Abstract
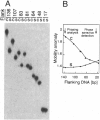
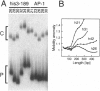
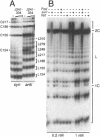
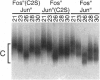
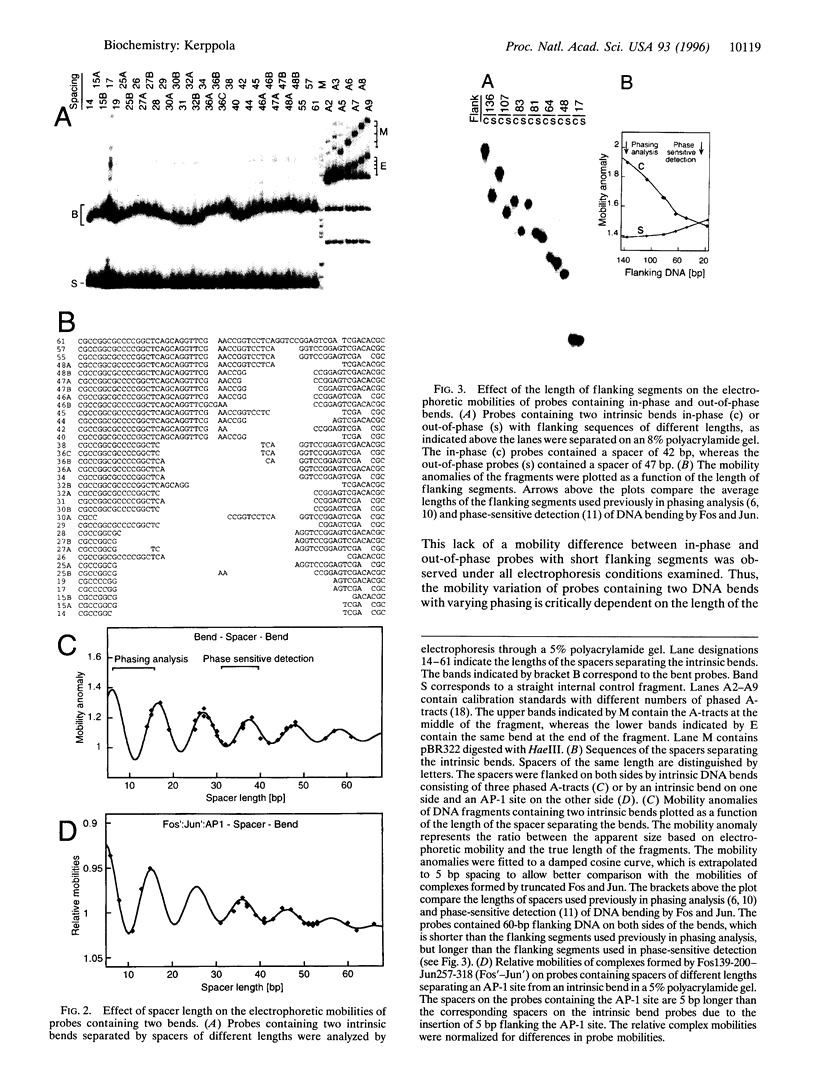
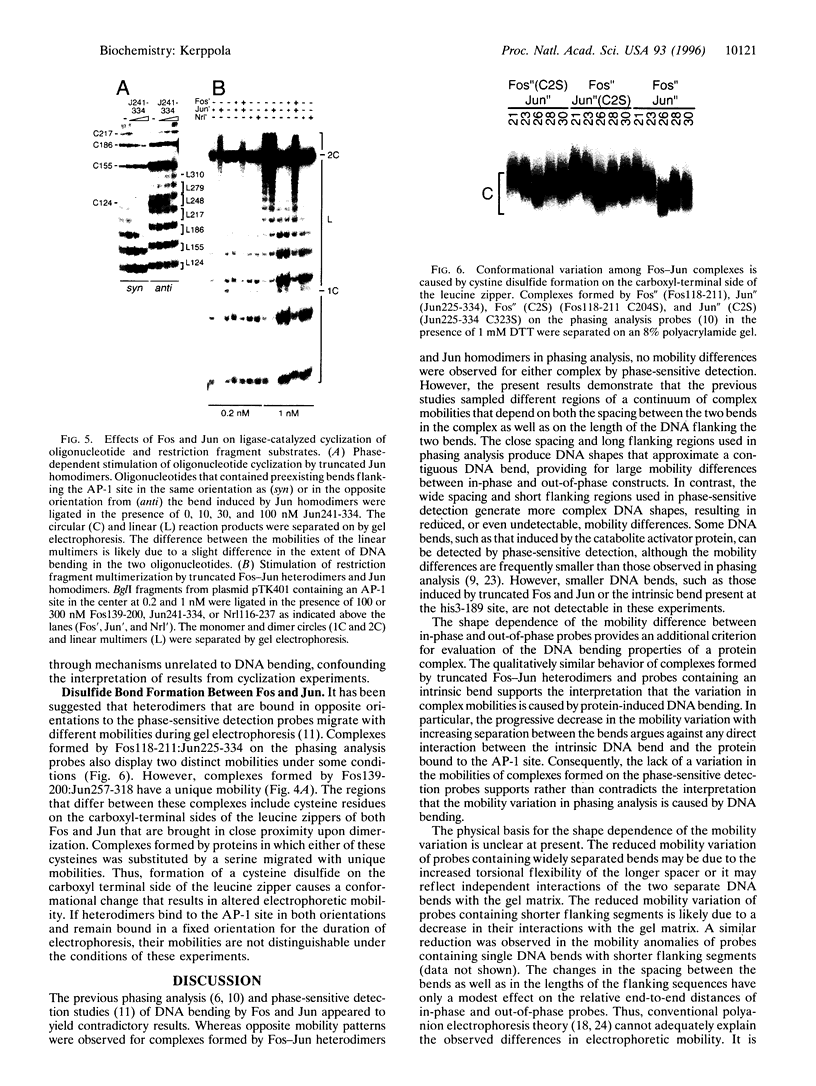
The effect of Fos and Jun binding on the structure of the AP-1 recognition site is controversial. Results from phasing analysis and phase-sensitive detection studies of DNA bending by Fos and Jun have led to opposite conclusions. The differences between these assays, the length of the spacer between two bends and the length of the sequences flanking the bends, are investigated here using intrinsic DNA bend standards. Both an increase in the spacer length as well as a decrease in the length of flanking sequences resulted in a reduction in the phase-dependent variation in electrophoretic mobilities. Probes with a wide separation between the bends and short flanking sequences, such as those used in the phase-sensitive detection studies, displayed no phase-dependent mobility variation. This shape-dependent variation in electrophoretic mobilities was reproduced by complexes formed by truncated Fos and Jun. Results from ligase-catalyzed cyclization experiments have been interpreted to indicate the absence of DNA bending in the Fos-Jun-AP-1 complex. However, truncated Fos and Jun can alter the relative rates of inter- and intramolecular ligation through mechanisms unrelated to DNA bending, confounding the interpretation of cyclization data. The analogous phase- and shape-dependence of the electrophoretic mobilities of the Fos-Jun-AP-1 complex and an intrinsic DNA bend confirm that Fos and Jun bend DNA, which may contribute to their functions in transcription regulation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Patel L., Rauscher F. J., 3rd, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990 Sep 7;249(4973):1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- Avitahl N., Calame K. The C/EBP family of proteins distorts DNA upon binding but does not introduce a large directed bend. J Biol Chem. 1994 Sep 23;269(38):23553–23562. [PubMed] [Google Scholar]
- Becker J. C., Nikroo A., Brabletz T., Reisfeld R. A. DNA loops induced by cooperative binding of transcriptional activator proteins and preinitiation complexes. Proc Natl Acad Sci U S A. 1995 Oct 10;92(21):9727–9731. doi: 10.1073/pnas.92.21.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I., Susic S., Dlakic M., Savic A., Pongor S. Physiological concentration of magnesium ions induces a strong macroscopic curvature in GGGCCC-containing DNA. J Mol Biol. 1994 Feb 11;236(1):26–32. doi: 10.1006/jmbi.1994.1115. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Drak J., Kahn J. D., Levene S. D. DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- Dlakic M., Harrington R. E. Bending and torsional flexibility of G/C-rich sequences as determined by cyclization assays. J Biol Chem. 1995 Dec 15;270(50):29945–29952. doi: 10.1074/jbc.270.50.29945. [DOI] [PubMed] [Google Scholar]
- Falvo J. V., Thanos D., Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y). Cell. 1995 Dec 29;83(7):1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993 May 6;363(6424):38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Parent L. A., Sharp P. A. Myc/Max and other helix-loop-helix/leucine zipper proteins bend DNA toward the minor groove. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11779–11783. doi: 10.1073/pnas.89.24.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Giese K., Cox J., Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992 Apr 3;69(1):185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Glover J. N., Harrison S. C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995 Jan 19;373(6511):257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Kahn J. D., Crothers D. M. Protein-induced bending and DNA cyclization. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6343–6347. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene. 1994 Nov;9(11):3149–3158. [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991 Nov 22;254(5035):1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991 Jul 26;66(2):317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- Kerppola T., Curran T. Transcription. Zen and the art of Fos and Jun. Nature. 1995 Jan 19;373(6511):199–200. doi: 10.1038/373199a0. [DOI] [PubMed] [Google Scholar]
- Kuprash D. V., Rice N. R., Nedospasov S. A. Homodimer of p50 (NF kappa B1) does not introduce a substantial directed bend into DNA according to three different experimental assays. Nucleic Acids Res. 1995 Feb 11;23(3):427–433. doi: 10.1093/nar/23.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene S. D., Zimm B. H. Understanding the anomalous electrophoresis of bent DNA molecules: a reptation model. Science. 1989 Jul 28;245(4916):396–399. doi: 10.1126/science.2756426. [DOI] [PubMed] [Google Scholar]
- Lumpkin O. J., Déjardin P., Zimm B. H. Theory of gel electrophoresis of DNA. Biopolymers. 1985 Aug;24(8):1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- McCaffrey P. G., Luo C., Kerppola T. K., Jain J., Badalian T. M., Ho A. M., Burgeon E., Lane W. S., Lambert J. N., Curran T. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993 Oct 29;262(5134):750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Kim S., Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989 Jun 23;244(4911):1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J Mol Biol. 1990 Nov 5;216(1):69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- Sitlani A., Crothers D. M. Fos and Jun do not bend the AP-1 recognition site. Proc Natl Acad Sci U S A. 1996 Apr 16;93(8):3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprous D., Zacharias W., Wood Z. A., Harvey S. C. Dehydrating agents sharply reduce curvature in DNAs containing A tracts. Nucleic Acids Res. 1995 May 25;23(10):1816–1821. doi: 10.1093/nar/23.10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. S., Dang C. V. Opposite orientations of DNA bending by c-Myc and Max. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7635–7639. doi: 10.1073/pnas.89.16.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. Catabolite activator protein-induced DNA bending in transcription initiation. J Mol Biol. 1991 May 20;219(2):201–215. doi: 10.1016/0022-2836(91)90562-k. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]