Abstract
Retinoic acid (RA) is involved in multifarious and complex functions necessary for vertebrate development. RA signaling is reliant on strict enzymatic regulation of RA synthesis and metabolism. Improper spatiotemporal expression of RA during development can result in vertebrate axis defects. microRNAs (miRNAs) are also pivotal in orchestrating developmental processes. While mechanistic links between miRNAs and axial development are established, the role of miRNAs in regulating metabolic enzymes responsible for RA abundance during axis formation has yet to be elucidated. Our results uncovered a role of miR-19 family members in controlling RA metabolism through the regulation of CYP26A1 during vertebrate axis formation. Global miRNA expression profiling showed that developmental RA exposure suppressed the expression of miR-19 family members during zebrafish somitogenesis. A reporter assay confirmed that cyp26a1 is a bona fide target of miR-19 in vivo. Transient knockdown of miR-19 phenocopied axis defects caused by RA exposure. Exogenous miR-19 rescued the axis defects induced by RA exposure. Taken together, these results indicate that the teratogenic effects of RA exposure result, in part, from repression of miR-19 expression and subsequent misregulation of cyp26a1. This highlights a previously unidentified role of miR-19 in facilitating vertebrate axis development via regulation of RA signaling.—Franzosa, J. A., Bugel, S. M., Tal, T. L., La Du, J. K., Tilton, S. C., Waters, K. M., Tanguay, R. L. Retinoic acid-dependent regulation of miR-19 expression elicits vertebrate axis defects.
Keywords: microRNAs, somitogenesis, zebrafish
Vitamin a (retinol) is an essential nutrient necessary for normal vertebrate growth and development. Retinoic acid (RA) is an active product of vitamin A synthesis and a critical signaling molecule in morphogenesis. In vertebrates, numerous studies have identified a diverse range of functions of RA signaling, including early axial development and patterning, regional patterning of the central nervous system, regulation of neurogenesis and limb development, and a variety of roles during organogenesis (reviewed in refs. 1–3). Both excesses and deficiencies in RA levels during development result in teratogenic effects (1, 4). Vitamin A deficiency induces congenital malformations in the cardiac, respiratory, ocular, and urogenital systems (5, 6), whereas excessive amounts of vitamin A can cause abnormal morphological development of the central nervous system, skeletal system, liver, and skin (4, 7). It is over 60 years since Sir Edward Mellanby and Dame Honor Fell discovered one of the first known effects of retinol in skeletal tissue (8), yet the molecular mechanisms underlying RA-induced teratogenicity are not fully understood.
The pleiotropic functions of RA signaling require strict control over tissue distribution during development (3). Two important tissues for RA production during embryonic development in vertebrates include the presomatic mesoderm (PSM) or mesenchymal tissue that lines both sides of the neural tube (reviewed in ref. 9). In addition, the somites, which are epithelial spheres of mesoderm generated in a rhythmic pattern from the PSM, give rise to the vertebrae and skeletal muscles in an RA-dependent fashion (9–12). RA signaling is required to orchestrate the directional, periodic, and synchronous segmentation of the PSM into somites through control of somite positioning along the anterior-posterior (A-P) body axis in a “clock and wavefront”-type model (11, 13–15). RA signaling also controls symmetric somite formation along the left-right (LR) body axis through the segmentation clock (10, 12, 16–21). Perturbations of RA signaling during somitogenesis desynchronize somite formation in quail, zebrafish, mice, and chickens (12, 19, 20, 22), mimicking RA-related defects of the human vertebral column (23–25). RA mediates its key role in vertebrate segmentation by antagonizing the traveling fibroblast growth factor (FGF)/WNT signaling gradient that controls the mechanism for somite spacing (26). In addition to mesoderm segmentation, evidence suggests that the RA/FGF mutually negative feedback loop regulates neurogenesis and growth and differentiation of the elongating embryonic axis (reviewed in ref. 1).
Although tight control over RA synthesis is essential during vertebrate segmentation, regulation of RA catabolism is equally important. Cytochrome P450 26 (Cyp26) enzymes, members of the RA cytochrome P450 family, convert all-trans-RA to polar metabolites (4-hydroxy-RA, 4-oxo-RA, 18-hyrdroxy-RA and 5,8-epoxy-RA) that are subject to conjugation and elimination (1, 2, 27). In most vertebrates, the Cyp26 family of enzymes includes Cyp26a1, Cyp26b1, and Cyp26c1, each with its own distinct expression domains during development (reviewed in refs. 2, 28). Targeted manipulation of Cyp26a1 and Cyp26b1 results in abnormalities that mimic the teratogenic effects of exogenous RA exposure (29, 30), suggesting that Cyp26 enzymes function as primary players in RA detoxification. This supports the critical role of these enzymes in regulating the spatiotemporal distribution of RA and preventing inappropriate RA signaling during development (31, 32).
In addition to somite formation, RA has a pivotal function in protecting the paraxial mesoderm from molecular signals directing LR patterning and, thus, maintaining metamery in the PSM (12, 19–21). Disruptions in RA signaling cause asymmetry in somite formation (19). In a seminal paper on the topic, the role of key RA synthesizing and metabolizing enzymes (cyp26a1, cyp26b1, and cyp26c1) in the mechanism that controls lateralization of RA in response to the LR information cascade was investigated. The expression of these enzymes was evaluated after experimental manipulation of the LR signaling network, and no disparate asymmetries in transcript expression patterns were observed (19). Therefore, it was speculated that regulation of RA signaling by the LR information cascade is likely to be post-transcriptional (19).
microRNAs (miRNAs) are critical post-transcriptional regulators of developmental timing in vertebrates (33–35). Mature miRNAs are ∼22-nt-long endogenous noncoding RNAs that bind the 3′ UTR of target mRNAs, resulting in mRNA destabilization and/or translational repression (36–39). Somitogenesis requires precise temporal regulation and is governed in part by miRNAs (40–47). Maternal zygotic Dicer zebrafish mutants that lack the Dicer enzyme necessary to generate mature miRNAs have severe defects in somitogenesis, gastrulation, and heart and brain development (41). These mutants exhibit truncated tails resultant from reduced axis extension (41). Similarly, inactivation of Dicer during mesoderm development in mice elicited reduction in somite size, a reduced A-P axis, and caudalization of somites (47). Numerous studies have revealed the importance of miRNAs in somite derived sclerotome and dermomyotome development in mice (48–50), chicken (51, 52), zebrafish (40, 53, 54), xenopus (55), and segment formation in Drosophila (56). Taken together, these findings suggest miRNAs importance as post-transcriptional regulators of somitogenesis.
Several reports describe a mechanistic link between miRNAs, early embryonic patterning, and somitogenesis. For example, in developing zebrafish, misregulation of miR-92 resulted in aberrant LR patterning (57). miR-In300 targets a gene involved in Wnt signaling, repressing the zebrafish myf5 promoter activity during somite formation (44). Delta-like-1, a ligand of the Notch signaling pathway, an important regulator of cyclic gene expression that dictates rhythmic somite production, is targeted by numerous miRNAs during somite development (43). miRNAs also modulate the expression of Hox cluster genes that participate in the patterning of the A-P axis (42, 45, 58–60). RA also directly regulates the transcription of several Hox genes (61–63), further suggestive of the importance of RA-mediated miRNA regulation of somitogenesis. For example, miR-196 influences axial patterning through putative regulation of Hox genes expression, and it influences pectoral appendage development, specifically through direct regulation of the RA receptor gene, rarb (42). In addition, high-throughput sequencing revealed the presence of novel miRNAs in chicken somite tissue (46). Despite the critical involvement of RA signaling in somite formation and vertebrate symmetry, the role of RA-controlled miRNAs in these developmental processes has yet to be investigated.
In this study we investigated the role of miRNAs in orchestrating teratogenic axis defects elicited by developmental exposure to RA. Our unbiased global miRNA expression profiling revealed that RA exposure dysregulated miR-19 expression during the early stages of zebrafish somitogenesis. Empirical experiments confirmed the role of miR-19 family members in cyp26a1 regulation and disruption of the RA signaling gradient. These findings demonstrate the necessity for miR-19 in post-transcriptional regulation of RA metabolism and define the role for miR-19 in facilitating somitogenesis and normal vertebrate development.
MATERIALS AND METHODS
Fish care and husbandry
All zebrafish (Danio rerio) were reared according to Institutional Animal Care and Use Committee protocols at the Sinnhuber Aquatic Research Laboratory (Oregon State University). The Tropical 5D strain was used for the described experiments. Adults were raised on a recirculating water system (28±1°C) with a 14:10-h light-dark schedule. Spawning and embryo collection procedures were followed as described previously (64).
RA exposure
All-trans-RA (catalog no. 554720; EMD Chemicals, Billerica, MA, USA) was dissolved in DMSO. A range-finding experiment was conducted by batch-exposing embryos (50–75 embryos) to 1–1000 nM RA in buffered embryo medium (100 μl/embryo; ref. 65) in 20-ml glass vials with Teflon-lined lids (VWR International, Radnor, PA, USA). Each vial was treated as a single replicate. Embryos were exposed to RA or embryo medium control at 6 h postfertilization (hpf). The vials were covered with aluminum foil to prevent photodegradation and incubated at 28 ± 1°C. At 48 hpf, the embryos were assessed for mortality and morphological defects. The concentration that elicited no mortality but produced a predominant occurrence of body axis defects was determined to be 5 nM. This concentration was used for the remaining experiments. Depending on experiment, fish were humanely euthanized using MS-222 overdose (65) at various time points (12, 24, 36, 48, or 120 hpf). Subgroups were fixed with 4% paraformaldehyde (J.T. Baker, Phillipsburg, NJ, USA) for in situ hybridization or immunohistochemistry experiments or homogenized in QIAzol Lysis Reagent (Qiagen, Valencia, CA, USA) for gene expression analyses.
miRNA microarray assay and bioinformatics
Total RNA was isolated from pooled tissue that was harvested from 75 embryos batch exposed to either 5 nM RA or embryo medium at 12, 24, 36, and 48 hpf (n=2) using miRNEasy Kits (Qiagen). LC Sciences (Houston, TX, USA) performed the microarray assay on miR Zebrafish V12 Chips that contained probes for all the sequences present in the miRBase Sequence Database (V12) as described previously (66). The data were first background-subtracted and normalized by LOWESS (locally weighted regression; ref. 67); t tests were conducted for differential expression (P<0.05). The array dataset is available through the U.S. National Center for Biotechnology Information Gene Expression Omnibus (68), series accession number GSE449917 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49917). Bioinformatics tools, Bioinformatics Resource Manager (BRM; http://www.sysbio.org/research/bsi/bioanalytics/Bioinformatics.stm) and EMBL-EBI MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), were used to predict putative mRNA targets of miRNA significantly expressed in RA-exposed embryos in comparison to controls.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from whole body homogenate of pooled embryos using miRNEasy Kits (Qiagen), and oligo(dT)-primed cDNA was synthesized using Superscript III (Invitrogen) (n=3–4). qRT-PCR was performed with gene-specific primers for cyp26a1 and β-actin (Eurofins MGW Operon, Ebersburg, Germany) listed in Supplemental Table S2 using the DyNAmo SYBR Green qPCR kit (Finnzymes, Oy, Finland) on the Opticon 2 real-time detection system (MJ Research, St. Bruno, QC, Canada). All samples were normalized to β-actin. To quantify miRNA expression, the miRCURY LNA microRNA PCR System (Exiqon, Woburn, MA, USA) was used. Primer sets for miR-19 (PN 204781) and U6 small nucleolar RNA (snoRNA; control, PN 203907) were used to assess miR-19 expression on a Step One Plus Instrument (Applied Biosystems, Foster City, CA, USA).
Whole-mount in situ hybridization
In situ hybridization was performed as described previously (69) using a digoxygenin-labeled antisense RNA probe to cyp26a1. To synthesize the probe, the cyp26a1 transcript was cloned using qRT-PCR gene-specific primers (Supplemental Table S2; Eurofins MGW Operon), and cDNA was prepared from RNA isolated as described above from whole zebrafish at 24 hpf. Digital images were captured using a Nikon Coolpix E500 digital camera mounted on a Nikon SMZ 1500 stereomicroscope (Nikon Co., Tokyo, Japan).
Immunohistochemistry
Embryos were fixed at 24 hpf in 4% paraformaldehyde overnight at 4°C. Rabbit α-zebrafish Cyp26a1 (dilution: 1:1000, cat. no. 55733; AnaSpec, Fremont, CA, USA) primary antibody and AlexaFluor 555 goat α-rabbit (dilution: 1:1000, Molecular Probes, Eugene, OR, USA) secondary antibody were used. Briefly, fixed embryos were washed with PBST followed by Milli-Q water (Millipore, Billerica, MA, USA) for 1 h. A 10-min collagenase (0.0001 g/ml PBST, C9891; Sigma-Aldrich, St. Louis, MO, USA) treatment was performed to permeabilize the embryos, followed by a 30-min rinse with PBST. The embryos were blocked with 10% normal goat serum (Sigma, G6767) for 1 h at room temperature prior to adding the primary antibody, in which the samples were incubated overnight at 4°C. The following day, all samples were rinsed in PBST for 1.5 h, incubated with secondary antibody for 1.5 h, and washed 2 times for 15 min, followed by 3 times for 30 min with PBST. Embryos were imaged on an inverted Zeiss Axiovert 200 M epifluorescence microscope using a Zeiss Axiocam HRm camera (Carl Zeiss, Oberkochen, Germany).
Microinjections
An antisense morpholino oligonucleotide (MO) designed against both the guide strand and dicer cleavage site of Danio rerio (dre)-miR-19 or a control 3′ fluorescein-tagged MO (Gene Tools, Philomath, OR, USA; Supplemental Table S3) were injected into single-cell-stage embryos. Approximately 2 nl of 1.5 mM morpholino in ultrapure water with 0.5% phenol red was microinjected into each embryo. At 24 hpf, the fish were screened for uniform incorporation of the MO by fluorescein visualization under ultraviolet light. For the reporter assay, a synthetic miR-19 or control Dharmacon miRIDIAN mimic was injected at 10 μM (C-300488-03-0005; Thermo Scientific, Waltham, MA, USA). Approximately 50 pg of wild-type or mutated cyp26a1 3′ UTR GFP mRNA was coinjected with either the synthetic miR-19 or control mimic (10 μM) into the yolk at the single-cell stage (1 nl injection volume). Following microinjection of miR-19 MO or mimic, qRT-PCR was performed to examine miR-19 and cyp26a1 expression at 12 hpf. For the rescue experiment, single-cell-stage embryos were injected with 5, 10, or 20 μM miR-19 or CT mimic and subsequently exposed to RA (5 nM) at 6 hpf. The presence or absence of posterior axis defects was evaluated at 24 hpf.
GFP reporter assay
The zebrafish cyp26a1 3′ UTR sequence was purchased as gBlock gene fragments from Integrated DNA Technologies (Coralville, IA, USA). Two cyp26a1 3′ UTR gBlock fragments were used: a wild-type cyp26a1 3′ UTR containing 2 putative miR-19 binding sites and a mutated cyp26a1 3′ UTR in which the nucleotides predicted to be involved in miR-19 binding were mutated (G↔T and C↔A substitutions). The wild-type and mutated cyp26a1 3′ UTR gBlock fragments were cloned downstream of the GFP open reading frame that was inserted into pCS2+ vector (70). Constructs were purified and sequence verified, and capped mRNA was synthesized using the Ambion mMessage mMachine High Yield Capped RNA Transcript Kit (Austin, TX, USA). To determine whether miR-19 represses translation of GFP, the wild-type and mutated GFP-cyp26a1 3′ UTR transcripts were coinjected into single-celled embryos along with either a control mimic or miR-19 mimic. At 24 hpf, reporter injected animals were placed into 96-well polystyrene microplates and imaged using fluorescent microscopy with Image Xpress Micro (Molecular Devices). Measurements of average fluorescent intensity per well (n=16) were obtained from wells containing 7 embryos/well (112 animals/treatment group).
Statistical analysis
To analyze differences between treatment and control groups, a Student's t test, 1-way ANOVA or 2-way ANOVA with a Dunnett's or Tukey's multiple-comparison post hoc tests, or Fisher's exact test was conducted. Data shown represent means ± se. Values of P ≤ 0.05 were considered statistically significant. Results were calculated using Prism 5.01 (GraphPad, San Diego, CA, USA).
RESULTS
RA exposure misregulates miRNA expression during zebrafish somitogenesis
Exposure to 5 nM RA from 6 to 24 hpf resulted in posterior curved body axis defects in the majority of the larvae at 48 hpf (Fig. 1B) in comparison to control (Fig. 1A). To determine whether misexpression of miRNAs may play a role in observed body axis defects, a miRNA microarray was performed with whole-embryo homogenate collected at 4 time points spanning early zebrafish organogenesis (12, 24, 36, and 48 hpf). Numerous miRNAs were significantly differentially expressed in comparison to vehicle-exposed controls (Fig. 1C and Supplemental Table S1). The greatest number of misexpressed miRNAs was observed at 12 hpf, which falls within the early stages of zebrafish somitogenesis (10–24 hpf). Three members of the zebrafish miR-19 family (miR-19a, miR-19c, and miR-19d) were significantly decreased relative to the control (Fig. 1C, Table 1, and Supplemental Table S1). In addition to the miR-19 family members, the expression of miR-22a was decreased 1.5-fold in RA-exposed embryos (Table 1 and Supplemental Table S1). Validation of the miRNA microarray results by qRT-PCR confirmed repression of miR-19 expression by RA at 12 hpf (Fig. 1D).
Figure 1.
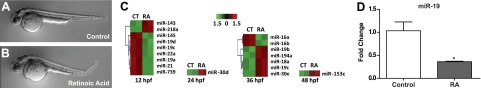
Developmental RA exposure results in axial defects and suppression of miR-19 expression. Embryos were exposed to 5 nM RA from 6 to 24 hpf. A, B) Representative images of control (A) and RA-exposed (B) animals at 48 hpf. C) Results of miRNA microarray analysis performed on samples obtained at 12, 24, 36, or 48 hpf from embryos batch-exposed to 5 nM RA from 6 to 24 hpf (n=2). Heat map represents bidirectional hierarchical clustering of significant miRNA genes (Data represent fold change relative to control-exposed embryos, n=2, P<0.05, 5% FDR, t test.) miR-739 was removed from the miRBase database due to its prediction as a fragment of rRNA. D) miR-19 expression levels were measured by qRT-PCR at 12 hpf in pools of 25 embryos exposed to 5 nM RA. (Values represent fold change compared to unexposed control embryos, means±se, n=3.) *P < 0.05, independent samples t test.
Table 1.
miRNA microarray results at 12 hpf
| miRNA | P | Log2 |
|---|---|---|
| dre-miR-22a | 2.52E-02 | −1.98 |
| dre-miR-19c | 2.95E-02 | −2.40 |
| dre-miR-21 | 3.05E-02 | −0.52 |
| dre-miR-19a | 4.64E-02 | −1.71 |
| dre-miR-19d | 4.78E-02 | −2.74 |
| dre-miR-145 | 3.13E-02 | −0.55 |
| dre-miR-143 | 4.50E-02 | 0.55 |
| dre-miR-218a | 6.06E-03 | 0.55 |
RA exposure disrupts Cyp26a1 expression at the onset of somitogenesis
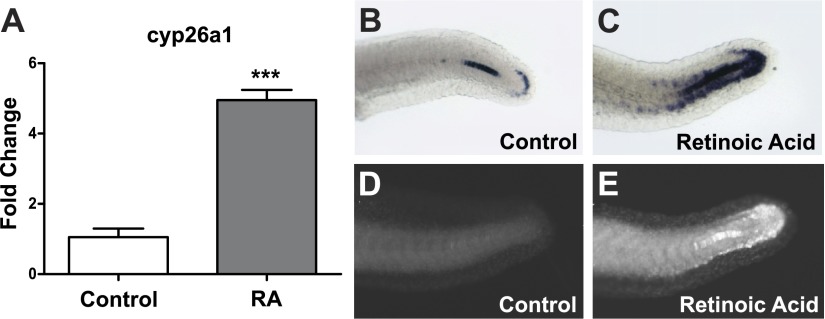
The posterior body axis curvature elicited by RA exposure is consistent with the asymmetrical patterning and irregular somite sizes that result during misregulation of somitogenesis (12, 19, 20). Bioinformatics analysis was conducted with the miRNA tools incorporated in BRM (71) to uncover potential targets that are both related to RA signaling and somitogenesis, and targeted by misexpressed miRNA at 12 hpf. The 3′ UTR of zebrafish cyp26a1, the primary enzyme responsible for converting active RA into its inactive polar metabolites, is predicted by EMBL-EBI MicroCosm Targets to be targeted by miR-19a, miR-19c, and miR-19d, miRNAs that were significantly repressed by RA exposure at 12 hpf (Table 1). Of all miRNAs examined following transient developmental RA exposure, these miR-19 family members exhibited the largest magnitude decreased expression at 12 hpf in comparison to control (Table 1). In contrast, a fourth miR-19 family member, miR-19b, was not predicted to target the cyp26a1 3′ UTR nor was it significantly down-regulated at 12 hpf. Consistent with the concept that miR-19 family members post-transcriptionally regulate cyp26a1, there was a concomitant increase in cyp26a1 transcript expression relative to controls at 12 hpf in embryos exposed to RA (Fig. 2A). To investigate the spatial distribution of cyp26a1 transcript expression during the later stages of somitogenesis, whole-mount in situ hybridization analysis was employed at 24 hpf and indicated that in the majority of embryos there was an expansion of cyp26a1 transcript expression in the dorsal tip of the tailbud and along the ventral side of the tail of RA-exposed embryos compared to controls (Fig. 2B, C). Immunohistochemical analysis showed that RA-induced CYP26A1 protein expression at 24 hpf is also increased around the dorsal tip of the tailbud in comparison to controls (Fig. 2D, E).
Figure 2.
Transient developmental exposure to RA disrupts cyp26a1 expression during somitogenesis. A) Embryos were exposed to 5 nM RA from 6 to 12 hpf, and RNA was collected at 12 hpf. cyp26a1 expression levels at 12 hpf in embryos exposed to 5 nM RA. (Data represent fold change relative to control exposed embryos, means±se, n=3.) ***P < 0.001, independent samples t test. B–E) Representative images at 24 hpf showing measurements of cyp26a1 transcript detected by in situ hybridization and protein levels detected by IHC in controls (B, D) as compared to RA-exposed embryos (C, E).
Transient knockdown of miR-19 recapitulates axis defects induced by RA exposure
To determine whether decreased miR-19 was sufficient to produce the developmental axis defect shown in Fig. 1A, a single MO was designed to knock down miR-19a, miR-19-c, and miR-19-d during development (Fig. 3A). A majority of the embryos injected with the miR-19 MO exhibited the posterior curved body axis defect observed in embryos exposed to RA (Fig. 3B, C). qRT-PCR analysis was used to confirm miR-19 knockdown (Fig. 3D) and demonstrated that miR-19 repression causes increased cyp26a1 expression at 12 hpf (Fig. 3E). Although not as robust as observed on RA exposure, miR-19 repression caused expansion of cyp26a1 transcript expression along the dorsal end of the tailbud tip (Fig. 3F, G). CYP26A1 protein expression (Supplemental Fig. S1) was increased in dorsal tip of the tailbud and portions of the notochord in miR-19 MO injected embryos relative to controls at 24 hpf.
Figure 3.
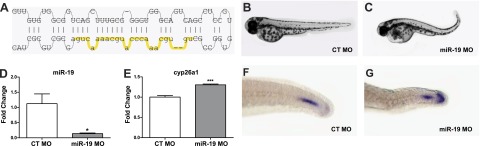
miR-19 disrupts axial patterning and development through post-transcriptional regulation of cyp26a1. A) Schematic of the miR-19 MO target site on the pre-miR-19d sequence that was injected into single-cell-stage embryos. B, C) Representative images of the axis defects elicited by miR-19 MO in 48 hpf embryos (C) in comparison to control (CT) MO-injected embryos (B). D) Effective knockdown of miR-19 transcript levels confirmed in miR-19 MO-injected embryos at 12 hpf via qRT-PCR. E) cyp26a1 transcript levels measured by qRT-PCR at 12 hpf in miR-19 and control morphants. (Values reflect fold change relative to control MO injected embryos, means±se, n=4.) *P < 0.05, ***P < 0.001, independent samples t test. G, F) Representative images of cyp26a1 transcript and protein expression at 24 hpf in controls (G) relative to miR-19 morphants (F), as demonstrated by in situ hybridization.
miR-19 targets the 3′ UTR of cyp26a1 in vivo
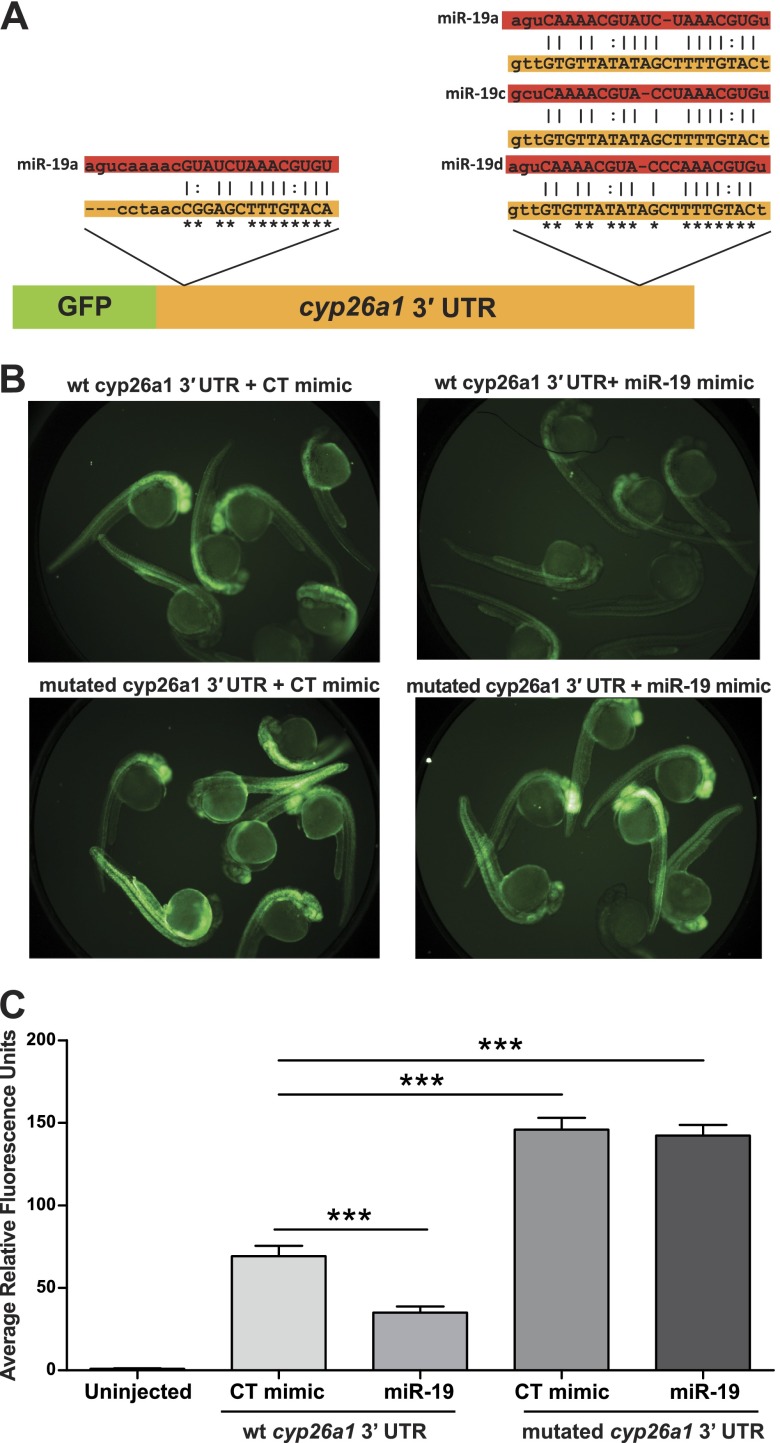
To confirm that cyp26a1 is targeted by miR-19, an mRNA reporter was designed that contained the 3′ UTR of cyp26a1 inserted downstream of GFP (GFP-cyp26a1-3′ UTR, Fig. 4A). Two reporters were used: a wild-type cyp26a1 3′ UTR sequence containing 2 putative miR-19 miRNA recognition elements (MREs), and a mutated cyp26a1 3′ UTR where the miR-19 binding sites were mutated using G↔T and C↔A substitutions. Each reporter (wild-type or mutated 3′UTR) was coinjected as purified mRNA into single-cell embryos with either a miR-19 mimic or a control mimic, and GFP expression was quantified at 24 hpf. Coinjection of the GFP-cyp26a1-3′ UTR reporter (wild-type) and miR-19 mimic produced a significant knockdown in GFP expression relative to animals coinjected with the control mimic (Fig. 4B, C). In comparison, animals injected with the GFP-cyp26a1-3′ UTR reporter containing mutated miR-19 binding had significant fluorescence that was not a decreased when coinjected with the miR-19 mimic (Fig. 4B, C).
Figure 4.
Physiological mRNA reporter assay confirms that cyp26a1 is a bona fide target of miR-19 in vivo. A) Simplified schematic of cyp26a1 3′ UTR reporter assay along with predicted miR-19a, miR-19c, and miR-19d MREs. Single-cell-stage embryos were injected with either a wild-type GFP-cyp26a1 3′ UTR mRNA reporter or a mutated GFP-cyp26a1 3′ UTR mRNA reporter in which the miR-19 binding sites have been mutated with G↔T and C↔A substitutions (asterisks). Each mRNA reporter was coinjected with either a miR-19 mimic or control (CT) mimic. B, C) Representative images of each group at 24 hpf (B) and the quantification of fluorescence (C). (Significance represents coinjected GFP-cyp26a1 3′ UTR reporter and miR-19 mimic in comparison to reporter and control mimic or reporter and mimic coinjected animals in comparison to uninjected controls. Data are reported as fluorescence units relative to uninjected controls, means±se, n=16 well, 7 embryos/well.) ***P < 0.001, 2-way ANOVA with Tukey's multiple comparisons tests.
Exogenous miR-19 represses cyp26a1 and rescues RA-induced axial defects
To determine whether rescue of miR-19 repression is sufficient to prevent axial defects induced by RA exposure, embryos were injected with an exogenous miR-19 mimic and then exposed to RA from 6 to 24 hpf. Injection of synthetic miR-19 mimic significantly increased the level of miR-19 transcript present (Fig. 5A) and decreased cyp26a1 transcript expression (Fig. 5B) at 12 hpf. In situ hybridization revealed that miR-19-mimic embryos had reduced cyp26a1 transcript expression in the tailbud region during the later stages of somitogenesis (24 hpf) as compared to the control mimic injected embryos (Supplemental Fig. S2). Injection of exogenous miR-19 rescued the RA-induced body axis defects in ∼25% of the embryos treated with 5 μM RA (Fig. 5C–E).
Figure 5.
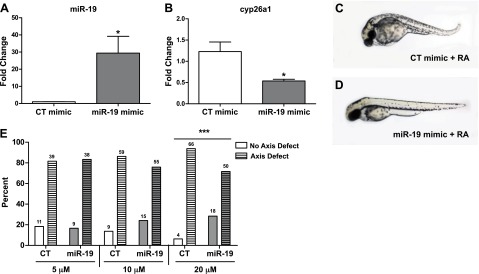
Exogenous miR-19 reduces cyp26a1 expression and rescues the body axis defects elicited by RA exposure. A) miR-19 transcript expression levels at 12 hpf in pools of embryos injected with miR-19 or control mimics. (Values represent fold change compared to control mimic injected embryos, means±se, n=3.) *P < 0.05, independent samples t test. B) cyp26a1 expression levels measured by qRT-PCR in 12 hpf embryos injected with exogenous miR-19 mimic. (Data represent fold change relative to control exposed embryos, means±se, n=3). *P < 0.05, independent samples t test. C, D) Embryos injected with control mimic (C) or miR-19 mimic (D) at 5, 10, and 20 μM were exposed to RA from 6 to 24 hpf. Representative images of CT and miR-19 mimic (20 μM) show that miR-19 rescues RA-induced body axis defects. E) Percentage of animals that either exhibited posterior axis defects (axis defect) or for which the defect was rescued (no axis defect) following RA exposure in embryos injected with 5, 10, or 20 μM CT or miR-19 mimic. (N values represent the sum of animals from 2 independent experiments.) ***P < 0.001, Fisher's exact test.
DISCUSSION
Perturbations in developmental RA signaling result in teratogenic effects (reviewed in refs. 1, 3) and miRNAs are known to absolve transcriptional programs from previous developmental stages (reviewed in refs. 72–74). Therefore, we sought to determine whether dysregulation of miRNA signaling resulted in the hallmark curved body axis phenotype triggered by RA exposure. Studies suggest that the developmental role of RA is strongly related to the enzymatic regulation of its synthesis and tissue-specific metabolism (reviewed in ref. 1). Therefore, understanding how miRNAs might regulate enzymes responsible for RA abundance that, in turn, affect the expression of RA-responsive genes is an important and unexplored area. During somitogenesis, CYP26A1 is necessary for maintaining homeostasis of RA activity in the PSM (reviewed in refs. 1, 3). The importance of CYP26A1 in vertebrate axis formation is well established (29, 75). Our experimental results uncovered a role for several miR-19 family members in controlling metabolism of RA, specifically through the regulation of CYP26A1 expression during the early stages of zebrafish somitogenesis.
Our data indicate that developmental exposure to RA represses the expression of miR-19 family members (miR-19a, miR-19c, and miR-19d) during early somitogenesis (12 hpf) but not at 24 hpf (Fig. 1C; and Supplemental Table S1). This is consistent with previous findings that RA is necessary only during the early stages of somitogenesis (somites 1–6 in mice) to assure proper and synchronous development of the remaining somites (7, 76). We showed that the 3′ UTR of cyp26a1 is a bona fide target of miR-19 using in vivo reporter analysis (Fig. 4A–C). Coexpressed miRNAs are known to act cooperatively to regulate the 3′ UTR of a common mRNA (77), supporting the concept that RA-induced repression of miR-19 family members likely acts as a compensatory mechanism to increase the expression of one of the molecule's main detoxifying enzymes. In addition, repression of miR-19 in vivo largely recapitulated the distinct posterior curved body axis morphology (Fig. 3C) and exogenous miR-19 rescued body axis defects associated with RA exposure (Fig. 5D). These findings demonstrate a direct post-transcriptional miRNA interaction with an enzyme indispensable for RA metabolism and its consequences on somitogenesis and, more broadly, the regulation of vertebrate axis formation.
The morphological defects exhibited by zebrafish developmentally exposed to RA are similar to those demonstrated by both transient and conditional knockdown of CYP26A1 and retinal dehydrogenase 2 (RALDH2) in diverse species (29, 78). CYP26A1-null mutants display morphological defects that are strikingly similar to those exhibited by exposure to excess RA, including spina bifida and posterior truncation of the tail and lumbosacral region (29, 79). Similar to the morphological abnormalities we documented in embryonic zebrafish following developmental exposure to RA, CYP26A1-null embryos also presented with abnormally curved notochords (29). Lack of CYP26A1 expression in the tailbud prevents the clearance of RA from the rostrally adjacent mesoderm (29). Adu-Abed et al. (29) hypothesized that the resulting dearth of CYP26A1 creates a state of RA “endogenous” teratogenicity that recapitulates the effects of excess exogenous RA exposure. RALDH2−/− mutants that are deficient in endogenous RA also displayed shortened body axes and abnormally small somites that can be rescued by maternal administration of RA (78). In this study, exposure to RA suppressed miR-19 expression. Transient knockdown of miR-19 partially mimicked RA-induced body axis defects and resulted in increased cyp26a1 expression localized to the dorsal end of the tailbud (Fig. 3B–G). Repression of miR-19 did not elicit as robust of an increase in cyp26a1 expression as observed on exposure to RA. The cyp26a1 3′ UTR is predicted (http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/) to have 10 MREs and be targeted by 17 or more miRNAs. Therefore, it is highly probable that other miRNAs are contributing to the observed expression pattern in the tailbud. Although many miRNA knockouts fail to confer a gross phenotype (reviewed in ref. 80), our knockdown of all the miR-19 seed family members predicted to target cyp26a1 resulted in observable morphological defects in the whole animal. Elevated levels of miR-19 also reduced cyp26a1 expression (Fig. 5A–D). In addition, in our rescue experiment, the body axis curvature caused by exogenous RA was partially rescued by artificially increased miR-19 levels and subsequent suppression of CYP26A1 expression (Fig. 5E). Hence, we postulate a negative feedback loop in which exogenous RA exposure during development down-regulates miRNAs that negatively regulate CYP26A1, leading to increased CYP26A1 expression and decreases in endogenous RA expression. This builds on a recent article that purposed a paradoxical mechanism for RA teratogenicity by demonstrating that adverse effects observed by RA exposure are a result of local RA deficiency (81). Thus, our findings place post-transcriptional regulation of cyp26a1 by a conserved class of miRNAs at the nexus of RA-induced teratogenicity.
miRNA regulation of multiple genes in a shared pathway strengthens their effect (reviewed in ref. 80). During somitogenesis, opposing RA and FGF pathways act antagonistically in the elongating body axis (26). Our miRNA array data also indicated that at 12 hpf, early developmental exposure caused a significant decrease in miR-22a (Table 1), the only miRNA predicted by EMBL-EBI MicroCosm Targets to target the 3′ UTR of fgf8, suggesting that FGF signaling may also be controlled by RA-sensitive miRNAs during early somitogenesis. RA reportedly attenuates FGF8 levels by transcriptional repression or mRNA decay (11). In addition, FGF8 represses the onset of RALDH2 expression in the PSM and down-regulation of FGF8 is necessary for normal axis formation (26, 82). Fgf8 transcript expression is nonexistent in CYP26A1-deficient mice and is shown to have increased expression in response to decreased RA signaling molecules (26, 29). Changes in FGF8 expression in RA-deficient embryos result in aberrations in the determination wavefront that controls somite size, further demonstrating the mutual inhibitory role of RA and FGF signaling in axis development (1, 11, 20, 26, 76). Our data raise the possibility that RA-sensitive miRNAs that target FGF signaling might also play a role in RA toxicity. Much remains to be learned about the post-transcriptional regulation of RA and FGF interactions during vertebrate segmentation.
Beyond cyp26a1, our findings have broader implications to altered temporal-spatial expression of genes transcriptionally regulated by RA or induced by FGF expression. These targets include members of the Hox gene family that regulate axis patterning, specification, and LR symmetry (82). RA acts as a transcriptional activator for several Hox genes and Notch signaling (59, 61–63). In addition, FGF signaling is functionally associated with Hox gene expression and axis patterning (reviewed in ref. 83). Several studies have explored the role of miRNAs conserved within vertebrate Hox clusters in regulating Hox gene expression and subsequently affecting axial development and patterning (42, 45, 58, 60). Furthermore, specific Hox gene expression changes can be partially modulated by miR-196 regulation of the retinoic acid receptor gene, rarb (42). This supports our findings on the role of miRNAs in controlling RA abundance during morphogenesis and further confirms the necessity of miRNAs in the precise regulation of RA signaling during vertebrate axis development.
In addition to the role of miR-19 in RA metabolism, our finding that miR-19 family members regulate vertebrate axis formation provides compelling insight into their evolutionary significance. The miR-19 family has no known homologs in invertebrates (84, 85). The introduction of miRNA families correlates with drastic increases in morphological complexity (85–87). This is attributed to miRNAs role in stabilizing gene expression, and, in turn, rendering phenotypic traits influenced by miRNA-targets more likely to evolve (85, 88). It is hypothesized that the continuous addition of novel miRNAs to metazoan genomes is influential in the canalization, or evolved robustness, of a trait and contributes to the production of distinct developmental outcomes (85, 88). This in conjunction with our experimental data suggests the evolution and necessity for miR-19 in vertebrate identity. Taken together, the findings presented stress the evolutionary significance of miR-19 in somitogenesis, a developmental phenomenon common to all vertebrates.
Beyond its importance as miRNA family acquired by vertebrates, miR-19 is a member of the evolutionarily conserved miR-17–92 cluster (reviewed in ref. 89). In zebrafish, the miR-19 family members are located in 3 conserved clusters miR-17–92, miR-106a-363, and miR-106b-2 (Supplemental Table S4). The high conservation of these clusters underscores their functional relevance, while the unique sequences of the mature miRNAs direct their interactions with target mRNAs and ultimately govern their function (90). The miR-17–92 cluster encodes the miRNA genes miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a1 in humans, mice, zebrafish, and >25 other known vertebrate species (miRBase). To date, miR-19c and miR-19d miRNAs have been identified only in lamprey and medaka, respectively, in addition to zebrafish (miRBase). This is likely due to the genome duplication events leading to teleost fish (88, 91, 92). The emergence of miR-19 in the miR-17–92 cluster and its paralogs supports the speculation that tandem duplication events play a pivotal evolutionary role in the emergence of novel miRNA (84, 88). In addition, the origin of novel miRNAs in preestablished clusters eliminates the necessity for the evolution of separate promoters for the new miRNAs (88).
An elegant study demonstrated an essential function of miR-17–92 for normal development in mice (93). The results also demonstrated a functional cooperation between miR-17–92 and miR-106b-25 clusters. In addition, it was speculated that among cluster members, it was the loss of miR-19 family members and miR-18 that were responsible for the observed developmental phenotypes in mice (93). The teratogenic effects we observed due to developmental RA exposure were notably accompanied by a decrease in the expression of 3 miR-19 family members, miR-19a, miR-19c, miR-19d, each located in one of the 3 miR-17–92 paralog clusters. This also suggests a plausible functional synergy between the clusters dictating the observed developmental axis defects.
The only miR-19 family member that was not misregulated by developmental RA exposure in this study was miR-19b, which is located in the miR-17–92 cluster with miR-19a (Table 1 and Supplmental Table S4). Previous research highlighted differences in miR-19a and miR-19b expression, suggesting their functional divergences even though they evolved from the same cluster (94). Structural analysis of the miR-17–92 cluster in humans suggests that efficient enzymatic (Drosha) processing of miR-19b is sterically hindered (95). A similar structural limitation in the zebrafish miR-17–92 cluster may explain why miR-19b expression was not affected by RA exposure in stark contrast to its homologue, miR-19a.
In support of our findings implicating miR-17–92 cluster members in vertebrate axis formation, a germline deletion of the miR-17–92 cluster was found to elicit skeletal abnormalities in humans (96). This was the first report of a miRNA mutation causing a hereditary condition responsible for developmental defects in humans. While the cluster regulates TGF-β and sonic hedgehog signaling, two pathways influential in skeletal development, the authors state that other targets of the miR-17–92 cluster that control skeletal growth and patterning likely exist (96). Our data expand on these findings to describe a mechanism through which specific miR-17–92 cluster members controls the proper development of somites, precursors to skeletal formation.
Together, this study highlights a role for miR-19 in facilitating normal vertebrate development by serving as a RA-sensitive switch to promote CYP26A1-mediated RA turnover during somitogenesis. Our findings suggest a previously undescribed miRNA-driven compensatory mechanism initiated to increase the expression of CYP26A1 during a period in which spatial maintenance of endogenous RA abundance is critical. Furthermore, these data underscore the evolutionary significance and plausible relevance of miR-19 as a vertebrate innovation.
Supplementary Material
Acknowledgments
The authors thank Leah Wehmas for her experimental and technical assistance. The authors are grateful to Cari Buchner, Carrie Barton, and the staff at the Sinnhuber Aquatic Research Laboratory (Oregon State University) for exemplary fish husbandry and technical expertise. The authors thank Dr. James Patton (Vanderbilt University, Nashville, TN, USA) for sharing the pCS2-GFP vector. The authors appreciate the critical comments on the manuscript provided by Britton Goodale, Katerine Saili, Michael Simonich, and R.L.T. laboratory members.
This work was supported by U.S. National Institute of Environmental Health Sciences (NIEHS) Environmental Health Sciences Core Center grant P30 ES000210, NIEHS Training grant T32 ES007060, and an Oregon State University Linus Pauling Institute grant to R.L.T., and by NIEHS Superfund Basic Research Program grant P42 ES016465 to R.L.T. and K.M.W.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflicts of interest. J.A.F., S.M.B., T.L.T., and R.L.T designed the research; J.A.F., S.M.B., J.K.L., and T.L.T. performed the research; S.C.T. and K.M.W. contributed analytic tools; J.A.F., S.M.B., T.L.T., S.C.T., K.M.W., and R.L.T. analyzed data; and J.A.F. and R.L.T. wrote the paper.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- A-P
- anterior-posterior
- BRM
- Bioinformatics Resource Manager
- Cyp26
- cytochrome P450 26
- dre
- Danio rerio
- FGF
- fibroblast growth factor
- hpf
- hours postfertilization
- LR
- left-right
- miR
- microRNA
- miRNA
- microRNA
- MO
- morpholino oligonucleotide
- MRE
- microRNA recognition element
- PSM
- presomatic mesoderm
- RA
- retinoic acid
- RALDH2
- retinal dehydrogenase 2
- snoRNA
- small nucleolar RNA
REFERENCES
- 1. Niederreither K., Dolle P. (2008) Retinoic acid in development: towards an integrated view. Nat. Rev. Genet. 9, 541–553 [DOI] [PubMed] [Google Scholar]
- 2. Pennimpede T., Cameron D. A., MacLean G. A., Li H., Abu-Abed S., Petkovich M. (2010) The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth Defects Res. A Clin. Mol. Teratol. 88, 883–894 [DOI] [PubMed] [Google Scholar]
- 3. Rhinn M., Dolle P. (2012) Retinoic acid signalling during development. Development 139, 843–858 [DOI] [PubMed] [Google Scholar]
- 4. Collins M. D., Mao G. E. (1999) Teratology of retinoids. Annu. Rev. Pharmacol. Toxicol. 39, 399–430 [DOI] [PubMed] [Google Scholar]
- 5. Mark M., Ghyselinck N. B., Chambon P. (2006) Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 46, 451–480 [DOI] [PubMed] [Google Scholar]
- 6. Wilson J. G., Roth C. B., Warkany J. (1953) An analysis of the syndrome of malformations induced by maternal vitamin A deficiency. Effects of restoration of vitamin A at various times during gestation. Am. J. Anat. 92, 189–217 [DOI] [PubMed] [Google Scholar]
- 7. Duester G. (2007) Retinoic acid regulation of the somitogenesis clock. Birth Defects Res. C 81, 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fell H. B., Mellanby E. (1952) The effect of hypervitaminosis A on embryonic limb bones cultivated in vitro. J. Physiol. 116, 320–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dequeant M. L., Pourquie O. (2008) Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9, 370–382 [DOI] [PubMed] [Google Scholar]
- 10. Campo-Paysaa F., Marletaz F., Laudet V., Schubert M. (2008) Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis 46, 640–656 [DOI] [PubMed] [Google Scholar]
- 11. Dubrulle J., Pourquie O. (2004) Coupling segmentation to axis formation. Development 131, 5783–5793 [DOI] [PubMed] [Google Scholar]
- 12. Vermot J., Pourquie O. (2005) Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature 435, 215–220 [DOI] [PubMed] [Google Scholar]
- 13. Cooke J. (1975) Control of somite number during morphogenesis of a vertebrate, Xenopus laevis. Nature 254, 196–199 [DOI] [PubMed] [Google Scholar]
- 14. Saga Y., Takeda H. (2001) The making of the somite: molecular events in vertebrate segmentation. Nat. Rev. Genet. 2, 835–845 [DOI] [PubMed] [Google Scholar]
- 15. Aulehla A., Herrmann B. G. (2004) Segmentation in vertebrates: clock and gradient finally joined. Genes Dev. 18, 2060–2067 [DOI] [PubMed] [Google Scholar]
- 16. Hamada H., Meno C., Watanabe D., Saijoh Y. (2002) Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3, 103–113 [DOI] [PubMed] [Google Scholar]
- 17. Levin M. (2005) Left-right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 122, 3–25 [DOI] [PubMed] [Google Scholar]
- 18. Raya A., Izpisua Belmonte J. C. (2004) Sequential transfer of left-right information during vertebrate embryo development. Curr. Opin. Genet. Dev. 14, 575–581 [DOI] [PubMed] [Google Scholar]
- 19. Kawakami Y., Raya A., Raya R. M., Rodriguez-Esteban C., Belmonte J. C. (2005) Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature 435, 165–171 [DOI] [PubMed] [Google Scholar]
- 20. Vermot J., Gallego Llamas J., Fraulob V., Niederreither K., Chambon P., Dolle P. (2005) Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science 308, 563–566 [DOI] [PubMed] [Google Scholar]
- 21. Brend T., Holley S. A. (2009) Balancing segmentation and laterality during vertebrate development. Semin. Cell Dev. Biol. 20, 472–478 [DOI] [PubMed] [Google Scholar]
- 22. Halilagic A., Ribes V., Ghyselinck N. B., Zile M. H., Dolle P., Studer M. (2007) Retinoids control anterior and dorsal properties in the developing forebrain. Dev. Biol. 303, 362–375 [DOI] [PubMed] [Google Scholar]
- 23. Eckalbar W. L., Fisher R. E., Rawls A., Kusumi K. (2012) Scoliosis and segmentation defects of the vertebrae. WIREs Dev. Biol. 1, 401–423 [DOI] [PubMed] [Google Scholar]
- 24. Giampietro P. F., Dunwoodie S. L., Kusumi K., Pourquie O., Tassy O., Offiah A. C., Cornier A. S., Alman B. A., Blank R. D., Raggio C. L., Glurich I., Turnpenny P. D. (2009) Progress in the understanding of the genetic etiology of vertebral segmentation disorders in humans. Ann. N. Y. Acad. Sci. 1151, 38–67 [DOI] [PubMed] [Google Scholar]
- 25. Pourquie O. (2011) Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003) Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 65–79 [DOI] [PubMed] [Google Scholar]
- 27. Emoto Y., Wada H., Okamoto H., Kudo A., Imai Y. (2005) Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev. Biol. 278, 415–427 [DOI] [PubMed] [Google Scholar]
- 28. Thatcher J. E., Isoherranen N. (2009) The role of CYP26 enzymes in retinoic acid clearance. Expert Opin. Drug Metab. Toxicol. 5, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abu-Abed S., Dolle P., Metzger D., Beckett B., Chambon P., Petkovich M. (2001) The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 15, 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hernandez R. E., Putzke A. P., Myers J. P., Margaretha L., Moens C. B. (2007) Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu P., Tian M., Bao J., Xing G., Gu X., Gao X., Linney E., Zhao Q. (2008) Retinoid regulation of the zebrafish cyp26a1 promoter. Dev. Dyn. 237, 3798–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spoorendonk K. M., Peterson-Maduro J., Renn J., Trowe T., Kranenbarg S., Winkler C., Schulte-Merker S. (2008) Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development 135, 3765–3774 [DOI] [PubMed] [Google Scholar]
- 33. Stefani G., Slack F. J. (2008) Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 [DOI] [PubMed] [Google Scholar]
- 34. Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., Plasterk R. H. (2005) MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 [DOI] [PubMed] [Google Scholar]
- 35. Darnell D. K., Kaur S., Stanislaw S., Konieczka J. H., Yatskievych T. A., Antin P. B. (2006) MicroRNA expression during chick embryo development. Dev. Dyn. 235, 3156–3165 [DOI] [PubMed] [Google Scholar]
- 36. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 37. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 38. He L., Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 39. Kim V. N., Han J., Siomi M. C. (2009) Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 40. Flynt A. S., Li N., Thatcher E. J., Solnica-Krezel L., Patton J. G. (2007) Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39, 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P., Schier A. F. (2005) MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838 [DOI] [PubMed] [Google Scholar]
- 42. He X., Yan Y. L., Eberhart J. K., Herpin A., Wagner T. U., Schartl M., Postlethwait J. H. (2011) miR-196 regulates axial patterning and pectoral appendage initiation. Dev. Biol. 357, 463–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoesel B., Bhujabal Z., Przemeck G. K., Kurz-Drexler A., Weisenhorn D. M., Angelis M. H., Beckers J. (2010) Combination of in silico and in situ hybridisation approaches to identify potential Dll1 associated miRNAs during mouse embryogenesis. Gene Expr. Patterns 10, 265–273 [DOI] [PubMed] [Google Scholar]
- 44. Hsu R. J., Lin C. Y., Hoi H. S., Zheng S. K., Lin C. C., Tsai H. J. (2010) Novel intronic microRNA represses zebrafish myf5 promoter activity through silencing dickkopf-3 gene. Nucleic Acids Res. 38, 4384–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McGlinn E., Yekta S., Mansfield J. H., Soutschek J., Bartel D. P., Tabin C. J. (2009) In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc. Natl. Acad. Sci. U. S. A. 106, 18610–18615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rathjen T., Pais H., Sweetman D., Moulton V., Munsterberg A., Dalmay T. (2009) High throughput sequencing of microRNAs in chicken somites. FEBS Lett. 583, 1422–1426 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Z., O'Rourke J. R., McManus M. T., Lewandoski M., Harfe B. D., Sun X. (2011) The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning. Dev. Biol. 351, 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Rourke J. R., Georges S. A., Seay H. R., Tapscott S. J., McManus M. T., Goldhamer D. J., Swanson M. S., Harfe B. D. (2007) Essential role for Dicer during skeletal muscle development. Dev. Biol. 311, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu N., Williams A. H., Kim Y., McAnally J., Bezprozvannaya S., Sutherland L. B., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. U. S. A. 104, 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Crist C. G., Montarras D., Pallafacchina G., Rocancourt D., Cumano A., Conway S. J., Buckingham M. (2009) Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. U. S. A. 106, 13383–13387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sweetman D., Rathjen T., Jefferson M., Wheeler G., Smith T. G., Wheeler G. N., Munsterberg A., Dalmay T. (2006) FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev. Dyn. 235, 2185–2191 [DOI] [PubMed] [Google Scholar]
- 52. Goljanek-Whysall K., Sweetman D., Abu-Elmagd M., Chapnik E., Dalmay T., Hornstein E., Munsterberg A. (2011) MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. Proc. Natl. Acad. Sci. U. S. A. 108, 11936–11941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mishima Y., Abreu-Goodger C., Staton A. A., Stahlhut C., Shou C., Cheng C., Gerstein M., Enright A. J., Giraldez A. J. (2009) Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 23, 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shkumatava A., Stark A., Sive H., Bartel D. P. (2009) Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 23, 466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sokol N. S., Ambros V. (2005) Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li N., Wei C., Olena A. F., Patton J. G. (2011) Regulation of endoderm formation and left-right asymmetry by miR-92 during early zebrafish development. Development 138, 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hornstein E., Mansfield J. H., Yekta S., Hu J. K., Harfe B. D., McManus M. T., Baskerville S., Bartel D. P., Tabin C. J. (2005) The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature 438, 671–674 [DOI] [PubMed] [Google Scholar]
- 59. Yekta S., Tabin C. J., Bartel D. P. (2008) MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat. Rev. Genet. 9, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Woltering J. M., Durston A. J. (2008) MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE 3, e1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lohnes D., Mark M., Mendelsohn C., Dolle P., Dierich A., Gorry P., Gansmuller A., Chambon P. (1994) Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development 120, 2723–2748 [DOI] [PubMed] [Google Scholar]
- 62. Cordes R., Schuster-Gossler K., Serth K., Gossler A. (2004) Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development 131, 1221–1233 [DOI] [PubMed] [Google Scholar]
- 63. Zakany J., Kmita M., Alarcon P., de la Pompa J. L., Duboule D. (2001) Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 106, 207–217 [DOI] [PubMed] [Google Scholar]
- 64. Mathew L. K., Sengupta S., Kawakami A., Andreasen E. A., Lohr C. V., Loynes C. A., Renshaw S. A., Peterson R. T., Tanguay R. L. (2007) Unraveling tissue regeneration pathways using chemical genetics. J. Biol. Chem. 282, 35202–35210 [DOI] [PubMed] [Google Scholar]
- 65. Westerfield M. (2000) The Zebrafish Book, University of Oregon Press, Eugene, OR, USA [Google Scholar]
- 66. Tal T. L., Franzosa J. A., Tilton S. C., Philbrick K. A., Iwaniec U. T., Turner R. T., Waters K. M., Tanguay R. L. (2012) MicroRNAs control neurobehavioral development and function in zebrafish. FASEB J. 26, 1452–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 68. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Thisse C., Thisse B. (2008) High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 70. Rupp R. A., Snider L., Weintraub H. (1994) Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8, 1311–1323 [DOI] [PubMed] [Google Scholar]
- 71. Shah A. R., Singhal M., Klicker K. R., Stephan E. G., Wiley H. S., Waters K. M. (2007) Enabling high-throughput data management for systems biology: the Bioinformatics Resource Manager. Bioinformatics 23, 906–909 [DOI] [PubMed] [Google Scholar]
- 72. Alvarez-Garcia I., Miska E. A. (2005) MicroRNA functions in animal development and human disease. Development 132, 4653–4662 [DOI] [PubMed] [Google Scholar]
- 73. Carrington J. C., Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301, 336–338 [DOI] [PubMed] [Google Scholar]
- 74. Wienholds E., Plasterk R. H. (2005) MicroRNA function in animal development. FEBS Lett. 579, 5911–5922 [DOI] [PubMed] [Google Scholar]
- 75. Sakai Y., Meno C., Fujii H., Nishino J., Shiratori H., Saijoh Y., Rossant J., Hamada H. (2001) The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 15, 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sirbu I. O., Duester G. (2006) Retinoic-acid signalling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nat. Cell Biol. 8, 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krek A., Grun D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 78. Niederreither K., Subbarayan V., Dolle P., Chambon P. (1999) Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 21, 444–448 [DOI] [PubMed] [Google Scholar]
- 79. Niederreither K., Abu-Abed S., Schuhbaur B., Petkovich M., Chambon P., Dolle P. (2002) Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat. Genet. 31, 84–88 [DOI] [PubMed] [Google Scholar]
- 80. Ebert M. S., Sharp P. A. (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee L. M., Leung C. Y., Tang W. W., Choi H. L., Leung Y. C., McCaffery P. J., Wang C. C., Woolf A. S., Shum A. S. (2012) A paradoxical teratogenic mechanism for retinoic acid. Proc. Natl. Acad. Sci. U. S. A. 109, 13668–13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Diez del Corral R., Storey K. (2004) Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays 26, 857–869 [DOI] [PubMed] [Google Scholar]
- 83. Aulehla A., Pourquie O. (2010) Signaling gradients during paraxial mesoderm development. Cold Spring Harb. Perspect. Biol. 2, a000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tanzer A., Stadler P. F. (2004) Molecular evolution of a microRNA cluster. J. Mol. Biol. 339, 327–335 [DOI] [PubMed] [Google Scholar]
- 85. Peterson K. J., Dietrich M. R., McPeek M. A. (2009) MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays 31, 736–747 [DOI] [PubMed] [Google Scholar]
- 86. Heimberg A. M., Sempere L. F., Moy V. N., Donoghue P. C., Peterson K. J. (2008) MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl. Acad. Sci. U. S. A. 105, 2946–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sempere L. F., Cole C. N., McPeek M. A., Peterson K. J. (2006) The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J. Exp. Zool. B Mol. Dev. Evol. 306, 575–588 [DOI] [PubMed] [Google Scholar]
- 88. Campo-Paysaa F., Semon M., Cameron R. A., Peterson K. J., Schubert M. (2011) microRNA complements in deuterostomes: origin and evolution of microRNAs. Evol. Dev. 13, 15–27 [DOI] [PubMed] [Google Scholar]
- 89. Mendell J. T. (2008) miRiad roles for the miR-17–92 cluster in development and disease. Cell 133, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bonauer A., Dimmeler S. (2009) The microRNA-17–92 cluster: still a miRacle? Cell Cycle 8, 3866–3873 [DOI] [PubMed] [Google Scholar]
- 91. Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., Mauceli E., Bouneau L., Fischer C., Ozouf-Costaz C., Bernot A., Nicaud S., Jaffe D., Fisher S., Lutfalla G., Dossat C., Segurens B., Dasilva C., Salanoubat M., Levy M., Boudet N., Castellano S., Anthouard V., Jubin C., Castelli V., Katinka M., Vacherie B., Biemont C., Skalli Z., Cattolico L., Poulain J., De Berardinis V., Cruaud C., Duprat S., Brottier P., Coutanceau J. P., Gouzy J., Parra G., Lardier G., Chapple C., McKernan K. J., McEwan P., Bosak S., Kellis M., Volff J. N., Guigo R., Zody M. C., Mesirov J., Lindblad-Toh K., Birren B., Nusbaum C., Kahn D., Robinson-Rechavi M., Laudet V., Schachter V., Quetier F., Saurin W., Scarpelli C., Wincker P., Lander E. S., Weissenbach J., Roest Crollius H. (2004) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431, 946–957 [DOI] [PubMed] [Google Scholar]
- 92. Meyer A., Van de Peer Y. (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27, 937–945 [DOI] [PubMed] [Google Scholar]
- 93. Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., Jaenisch R., Sharp P. A., Jacks T. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jevnaker A. M., Khuu C., Kjole E., Bryne M., Osmundsen H. (2010) Expression of members of the miRNA17-92 cluster during development and in carcinogenesis. J. Cell. Physiol. 226, 2257–2266 [DOI] [PubMed] [Google Scholar]
- 95. Chaulk S. G., Thede G. L., Kent O. A., Xu Z., Gesner E., Veldhoen R. A., Khanna S. K., Goping I. S., Macmillan A. M., Mendell J. T., Young H., Fahlman R. P., Glover J. N. (2011) Role of pri-miRNA tertiary structure in miR-17∼92 miRNA biogenesis. RNA Biol. 8, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 96. De Pontual L., Yao E., Callier P., Faivre L., Drouin V., Cariou S., Van Haeringen A., Genevieve D., Goldenberg A., Oufadem M., Manouvrier S., Munnich A., Vidigal J. A., Vekemans M., Lyonnet S., Henrion-Caude A., Ventura A., Amiel J. (2011) Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat. Genet. 43, 1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.