Abstract
Background
Life-threatening disorders of heart rhythm may arise during infancy and can result in the sudden and tragic death of a child. We performed exome sequencing on two unrelated infants presenting with recurrent cardiac arrest to discover a genetic cause.
Methods and Results
We ascertained two unrelated infants (probands) with recurrent cardiac arrest and dramatically prolonged QTc interval who were both born to healthy parents. The two parent-child trios were investigated using exome sequencing to search for de novo genetic variants. We then performed follow-up candidate gene screening on an independent cohort of 82 subjects with congenital long-QT syndrome without an identified genetic cause. Biochemical studies were performed to determine the functional consequences of mutations discovered in two genes encoding calmodulin. We discovered three heterozygous de novo mutations in either CALM1 or CALM2, two of the three human genes encoding calmodulin, in the two probands and in two additional subjects with recurrent cardiac arrest. All mutation carriers were infants who exhibited life-threatening ventricular arrhythmias combined variably with epilepsy and delayed neurodevelopment. Mutations altered residues in or adjacent to critical calcium binding loops in the calmodulin carboxyl-terminal domain. Recombinant mutant calmodulins exhibited several fold reductions in calcium binding affinity.
Conclusions
Human calmodulin mutations disrupt calcium ion binding to the protein and are associated with a life-threatening condition in early infancy. Defects in calmodulin function will disrupt important calcium signaling events in heart affecting membrane ion channels, a plausible molecular mechanism for potentially deadly disturbances in heart rhythm during infancy.
Keywords: arrhythmia, sudden cardiac death, exome, calcium signaling
INTRODUCTION
Sudden unexplained death during early development (prenatal period through infancy) may be caused by inborn errors including severe chromosomal abnormalities and monogenic conditions that predispose to life-threatening cardiac arrhythmias. Highly malignant arrhythmias may predispose to intrauterine fetal morbidity and mortality1,2 and sudden death of a neonate or infant as in the sudden infant death syndrome (SIDS).3–6 When a genetic cause has been identified in these early onset and highly malignant conditions, mutations are often de novo,3,7–9 but many cases do not have a clear genetic or molecular explanation.
In conditions such as the congenital long-QT syndrome (LQTS) and other inherited arrhythmia syndromes, the predisposition to sudden cardiac death is due to dysfunctional cardiac ion channels caused by mutations in genes encoding either pore-forming subunits or channel interacting proteins.10 The identification of novel arrhythmia susceptibility genes, particularly in clinically extreme cases, has great value for understanding the molecular basis of sudden cardiac death including unexplained infant mortality, and has the potential to inspire new therapeutic approaches.
Here we report that mutations in two genes encoding calmodulin, a ubiquitous and essential calcium signaling protein involved critically in a myriad of physiological events, are associated with life-threatening cardiac arrhythmias accompanied variably by neurological complications. We made this discovery by performing exome sequencing on two unrelated infants with recurrent cardiac arrest and subsequently by examining calmodulin genes for mutations in a cohort of LQTS cases without a defined genetic etiology. These findings suggest phenotypic and biochemical consequences of human calmodulin mutations, and offer a molecular basis for a novel life-threatening condition occurring in infancy.
METHODS
STUDY SUBJECTS
Study subjects were ascertained following informed consent procedures approved by the Ethics Review Board of the Fondazione IRCCS Policlinico San Matteo (Pavia, Italy), the Ethics Review Board of Klinikum Grosshadern (Munich, Germany), or the Institutional Review Boards of the Advocate Lutheran General Hospital and Cincinnati Children’s Hospital. Peripheral blood leukocytes were collected and used for isolation of genomic DNA. Study subjects included two probands (one Caucasian, one Hispanic) with highly malignant ventricular arrhythmia syndromes and their respective unaffected parents, and 82 additional unrelated cases of LQTS without an identified genetic cause. Control subjects were from two sources. A panel of Hispanic Americans (n=92) was obtained from the Coriell Institute for Medical Research, while a panel of Caucasian Europeans (n=1800) for whom exome data were available through the Institute of Human Genetics (Helmholtz Zentrum München).
EXOME SEQUENCING
Exome enrichment was performed with the Agilent SureSelect Human All Exon 50 Mb capture reagent used according to the supplier’s instructions. Paired-end (2 × 100 base pairs) sequencing was performed on the Illumina HiSeq2000 platform. Proband 1 was sequenced at the Institute of Human Genetics, Helmholtz Zentrum München, whereas proband 2 was sequenced by the Genome Sciences Resource, Vanderbilt University.
EXOME SEQUENCE DATA ANALYSIS
After removal of low quality reads, alignments to a reference human genome (UCSC assembly hg19) were performed using BWA (version 0.5.8),11 then sequences were processed using the Genome Analysis TookKit (GATK)12 to remove duplicate reads and to call variants. Default settings were used in the BWA alignments including a maximum of 2 mismatches in the ‘seed’ portion of reads (first 32 bp) and no more than 3 mismatches for the entire read. The threshold for detecting variants was set at a genotype quality score of 40.
Variants identified in the probands that were also found in dbSNP (v130), 1000Genomes, Exome Variant Server, and Helmholtz exome databases were excluded from further analyses. Synonymous and intronic (other than canonical splice sites) variants were also excluded. Variant annotation was performed using custom scripts. Further, based on the hypothesis that disease causing mutations in the probands are de novo, we excluded all inherited variants (i.e., that were observed in either parent). Non-excluded variants were validated in proband and parental DNAs using conventional Sanger sequencing, then further annotated based upon evolutionary nucleotide conservation (GERP),13 predicted impact on protein function (Polyphen2, SIFT).14,15
ADDITIONAL MUTATION DETECTION
Mutation screening was performed in 82 unrelated LQTS subjects (Schwartz Score ≥3.516 and/or resting QTc ≥480 ms) without a previously identified genetic cause to search for additional variants in the candidate disease-causing genes identified in the two probands. Specifically, the coding exons of CALM1, CALM2 and CALM3, three genes encoding identical calmodulin proteins, were amplified by polymerase chain reaction (PCR; primer sequences provided in supplemental Table S1) then sequenced using an automated capillary electrophoresis DNA sequencing platform (Applied Biosystems, Foster City, CA).
CALMODULIN GENE EXPRESSION AND FUNCTIONAL ANALYSES
The methods for determining calmodulin gene expression in human heart and biochemical studies of recombinant calmodulin proteins are described in detail in the online Data Supplement.
RESULTS
STUDY SUBJECTS
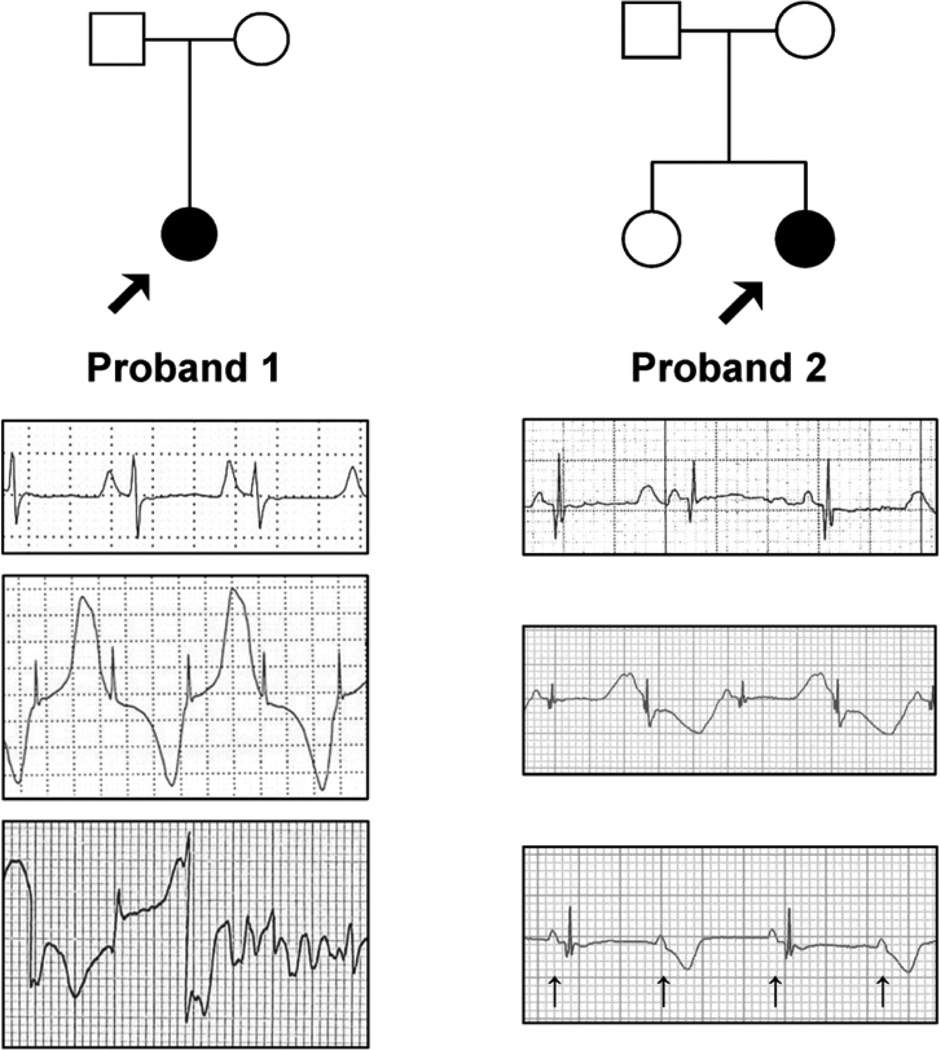
We ascertained two unrelated infants (probands) with recurrent cardiac arrest. Proband 1 was a Caucasian female from Italy who suffered cardiac arrest due to ventricular fibrillation (VF) at age 6 months. Her birth and prenatal history were unremarkable, family history was negative for sudden cardiac death, and both parents were asymptomatic with normal electrocardiograms (ECG). There had been no evidence of fetal bradycardia. Following successful external defibrillation, an ECG demonstrated a markedly prolonged QTc interval (630 ms), frequent episodes of T-wave alternans and intermittent 2:1 atrioventricular (AV) block (Fig. 1). Echocardiogram revealed normal cardiac anatomy and contractile function. The patient was treated with propranolol and an internal cardioverter defibrillator (ICD) was placed. Genetic testing for mutations in KCNQ1, KCNH2, SCN5A, KCNE1 and KCNE2 (Molecular Cardiology Laboratory, Fondazione IRCCS Policlinico S. Matteo, Pavia, Italy), the most frequently mutated genes in LQTS, was negative.
Figure 1.
Clinical phenotypes and pedigrees. Representative electrocardiographic recordings from two probands with early onset, life-threatening cardiac arrhythmias. Upper trace represents baseline ECG for both probands. Middle trace illustrates T-wave alternans. Lower trace for proband 1 illustrates onset of ventricular fibrillation following a period of T-wave alternans. Lower trace for proband 2 illustrates 2:1 AV block (arrows mark p-waves coincident with atrial depolarization).
In the following months, the child had multiple episodes of VF that were terminated by ICD shocks. Propranolol dosage was progressively increased to 10 mg/kg/day without arrhythmia suppression. Left cardiac sympathetic denervation (LCSD) was performed at age 12 months, but QTc remained prolonged (550–630 ms) and episodes of T-wave alternans continued to occur. A loading dose of mexiletine did not shorten the QT interval. Verapamil reduced the frequency of T-wave alternans, but persistent cardiac electrical instability prompted right cardiac sympathetic denervation (RCSD). Despite all treatments, there were 16 episodes of VF during the first 2 years of life, mostly induced by adrenergic stimulation and not pause-dependent or preceded by torsades de pointes (TdP). Verapamil was replaced with flecainide and in the following year she had only one VF episode triggered by strong emotional stress. A mild delay in language development was noted.
Proband 2 was an Hispanic female infant living in the United States who presented with sinus bradycardia, T-wave alternans, markedly prolonged QTc (690 ms) and 2:1 AV block occurring 2 hours after a normal delivery (Fig. 1). Fetal bradycardia (98–110 bpm) was first noted at 21 weeks’ gestation and lasted throughout the pregnancy. Fetal echocardiogram performed at 27 weeks’ gestation revealed normal cardiac anatomy and function except for a heart rate of 90–95 bpm. An episode of 2:1 AV block was noted at 28 weeks’ gestation. There was no family history of arrhythmia, miscarriages, sudden death, seizures or drowning, and ECGs from both parents and an older sister were normal. Postnatally, esmolol and propranolol treatment restored 1:1 AV conduction, but lidocaine did not shorten the QTc interval. Genetic testing was negative for mutations in KCNQ1, KCNH2, SCN5A, KCNE1 and KCNE2 (Familion®, Transgenomic Labs).
She was discharged from the hospital on postnatal day 8, but returned at age 3 weeks following cardiac arrest and multiple episodes of VF. She was successfully defibrillated but suffered a right parietal lobe cerebral infarction that was documented by brain MRI. Treatment at that time included propranolol, mexiletine and an ICD. In the first two years of life she was hospitalized numerous times for episodes of VF that were successfully terminated by ICD shocks. At 2 years of age she developed seizures that were attributed to the prior brain injury. At age 3, seizures were well controlled with levetiracetam and her QTc was 500–510 ms on a combination of mexiletine and propranolol, but she exhibited developmental delays (10–33%) in all categories of the Hawaii Early Learning Profile.
DISCOVERY OF CALMODULIN MUTATIONS BY EXOME SEQUENCING
Because both sets of parents were healthy with normal ECGs and there was no overt family history of sudden death or related symptoms suggesting an inherited cardiac arrhythmia syndrome, we hypothesized that de novo mutations were most likely. Further, because genetic testing for the major LQTS susceptibility genes was unrevealing in the probands, we predicted that a novel genetic basis for the clinical disease was plausible. Therefore, we performed exome sequencing on the two probands and their parents (parent-child trios), then searched for novel variants that were not inherited and predicted to have deleterious effects on protein structure or function. Exome sequencing of parent-child trios has emerged as a powerful approach for discovering de novo mutations in novel genes.17,18
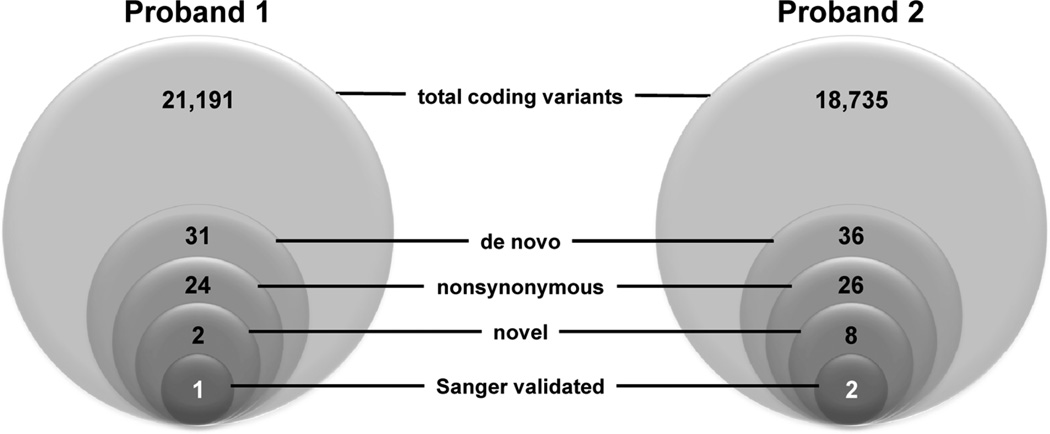
For proband 1 and her parents, the mean coverage depth across 50 Mb of captured sequence was 104-fold (104×) for the 3 samples, with 20× coverage depth for 90.5% of targeted bases and 88% of high quality reads mapped uniquely to the reference human genome (hg19 build). We detected 31 heterozygous de novo ‘coding’ variants (within exons or canonical splice sites) of which 24 were nonsynonmous (Fig. 2). However, only two of these variants were novel and only one was validated by Sanger DNA sequencing (see below).
Figure 2.
Exome variant filtering strategy. Sequential filters were applied to the pool of variants discovered by exome sequencing in the two probands. Total coding variants include all synonymous, nonsynonymous, nonsense, frameshift inducing insertions or deletions, and canonical splice site variants within captured exons, but exclude variants in 5’ and 3’ untranslated regions and introns. De novo variants are those found in proband but not in either parent. Novel variants were defined as those absent in public databases (dbSNP, 1000genomes). Novel variants in Proband 1 were also absent in 1202 exomes generated in the Institute of Human Genetics Helmholtz Zentrum München).
For proband 2, greater than 98% of high quality reads mapped uniquely to hg19 and the mean coverage depth was 58× with at least 20× coverage for 83% of targeted bases. Initially, we identified 36 de novo coding sequence variants of which 10 were synonymous. Of the remaining 26 nonsynonymous de novo variants, 8 were novel (absent in reference databases), but only two were validated by Sanger sequencing (Fig. 2). One of the two validated variants occurred in a non-cardiac expressed gene with no known function (C6orf108) at a nucleotide position with poor evolutionary conservation (E17K; GERP score −6.1), and therefore, was deemed unlikely to be pathogenic. The remaining novel, nonsynonymous de novo variant is discussed further below.
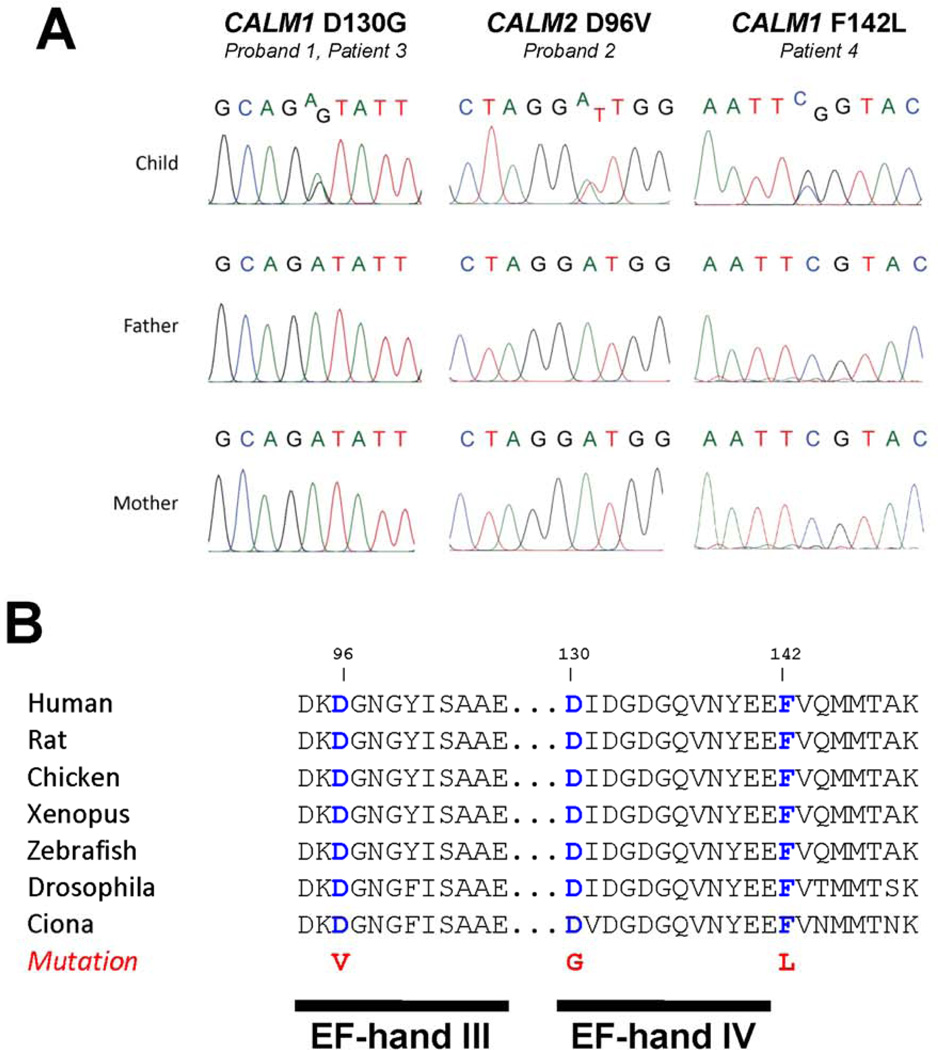
Among the validated, de novo variants discovered in the probands, two were in genes encoding the ubiquitous calcium signaling protein calmodulin (Fig. 3A). In proband 1, a missense mutation in CALM1 (chr 14q31) predicted replacement of a highly conserved aspartic acid residue at position 130 (first methionine residue assigned position 1) with glycine (D130G). In proband 2, a missense mutation in CALM2 (chr 2p21) predicted replacement of another highly conserved aspartic acid residue at position 96 with valine (D96V). Both mutations are predicted to be damaging by SIFT and PolyPhen2 analyses, and replace acidic residues within the carboxyl terminal domain (C-domain) (Fig. 3B). Neither mutation was found by direct screening of DNA from ethnically matched control subjects nor was found in publicly accessible databases of genetic variants (see Methods). We also did not observe either variant in 1800 exomes sequenced at the Institute of Human Genetics (Helmholtz Zentrum München) in which the mean coverage of CALM1 and CALM2 was >95×. To further illustrate the extreme rarity of calmodulin gene mutations, only two nonsynonymous coding variants in CALM1 (T10I, and L143V) and none in CALM2 were called in 8599 alleles of European ancestry by the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/). Futher, no CALM1 or CALM2 nonsynomous variants were identified in the Helmholtz exome data. Therefore, we concluded that mutations of CALM1 or CALM2 were likely responsible for the life-threatening syndrome observed in the probands.
Figure 3.
De novo calmodulin gene mutations in infants with severe cardiac arrhythmias. A, Nucleotide sequence traces indicating heterozygous calmodulin gene mutations in Proband 1 (same mutation as in Case 3), Proband 2 and Case 4. B, Amino acid sequence alignments for calmodulins from different species with location of missense mutations.
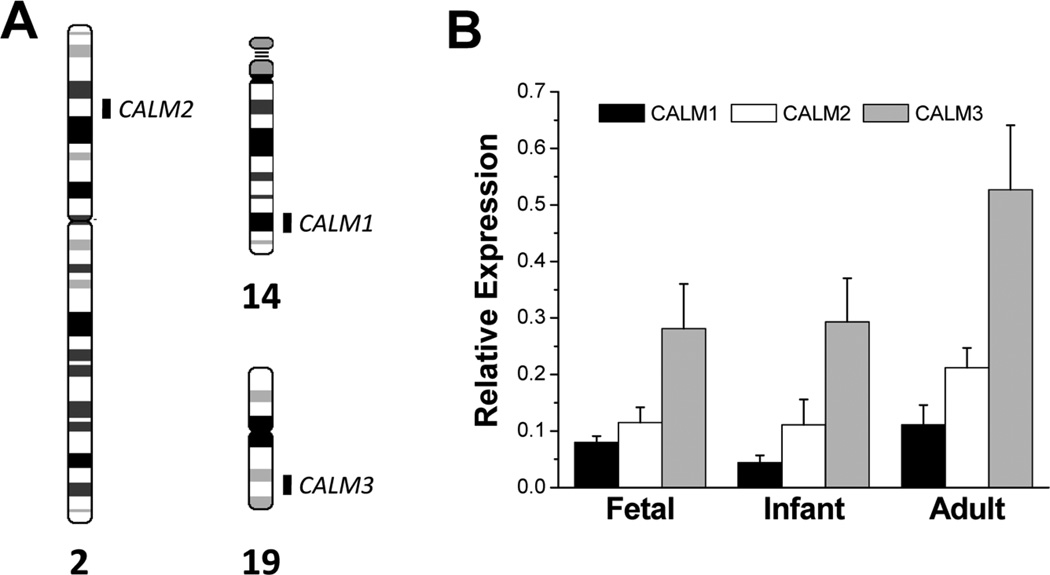
Human calmodulin is encoded by three separate genes each located on a different chromosome (Fig. 4A), but the protein products of each gene have identical amino acid sequences.19 We examined the expression of each calmodulin gene in human heart (left ventricle) from three developmental stages (fetal, infant, adult) using quantitative RT-PCR and gene-specific fluorogenic TaqMan probes. All calmodulin genes are expressed throughout development with the rank order of expression being CALM3 > CALM2 > CALM1 (Fig. 4B). These data demonstrate cardiac expression of the two calmodulin genes in which we discovered mutations.
Figure 4.
Expression of calmodulin genes in human heart. A, Chromosomal locations of the three human calmodulin genes indicated on ideograms representing G-banded chromosomes. B, Relative expression of mRNA for CALM1, CALM2, and CALM3 in normal human heart (left ventricle) normalized to expression of β-actin as determined by real-time quantitative RT-PCR using gene-specific Taqman probes. Human heart samples include fetal (n = 4), infant (n = 4), and adult (n = 8) developmental stages. Data plotted are mean ± SEM. Differences in expression among the three genes were significant (p<0.05; one-way ANOVA) in fetal, infant and adult hearts.
The discovery of mutations in calmodulin genes in the setting of a severe cardiac arrhythmia syndrome associated with markedly prolonged QT interval prompted us to examine a cohort of other cases of congenital long-QT syndrome (LQTS) for which no genetic cause had been found. We performed a directed search for mutations in CALM1, CALM2, and the closely related CALM3 in a cohort of 82 LQTS cases with no identified mutations. CALM1 mutations were discovered in two subjects within this cohort, both with early onset and severe clinical presentations.
The same mutation discovered in proband 1 (CALM1-D130G) was found in a 3 year-old Caucasian male from Greece (Case 3) who had suffered multiple cardiac arrests beginning at age 1 month. We also discovered a novel CALM1 missense mutation in an adopted 14 year-old Caucasian male from Italy (Case 4) with recurrent episodes of non-sustained ventricular tachycardia, T-wave alternans, markedly prolonged QTc interval and cardiac arrest due to ventricular fibrillation. The mutation discovered in this subject predicted substitution of a highly conserved phenylalanine residue at position 142 with leucine (F142L; Fig. 3A). This mutation was absent in Caucasian controls of western European ancestry and was not observed in the aforementioned databases of genetic variants (see Methods). Additional clinical information about these two additional mutation carriers is available in the online Data Supplement.
BIOCHEMICAL CONSEQUENCES OF CALMODULIN MUTATIONS
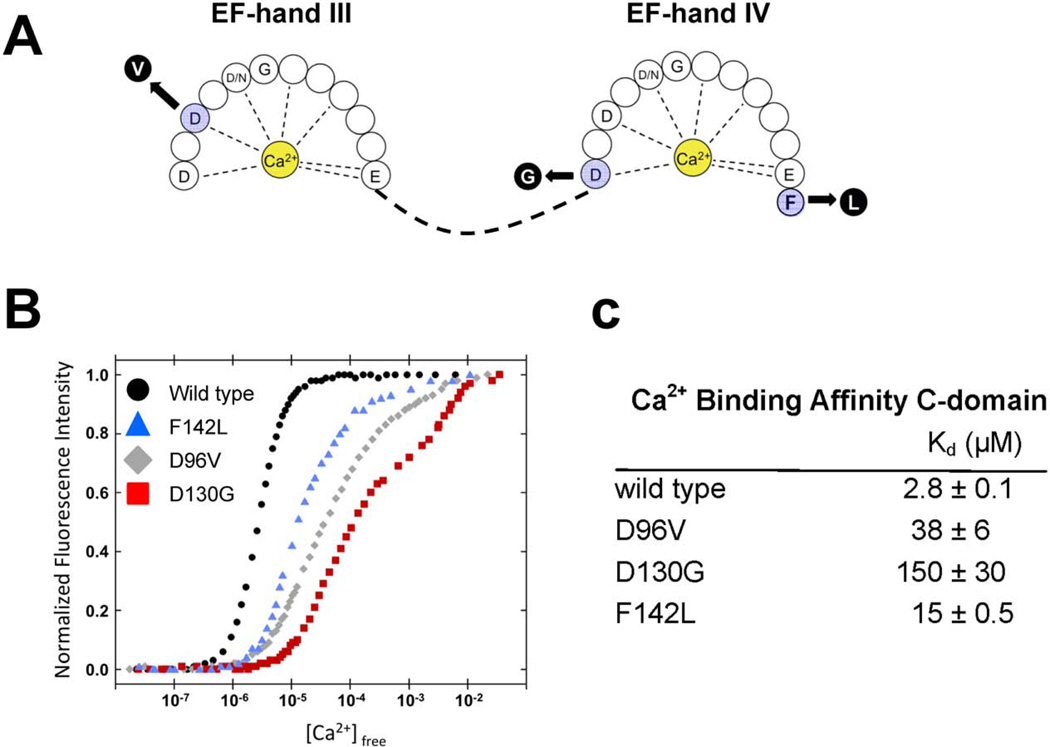
The correlation between the symptoms exhibited by these infants and the known involvement of calmodulin in modulating the activity of ion channels and other critical proteins in heart motivated investigation of the effect of the mutations on protein function. Two of the mutations we discovered (CALM1-D130G, CALM2-D96V) alter highly conserved aspartic acid residues that directly chelate Ca2+ ions in EF-hand domains IV and III, respectively (positions X and Y, respectively, in the pentagonal bipyrimidal coordination sphere;20 Fig. 5A) and were predicted to reduce Ca2+ affinity. The predicted effect of the CALM1-F142L mutation was an alteration of the energetic coupling of Ca2+ binding and the conformational change associated with calmodulin activation.21,22 To determine the functional consequences of calmodulin mutations, we generated wildtype and mutant recombinant calmodulin proteins in bacteria and performed in vitro Ca2+ binding studies by monitoring intrinsic tyrosine and phenylalanine fluorescence.23 The data revealed that all three mutations have reduced Ca2+ affinity in the C-domain (5–53 fold) (Fig. 5B,C), and no significant effect on N-domain Ca2+ affinity (data not shown). The structural integrity of the mutant proteins was then validated using heteronuclear nuclear magnetic resonance spectroscopy (see online Data Supplement, Fig. S1). These biochemical data predict a significant alteration in the ability of mutant calmodulins to transduce Ca2+ signals and perform essential physiological functions.
Figure 5.
Impaired Ca2+ binding by mutant calmodulin C-domains. A, Schematic model of the Ca2+ binding loops in the C-terminal EF-hand domains of calmodulin showing location of the mutations. B, Titration curves for Ca2+ binding to recombinant WT (black circles) and mutant calmodulins (F142L, blue triangles; D96V, grey diamonds; D130G, red squares). C, Calcium ion binding affinities for WT and mutant calmodulins.
DISCUSSION
We report the discovery of de novo calmodulin mutations in a severe, early onset cardiac arrhythmia syndrome with features of LQTS. Calmodulin is a ubiquitous, multifunctional Ca2+ binding protein essential for a myriad of intracellular signaling processes in eukaryotic cells.24 In electrically excitable tissues such as heart and brain, calmodulin transduces Ca2+ signals to influence activity of ion channels, kinases and other target proteins that contribute importantly to physiological functions of these organs.25–27 Calcium ion binding by four highly conserved EF-hand domains promotes conformational changes that are integral to calmodulin function.28 Given that calmodulin is essential to fundamental cell processes and that its protein sequence is perfectly conserved among vertebrates, complete absence of calmodulin is not expected to be compatible with survival.
The main clinical features of the conditions associated with calmodulin mutations are summarized in Table 1. The common cardiac features of this syndrome include life-threatening ventricular arrhythmias occurring very early in life, frequent episodes of T-wave alternans, markedly prolonged QTc interval (>600 ms), and intermittent 2:1 AV block. Ventricular fibrillation was typically triggered by adrenergic activation, and either occurred spontaneously or was preceded by a short period of polymorphic ventricular tachycardia that was not pause-dependent. Treatment with β-adrenergic antagonists was of some value albeit not sufficient to prevent all arrhythmic events. All mutation carriers were treated with a second anti-arrhythmic agent (most often mexiletine or flecainide) and either an implantable defibrillator, cardiac sympathetic denervation or both of these interventions.
Table 1 .
Summary of clinical features of calmodulin mutation carriers.
| Subject | Sex | Age at Diagnosis |
VF | QTc | TWA | 2:1 AVB | Seizures | Developmental delay |
Treatments | Mutation1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Proband 1 | F | 6 months | + | 630 ms | + | + | − | +/− | BB, MEX, VER, FLEC, ICD, LCSD, RCSD | CALM1-D130G |
| Proband 2 | F | prenatal | + | 690 ms | + | + | + | + | BB, MEX, ICD | CALM2-D96V |
| Case 3 | M | 1 month | + | 610 ms | + | + | + | + | BB, MEX, ICD | CALM1-D130G |
| Case 4 | M | ? neonatal | + | >600 ms | + | − | + | + | BB, MEX, LCSD | CALM1-F142L |
Mutation position is based on RefSeq NP_005175 and counting the predicted translational start codon (Met) as position 1.
VF, ventricular fibrillation; QTc, rate-corrected QT interval; TWA, T-wave alternans; AVB, atrioventricular block; BB, β-blocker (propranolol); MEX, mexiletine; VER, verapamil; FLEC, flecainide; ICD, implantable cardioverter defibrillator; LCSD, left cardiac sympathetic denervation; RCSD, right cardiac sympathetic denervation.
The clinical syndrome we describe bears many similarities with congenital LQTS, but the intensity and frequency of arrhythmic events as well as the very early age of onset are not entirely typical of LQTS. Further, all cases exhibited some degree of neurodevelopmental delay ranging from mild delay in language development (proband 1) to moderate and severe cognitive or motor impairment (proband 2, cases 3 and 4). Seizures were present in 3 of the 4 cases, and one subject (proband 2) suffered a cerebrovascular accident following her first cardiac arrest. Neurodevelopmental phenotypes and epilepsy can be attributed to brain injury secondary to cardiac arrest during early life as seen in young children with congenital heart disease.29,30 However, given that calmodulin is highly expressed in brain, we cannot completely exclude the possibility that calmodulin mutations confer increased susceptibility to neuronal injury in the setting of circulatory insufficiency thereby contributing to the high prevalence of neurological and neurodevelopmental deficits.
The impaired Ca2+ binding exhibited by calmodulin mutants suggests a molecular basis for the life-threatening condition experienced by the four mutation carriers we report here. Calmodulin serves as the Ca2+ sensor for Ca2+-dependent inactivation of L-type voltage-gated Ca2+ channels in cardiac myocytes.31,32 Over expression of calmodulin mutants with defective Ca2+ binding causes dramatic prolongation of ventricular action potentials in guinea pig myocytes,33 a plausible mechanistic link with prolonged QT interval, and predisposition to ventricular arrhythmia. Other essential heart proteins require functional calmodulin for normal cardiac repolarization. A voltage-gated potassium channel (KCNQ1 or KV7.1) responsible for the slow component of the delayed rectifier current (IKs) is needed for myocardial repolarization and requires calmodulin for activity.34,35 Inhibition of calmodulin or chelation of intracellular Ca2+ inhibits IKs and this would delay repolarization setting up conditions favoring early afterdepolarizations and triggered arrhythmias. Inactivation of cardiac sodium channels involves calmodulin and disrupting this interaction might evoke arrhythmogenic sodium channel dysfunction.36–39 Finally, disturbances in calmodulin-dependent kinase II activity can promote ventricular arrhythmogenesis by several mechanisms.40
Functional studies in native cardiac myocytes should be informative as to the major electrophysiological events disrupted by the calmodulin mutations we discovered and help explain the pathogenesis of the associated arrhythmia syndrome. We predict that expression of mutant calmodulins will adversely affect repolarization because of ion channel dysfunction such as impaired calcium channel inactivation. Understanding the manner in which these effects are generated will also help clarify how a single mutant allele can have such severe consequences despite the redundancy of calmodulins in heart. However, a thorough understanding of the molecular mechanisms responsible for the complex heart rhythm disturbances we report in association with calmodulin mutations will require extensive experimental work including studies of genetically engineered animals and is beyond the scope of this report.
A recent study demonstrated genetic linkage of autosomal dominant catecholaminergic polymorphic ventricular tachycardia (CPVT) in a RYR2 and CASQ2 mutation-negative Swedish family to chromosome 14q31–32, a locus that includes CALM1 among nearly 70 genes.41 Screening CALM1 revealed a missense mutation affecting a conserved asparagine residue within EF-hand domain II (reported as N53I). A subsequent search for CALM1 mutations in a cohort of 62 other mutation-negative CPVT cases identified a single de novo missense mutation (reported as N97S; EF-hand domain III) in an Iraqi child. Both mutations impair Ca2+ binding to recombinant calmodulin and disrupt binding to a peptide encompassing the calmodulin binding site on RYR2. There was no evidence of prolonged QT intervals in any of these mutation carriers. Combined with our observations, these findings suggest an intriguing genotype-phenotype correlation among calmodulin mutations, and further suggest different pathophysiological mechanisms.
The amino acid sequence of calmodulin is perfectly conserved among vertebrates. Furthermore, vertebrates have three genes encoding calmodulin, but the transcriptional regulation, tissue-specific expression and intracellular distribution of the three gene products are incompletely understood. We demonstrated the expression of all three calmodulin genes in human fetal, infant and adult left ventricle indicating that heterozygous mutations in CALM1 or CALM2 will be present in mutation carriers among all other wildtype protein alleles encoded by the three calmodulin genes. This suggests that haploinsufficiency is unlikely to account for the severe phenotype observed in heterozygous carriers of either CALM1 or CALM2 mutations and raises the possibility of a dominant-negative mechanism. Further, calmodulin mutations with such severe phenotypic consequences as we report here are less likely to be inherited and will therefore appear as sporadic cases but only if life-saving measures are successful. Finally, the reason why mutations in ubiquitously expressed calmodulin genes present predominantly with a cardiac phenotype, albeit with notable neurological deficits following cardiac arrest, is not clear. We speculate that the heart may simply have less physiological reserve with respect to Ca2+ signaling than other tissues.
In conclusion, we discovered calmodulin mutations that offer an explanation for recurrent cardiac arrest during early infancy with features of severe LQTS. The high degree of conservation and the absence of inherited mutations attest to the importance of calmodulin in transducing Ca2+ signals into essential cellular responses. Additional investigations to determine the contribution of mutant calmodulins to unexplained sudden death in early development are warranted.
Supplementary Material
CLINICAL PERSPECTIVE.
Calmodulin is a ubiquitous calcium binding protein essential for a myriad of intracellular signaling processes in eukaryotic cells. In heart, calmodulin transduces Ca2+ signals to influence activity of ion channels, kinases and other target proteins that contribute importantly to cardiac function. Given that calmodulin is essential to fundamental cell processes, mutations are expected to have severe consequences. Here we report calmodulin mutations in two unrelated infants with recurrent cardiac arrest and features of severe LQTS who were both negative for mutations in known arrhythmia predisposing genes. Using next-generation sequencing technologies to scan coding exons across the genome (e.g., exome), we discovered de novo mutations in two distinct genes (CALM1, CALM2) encoding identical calmodulin proteins. Screening of calmodulin genes in a cohort of LQTS cases without a defined genetic etiology identified two additional subjects with de novo mutations in CALM1. Mutation carriers had recurrent cardiac arrest secondary to ventricular tachyarrhythmias, severely prolonged QTc interval, and evidence of electrically unstable myocardium (T-wave alternans). Most carriers exhibited neurological deficits (epilepsy, neurodevelopmental delays) of variable severity that could be attributed to brain injury secondary to cardiac arrest or possibly to enhanced susceptibility to neuronal injury in the setting of circulatory insufficiency. Mutant calmodulin proteins exhibited reduced affinity for Ca2+ predicted to disrupt critical functions of calmodulin that would be arrhythmogenic. Our findings provide a genetic etiology for ventricular arrhythmias during infancy and illustrate the phenotypic and biochemical consequences of human calmodulin mutations. Calmodulin genes should perhaps be screened in severe, early onset cardiac arrhythmias.
Acknowledgments
The authors are grateful to Susanne Lindhof for technical support on genetic studies, to Madeline Shea for helpful advice on measurement and interpretation of Ca2+ binding affinities, and to Pinuccia De Tomasi for assistance with preparing figures.
Funding Sources: This work was supported by NIH grants HL083374, HL069712, T32-NS007941, RR028106, and HL065962; the German Federal Ministry of Education and Research (BMBF) funded Systems Biology of Metabotypes grant (SysMBO 031549A); the German Network for Mitochondrial Disorders (mitoNET 01GM0867 and 01GM0862); the BMBF funded German Center for Heart Research (Z76010017300 and Z56010015300); the European Commission 7th Framework Program (Project N261123 GEUVADIS), the German Ministry for Education and Research (01GR0804-4), Telethon Foundation (GGP09247), the Italian Ministry of Health (GR-2010-2305717), and the Vanderbilt Institute for Clinical and Translational Research (NIH grant RR024975).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures: The authors have no potential conflicts of interest related to the work reported in this manuscript.
Author Contributions: L.C., P.J.S. and A.L.G. conceived the experiments and wrote the manuscript. G.M.D.F., B.C., M.O., J.P., S.K., and D.W.B. ascertained study subjects and compiled clinical data. C.N.J., M.D.F., S.G.K, J.D.K., M.P., T.M.S., E.G., T.W., P.L., B.B., T.C., and C.S. performed critical experiments or performed data analysis. T.M., W.J.C. and A.L.G. planned and supervised critical experiments, and reviewed data.
References
- 1.Miller TE, Estrella E, Myerburg RJ, Garcia d V, Moreno N, Rusconi P, Ahearn ME, Baumbach L, Kurlansky P, Wolff G, Bishopric NH. Recurrent third-trimester fetal loss and maternal mosaicism for long-QT syndrome. Circulation. 2004;109:3029–3034. doi: 10.1161/01.CIR.0000130666.81539.9E. [DOI] [PubMed] [Google Scholar]
- 2.Murphy LL, Moon-Grady AJ, Cuneo BF, Wakai RT, Yu S, Kunic JD, Benson DW, George AL., Jr Developmentally regulated SCN5A splice variant potentiates dysfunction of a novel mutation associated with severe fetal arrhythmia. Heart Rhythm. 2011;9:590–597. doi: 10.1016/j.hrthm.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz PJ, Priori SG, Dumaine R, Napolitano C, Antzelevitch C, Stramba-Badiale M, Richard TA, Berti MR, Bloise R. A molecular link between the sudden infant death syndrome and the long-QT syndrome. N Engl J Med. 2000;343:262–267. doi: 10.1056/NEJM200007273430405. [DOI] [PubMed] [Google Scholar]
- 4.Ackerman MJ, Siu BL, Sturner WQ, Tester DJ, Valdivia CR, Makielski JC, Towbin JA. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 2001;286:2264–2269. doi: 10.1001/jama.286.18.2264. [DOI] [PubMed] [Google Scholar]
- 5.Wang DW, Desai RR, Crotti L, Arnestad M, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Rognum T, Schwartz PJ, George AL., Jr Cardiac sodium channel dysfunction in sudden infant death syndrome. Circulation. 2007;115:368–376. doi: 10.1161/CIRCULATIONAHA.106.646513. [DOI] [PubMed] [Google Scholar]
- 6.Arnestad M, Crotti L, Rognum TO, Insolia R, Pedrazzini M, Ferrandi C, Vege A, Wang DW, Rhodes TE, George AL, Jr, Schwartz PJ. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 7.Wang DW, Crotti L, Shimizu W, Pedrazzini M, Cantu F, De FP, Kishiki K, Miyazaki A, Ikeda T, Schwartz PJ, George AL., Jr Malignant perinatal variant of long-QT syndrome caused by a profoundly dysfunctional cardiac sodium channel. Circ Arrhythm Electrophysiol. 2008;1:370–378. doi: 10.1161/CIRCEP.108.788349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bankston JR, Yue M, Chung W, Spyres M, Pass RH, Silver E, Sampson KJ, Kass RS. A novel and lethal de novo LQT-3 mutation in a newborn with distinct molecular pharmacology and therapeutic response. PLoS ONE. 2007;2:e1258. doi: 10.1371/journal.pone.0001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz PJ, Priori SG, Bloise R, Napolitano C, Ronchetti E, Piccinini A, Goj C, Breithardt G, Schulze-Bahr E, Wedekind H, Nastoli J. Molecular diagnosis in a child with sudden infant death syndrome. Lancet. 2001;358:1342–1343. doi: 10.1016/S0140-6736(01)06450-9. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz PJ, Crotti L, Insolia R. Long-QT syndrome: from genetics to management. Circ Arrhythm Electrophysiol. 2012;5:868–877. doi: 10.1161/CIRCEP.111.962019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation. 1993;88:782–784. doi: 10.1161/01.cir.88.2.782. [DOI] [PubMed] [Google Scholar]
- 17.Vissers LE, de LJ, Gilissen C, Janssen I, Steehouwer M, de VP, van LB, Arts P, Wieskamp N, del RM, van Bon BW, Hoischen A, de Vries BB, Brunner HG, Veltman JA. A de novo paradigm for mental retardation. Nature Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 18.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, Rieder MJ, Nickerson DA, Bernier R, Fisher SE, Shendure J, Eichler EE. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer R, Koller M, Flura M, Mathews S, Strehler-Page MA, Krebs J, Penniston JT, Carafoli E, Strehler EE. Multiple divergent mRNAs code for a single human calmodulin. J Biol Chem. 1988;263:17055–17062. [PubMed] [Google Scholar]
- 20.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 Å resolution. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 21.Nelson MR, Chazin WJ. An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Sci. 1998;7:270–282. doi: 10.1002/pro.5560070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunick CG, Nelson MR, Mangahas S, Hunter MJ, Sheehan JH, Mizoue LS, Bunick GJ, Chazin WJ. Designing sequence to control protein function in an EF-hand protein. J Am Chem Soc. 2004;126:5990–5998. doi: 10.1021/ja0397456. [DOI] [PubMed] [Google Scholar]
- 23.VanScyoc WS, Sorensen BR, Rusinova E, Laws WR, Ross JB, Shea MA. Calcium binding to calmodulin mutants monitored by domain-specific intrinsic phenylalanine and tyrosine fluorescence. Biophys J. 2002;83:2767–2780. doi: 10.1016/S0006-3495(02)75286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 25.Halling DB, Aracena-Parks P, Hamilton SL. Regulation of voltage-gated Ca2+ channels by calmodulin. Sci STKE. 2005;2005:re15. doi: 10.1126/stke.3152005re15. [DOI] [PubMed] [Google Scholar]
- 26.Maier LS, Bers DM, Brown JH. Calmodulin and Ca2+/calmodulin kinases in the heart - physiology and pathophysiology. Cardiovasc Res. 2007;73:629–630. doi: 10.1016/j.cardiores.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nature Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Tanaka T, Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nature Struct Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
- 29.Nadkarni VM, Larkin GL, Peberdy MA, Carey SM, Kaye W, Mancini ME, Nichol G, Lane-Truitt T, Potts J, Ornato JP, Berg RA. First documented rhythm and clinical outcome from in-hospital cardiac arrest among children and adults. JAMA. 2006;295:50–57. doi: 10.1001/jama.295.1.50. [DOI] [PubMed] [Google Scholar]
- 30.Ortmann L, Prodhan P, Gossett J, Schexnayder S, Berg R, Nadkarni V, Bhutta A. Outcomes after in-hospital cardiac arrest in children with cardiac disease: a report from Get With the Guidelines--Resuscitation. Circulation. 2011;124:2329–2337. doi: 10.1161/CIRCULATIONAHA.110.013466. [DOI] [PubMed] [Google Scholar]
- 31.Zuhlke RD, Pitt GS, Deisseroth K, Tsien RW, Reuter H. Calmodulin supports both inactivation and facilitation of L-type calcium channels. Nature. 1999;399:159–162. doi: 10.1038/20200. [DOI] [PubMed] [Google Scholar]
- 32.Peterson BZ, DeMaria CD, Adelman JP, Yue DT. Calmodulin is the Ca2+ sensor for Ca2+ -dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 33.Alseikhan BA, DeMaria CD, Colecraft HM, Yue DT. Engineered calmodulins reveal the unexpected eminence of Ca2+ channel inactivation in controlling heart excitation. Proc Natl Acad Sci U S A. 2002;99:17185–17190. doi: 10.1073/pnas.262372999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shamgar L, Ma L, Schmitt N, Haitin Y, Peretz A, Wiener R, Hirsch J, Pongs O, Attali B. Calmodulin is essential for cardiac IKS channel gating and assembly: impaired function in long-QT mutations. Circ Res. 2006;98:1055–1063. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 35.Ciampa EJ, Welch RC, Vanoye CG, George AL. KCNE4 juxtamembrane region is required for interaction with calmodulin and for functional suppression of KCNQ1. J Biol Chem. 2010;286:4141–4149. doi: 10.1074/jbc.M110.158865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Ghosh S, Liu H, Tateyama M, Kass RS, Pitt GS. Calmodulin mediates Ca2+ sensitivity of sodium channels. J Biol Chem. 2004;279:45004–45012. doi: 10.1074/jbc.M407286200. [DOI] [PubMed] [Google Scholar]
- 37.Shah VN, Wingo TL, Weiss KL, Williams CK, Balser JR, Chazin WJ. Calcium-dependent regulation of the voltage-gated sodium channel hH1: intrinsic and extrinsic sensors use a common molecular switch. Proc Natl Acad Sci U S A. 2006;103:3592–3597. doi: 10.1073/pnas.0507397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potet F, Chagot B, Anghelescu M, Viswanathan PC, Stepanovic SZ, Kupershmidt S, Chazin WJ, Balser JR. Functional interactions between distinct sodium channel cytoplasmic domains through the action of calmodulin. J Biol Chem. 2009;284:8846–8854. doi: 10.1074/jbc.M806871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarhan MF, Tung CC, Van PF, Ahern CA. Crystallographic basis for calcium regulation of sodium channels. Proc Natl Acad Sci U S A. 2012;109:3558–3563. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson ME. Multiple downstream proarrhythmic targets for calmodulin kinase II: moving beyond an ion channel-centric focus. Cardiovasc Res. 2007;73:657–666. doi: 10.1016/j.cardiores.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Nyegaard M, Overgaard MT, Sondergaard MT, Vranas M, Behr ER, Hildebrandt LL, Lund J, Hedley PL, Camm AJ, Wettrell G, Fosdal I, Christiansen M, Borglum AD. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am J Hum Genet. 2012;91:703–712. doi: 10.1016/j.ajhg.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.