate retinal disease genes using EyeSAGE. Invest Ophthalmol Vis Sci. 2006;47:2305–16. doi: 10.1167/iovs.05-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Soest SS, de Wit GMJ, Essing AHW, ten Brink JB, Kamphuis W, de Jong PTVM, Bergen AAB. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol Vis. 2007;13:1608–17. [PubMed] [Google Scholar]
- 38.Bonilha VL, Rayborn ME, Bhattacharya SK, Gu X, Crabb JS, Crabb JW, Hollyfield JG. The retinal pigment epithelium apical microvilli and retinal function. Adv Exp Med Biol. 2006;572:519–24. doi: 10.1007/0-387-32442-9_72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hageman GS, Marmor MF, Yao XY, Johnson LV. The interphotoreceptor matrix mediates primate retinal adhesion. Arch Ophthalmol. 1995;113:655–60. doi: 10.1001/archopht.1995.01100050123041. [DOI] [PubMed] [Google Scholar]
- 40.Yuan X, Gu X, Crabb JS, Yue X, Shadrach K, Hollyfield JG, Crabb JW. Quantitative proteomics: comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol Cell Proteomics. 2010;9:1031–46. doi: 10.1074/mcp.M900523-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullins RF, Dewald AD, Streb LM, Wang K, Kuehn MH, Stone EM. Elevated membrane attack complex in human choroid with high risk complement factor H genotypes. Exp Eye Res. 2011;93:565–7. doi: 10.1016/j.exer.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koo B-H, Coe DM, Dixon LJ, Somerville RPT, Nelson CM, Wang LW, Young ME, Lindner DJ, Apte SS. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol. 2010;176:1494–504. doi: 10.2353/ajpath.2010.090655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The AMD Gene Consortium, Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Buitendijk GHS, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht A-M, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan C-C, Cheng C-Y, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdóttir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JPA, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis NN, Keilhauer C, Khan JC, Kim IK, Kiyohara Y, Klein BEK, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Saïd S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJT, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, Sahel J-A, Schaumberg DA, Scholl HPN, Schwartz SG, Scott WK, Shahid H, Sigurdsson H, Silvestri G, Sivakumaran TA, Smith RT, Sobrin L, Souied EH, Stambolian DE, Stefansson H, Sturgill-Short GM, Takahashi A, Tosakulwong N, Truitt BJ, Tsironi EE, Uitterlinden AG, van Duijn CM, Vijaya L, Vingerling JR, Vithana EN, Webster AR, Wichmann H-E, Winkler TW, Wong TY, Wright AF, Zelenika D, Zhang M, Zhao L, Zhang K, Klein ML, Hageman GS, Lathrop GM, Stefansson K, Allikmets R, Baird PN, Gorin MB, Wang JJ, Klaver CCW, Seddon JM, Pericak-Vance MA, Iyengar SK, Yates JRW, Swaroop A, Weber BHF, Kubo M, DeAngelis MM, Léveillard T, Thorsteinsdottir U, Haines JL, Farrer LA, Heid IM, Abecasis GR. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–9. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bevitt DJ, Mohamed J, Catterall JB, Li Z, Arris CE, Hiscott P, Sheridan C, Langton KP, Barker MD, Clarke MP, McKie N. Expression of ADAMTS metalloproteinases in the retinal pigment epithelium derived cell line ARPE-19: transcriptional regulation by TNFalpha. Biochim Biophys Acta. 2003;1626:83–91. doi: 10.1016/s0167-4781(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 45.Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–9. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 46.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Group AGCS, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–62. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yates JRW, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht A-M, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT, the Genetic Factors in AMD Study Group. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–61. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 48.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 49.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:14682–7. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards AO, Ritter R, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–4. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 51.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–21. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 53.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, Tan PL, Conley YP, Kanda A, Kopplin L, Li Y, Augustaitis KJ, Karoukis AJ, Scott WK, Agarwal A, Kovach JL, Schwartz SG, Postel EA, Brooks M, Baratz KH, Brown WL, Complications of Age-Related Macular Degeneration Prevention Trial (CAPT) Research Group, Brucker AJ, Orlin A, Brown G, Ho A, Regillo C, Donoso L, Tian L, Kaderli B, Hadley D, Hagstrom SA, Peachey NS, Klein R, Klein BEK, Gotoh N, Yamashiro K, Ferris Iii F, Fagerness JA, Reynolds R, Farrer LA, Kim IK, Miller JW, Cortón M, Carracedo A, Sanchez-Salorio M, Pugh EW, Doheny KF, Brion M, DeAngelis MM, Weeks DE, Zack DJ, Chew EY, Heckenlively JR, Yoshimura N, Iyengar SK, Francis PJ, Katsanis N, Seddon JM, Haines JL, Gorin MB, Abecasis GR, Swaroop A. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci USA. 2010;107:7401–6. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Clark ME, Crossman DK, Kojima K, Messinger JD, Mobley JA, Curcio CA. Abundant lipid and protein components of drusen. PLoS ONE. 2010;5:e10329. doi: 10.1371/journal.pone.0010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–4. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuo J, Ning B, Bojanowski CM, Lin Z-N, Ross RJ, Reed GF, Shen D, Jiao X, Zhou M, Chew EY, Kadlubar FF, Chan C-C. Synergic effect of polymorphisms in ERCC6 5′ flanking region and complement factor H on age-related macular degeneration predisposition. Proc Natl Acad Sci USA. 2006;103:9256–61. doi: 10.1073/pnas.0603485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu Y, Bhangale TR, Fagerness J, Ripke S, Thorleifsson G, Tan PL, Souied EH, Richardson AJ, Merriam JE, Buitendijk GHS, Reynolds R, Raychaudhuri S, Chin KA, Sobrin L, Evangelou E, Lee PH, Lee AY, Leveziel N, Zack DJ, Campochiaro B, Campochiaro P, Smith RT, Barile GR, Guymer RH, Hogg R, Chakravarthy U, Robman LD, Gustafsson O, Sigurdsson H, Ortmann W, Behrens TW, Stefansson K, Uitterlinden AG, van Duijn CM, Vingerling JR, Klaver CCW, Allikmets R, Brantley MA, Baird PN, Katsanis N, Thorsteinsdottir U, Ioannidis JPA, Daly MJ, Graham RR, Seddon JM. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20:3699–709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dewan A, Liu M, Hartman S, Zhang SS-M, Liu DTL, Zhao C, Tam POS, Chan WM, Lam DSC, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 60.Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–3. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- 61.Arakawa S, Takahashi A, Ashikawa K, Hosono N, Aoi T, Yasuda M, Oshima Y, Yoshida S, Enaida H, Tsuchihashi T, Mori K, Honda S, Negi A, Arakawa A, Kadonosono K, Kiyohara Y, Kamatani N, Nakamura Y, Ishibashi T, Kubo M. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43:1001–4. doi: 10.1038/ng.938. [DOI] [PubMed] [Google Scholar]
- 62.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, Postel EA, Olson L, Kenealy SJ, Hauser M, Gilbert JR, Pericak-Vance MA. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006;47:329–35. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 63.McLeod DS, Lefer DJ, Merges C, Lutty GA. Enhanced expression of intracellular adhesion molecule-1 and P-selectin in the diabetic human retina and choroid. Am J Pathol. 1995;147:642–53. [PMC free article] [PubMed] [Google Scholar]
- 64.Mullins RF, Skeie JM, Folk JC, Solivan-Timpe FM, Oetting TA, Huang J, Wang K, Stone EM, Fingert JH. Evaluation of variants in the selectin genes in age-related macular degeneration. BMC Med Genet. 2011;12:58. doi: 10.1186/1471-2350-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Almulki L, Noda K, Nakao S, Hisatomi T, Thomas KL, Hafezi-Moghadam A. Localization of vascular adhesion protein-1 (VAP-1) in the human eye. Exp Eye Res. 2010;90:26–32. doi: 10.1016/j.exer.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hageman GS, Zhu XL, Waheed A, Sly WS. Localization of carbonic anhydrase IV in a specific capillary bed of the human eye. Proc Natl Acad Sci USA. 1991;88:2716–20. doi: 10.1073/pnas.88.7.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otani A, Takagi H, Oh H, Koyama S, Matsumura M, Honda Y. Expressions of angiopoietins and Tie2 in human choroidal neovascular membranes. Invest Ophthalmol Vis Sci. 1999;40:1912–20. [PubMed] [Google Scholar]