Abstract
Motivation: Leveraging gene expression data through large-scale integrative analyses for multicellular organisms is challenging because most samples are not fully annotated to their tissue/cell-type of origin. A computational method to classify samples using their entire gene expression profiles is needed. Such a method must be applicable across thousands of independent studies, hundreds of gene expression technologies and hundreds of diverse human tissues and cell-types.
Results: We present Unveiling RNA Sample Annotation (URSA) that leverages the complex tissue/cell-type relationships and simultaneously estimates the probabilities associated with hundreds of tissues/cell-types for any given gene expression profile. URSA provides accurate and intuitive probability values for expression profiles across independent studies and outperforms other methods, irrespective of data preprocessing techniques. Moreover, without re-training, URSA can be used to classify samples from diverse microarray platforms and even from next-generation sequencing technology. Finally, we provide a molecular interpretation for the tissue and cell-type models as the biological basis for URSA’s classifications.
Availability and implementation: An interactive web interface for using URSA for gene expression analysis is available at: ursa.princeton.edu. The source code is available at https://bitbucket.org/youngl/ursa_backend.
Contact: ogt@cs.princeton.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Genome-scale expression profiling is an invaluable technique for quantifying gene-level activity under different experimental conditions. For more than a decade, researchers and clinicians have submitted their experimental data to public repositories such as NCBI’s Gene Expression Omnibus (GEO) (Barrett et al., 2011) and EBI’s ArrayExpress (Rustici et al., 2013). These repositories now include almost half a million human expression profiles from multiple laboratories and hospitals–only to further grow with the advent of next-generation sequencing technologies. Large but independent microarray datasets have been used to discover tissue-specific patterns (Lukk et al., 2010; Shyamsundar et al., 2005), establish breast cancer subtypes (Cancer Genome Atlas, 2012; Curtis et al., 2012) and delineate the transcriptome response to candidate drugs (Heiser et al., 2012; Lamb et al., 2006). Previous integrative studies have leveraged these independent datasets and have developed methods based on correlation (Hibbs et al., 2007), differential expression (Engreitz et al., 2011), supervised learning (Greene and Troyanskaya, 2011) and data integration (Wong et al., 2012). However, directly dealing with multicellularity is paramount for precisely defining human homeostasis, disease manifestation and pharmacokinetics/pharmacodynamics. To some effect, few studies have focused on certain sample characteristics such as disease or phenotype (Huang et al., 2010; Schmid et al., 2012). Yet, to take full advantage of the entire compendia in all the above contexts, we must explicitly uncover tissue/cell-type-specific signals in genome-wide expression data.
The current exponential rate of data submission nevertheless makes manual annotation impractical, leaving a curated annotation index for only a small fraction of samples (Supplementary Fig. S1). Text-mining sample descriptions are often unreliable due to the lack of standardized nomenclature and structured descriptive information (Krallinger et al., 2010). Furthermore, textual information may not reflect the potential specificity and heterogeneity that are concealed in the molecular-level expression measurements of these samples. Therefore, we need a scalable and robust computational method to discover the tissue/cell-type signals in each gene expression profile deposited in these large heterogeneous data compendia.
In practice, tissue/cell-type annotation of gene expression profiles relies on the expression of known biomarker genes. Although pervasive, this approach is limited by the number of sufficient (or often any) known discriminative expression biomarkers and ignores potential specific signals in the entire transcriptome. Machine learning methods that model genome-wide expression have emerged as promising alternatives (Li et al., 2004), but so far have only been applied in the context of classifying tumor subtypes (e.g. ALL versus AML) in single datasets (Juric et al., 2005; Ramaswamy et al., 2001; Tibshirani et al., 2002). Applying such methods across a large collection of datasets is impeded by the dataset, platform and technology biases (Leek et al., 2010; Rung and Brazma, 2013). The only successful attempt at addressing dataset biases is a nearest -neighbor (NN) classification method based on the barcode algorithm (McCall et al., 2011; Zilliox and Irizarry, 2007).
The task of indexing these large heterogeneous data collections by tissues/cell-types presents substantial challenges. First, a successful method for this task should be able to classify the variety of human tissues/cell-types, not just the better-studied large tissue classes. For example, classifying blood from brain is a relatively easy problem, but discriminating among different subtypes of blood is a much harder one. Second, the method should maintain consistency with the developmental and anatomical relationships between these tissues and cell-types. Third, the method must be robust across independent datasets to overcome study/laboratory biases. Finally, with emerging profiling technologies, the method should be readily applicable to novel platforms/technologies. No existing approach, to our knowledge, addresses all these challenges.
Here, we present a computational algorithm Unveiling RNA Sample Annotation (URSA) that is the first to leverage the relationships between tissues and cell-types (based on a tissue ontology) and accurately identifies specific tissue/cell-type signals present in a given gene expression profile. URSA constructs individual tissue/cell-type classifiers based on ontology-aware sample labels and uses Bayesian Network Correction (BNC) (Barutcuoglu et al., 2006) to integrate these individual classifiers. We demonstrate that URSA substantially outperforms barcode-based NN classification (the only prior approach to this problem) (Zilliox and Irizarry, 2007), as well as independent classifiers that do not use the tissue ontology. Furthermore, although URSA is trained on data from the single most popular microarray platform, it is able to make tissue/cell-type predictions (without re-training) for samples measured by other microarray platforms and even by next-generation RNA sequencing.
In the process of classification, our approach learns tissue/cell-type signals without the use of any tissue-specific gene database such as the human protein reference database (Prasad et al., 2009). Thus, by examining the biological pathways enriched among the feature-weights in each tissue/cell-type classifier, we are able to provide a molecular-level interpretation of URSA’s predictions.
2 METHODS
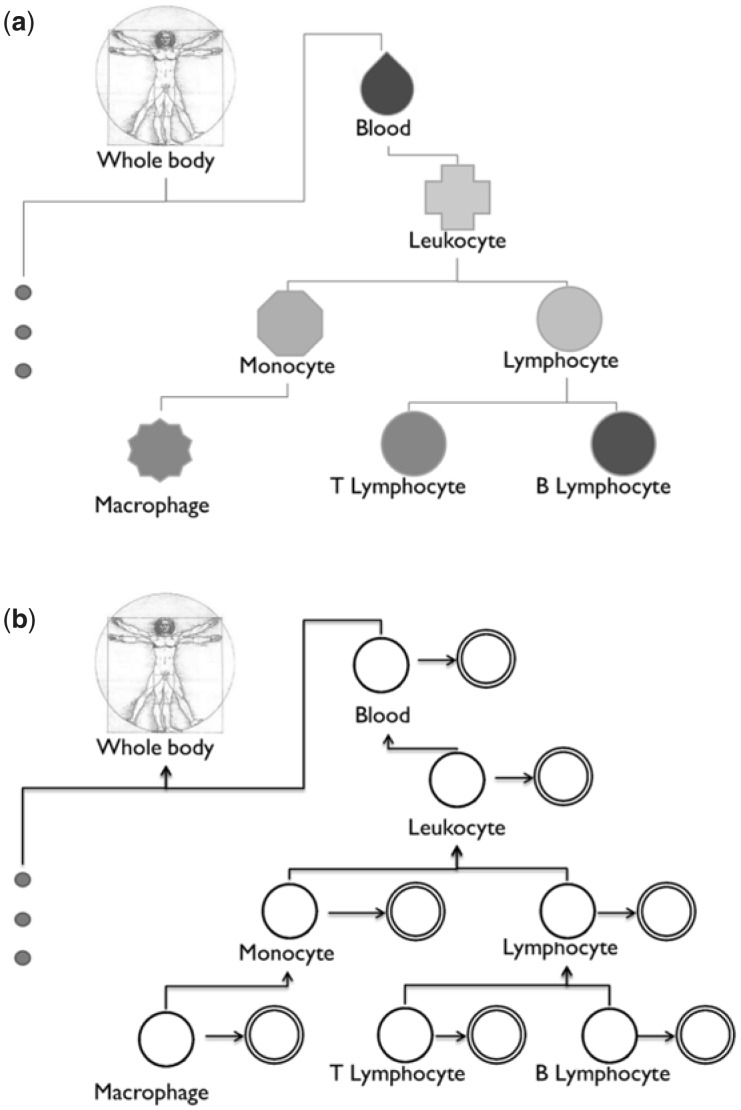
We setup the tissue/cell-type signal classification problem as a hierarchical multilabel classification problem. From a curated collection of samples, we first label samples into positives and negatives based on the tissue ontology to train an individual classifier for each tissue/cell-type. We then aggregate these individual classifiers (in a Bayesian framework) based on their ontological relationships (Fig. 1a and Supplementary Fig. S2) (Gremse et al., 2011). Each individual classifier identifies indicative features (i.e. genes) for that tissue or cell-type, and the Bayesian network models the probabilistic relationship between classifiers to refine those individual predictions. We have previously demonstrated that such BNC improves classification accuracy in other settings, including gene function prediction and geometric shape classification (Barutcuoglu and DeCoro, 2006; Barutcuoglu et al., 2006; Guan et al., 2008; Park et al., 2010). URSA uses the BNC approach to tackle the challenges in tissue and cell-type prediction: limited gold standards for many general (e.g. leukocyte) and specific (e.g. T-cell acute lymphoblastic leukemia cell and monocyte-derived dendritic cell) tissues/cell-types, and heterogeneity and diversity in large expression compendia.
Fig. 1.
Leveraging the complex relationship between tissues and cell-types. (a) A small sub-tree of the BTO. The full hematopoietic system sub-ontology is shown in Supplementary Figure S2. This complexity has yet been incorporated in tissue and cell-type-specific studies. (b) Our aggregation method uses this ontological structure to model the potential dependencies between individual cell-type models. The double circles indicate the noisy individual model predictions ŷi, and the single circles indicate the latent calibrated predictions yi
2.1 Gold standard generation by manual sample annotation
A set of high-quality tissue and cell-type annotations is needed for training accurate classifiers within the URSA framework. To this end, we manually annotated the cell-type(s) of >14 000 microarray experiments ranging over 500 GEO series/datasets from the hgu133plus2 platform. These annotations are based on the sample descriptions and other textual information available in GEO as well as the associated publications. Tissue and cell-type terms in the BRENDA Tissue Ontology (BTO) were used as the controlled vocabulary for sample annotation (Gremse et al., 2011). Detailed description of the manual sample annotation process is provided in the Supplementary Information. In our manual annotations, 71 tissue/cell-type terms were represented by at least 3 GEO series and 95 terms were represented in at least 2 GEO series. We excluded the term connective tissue from the ontology because it had many children terms, and thus appeared unresolved.
With an ontology of tissues, these manually curated annotations can be hierarchically propagated: e.g. monocyte samples can also annotated to leukocyte and blood (Fig. 1a and Supplementary Fig. S2). The minimal subgraph (i.e. directed acyclic graph) that is rooted at the whole body term and includes all cell-type terms covered in our manual annotations was identified, and our manual annotations for 95 tissues/cell-types were then propagated up to their ancestors based on the tissue ontology, hence providing examples for over 244 different tissue/cell-type terms.
2.2 Expression data preparation
The Supplementary raw CEL files of gene expression samples were downloaded from GEO, and their probes were mapped to Entrez GeneIDs using the BrainArray Custom CDF (Barrett et al., 2011; Dai et al., 2005). To compare methods across different preprocessing techniques, expression data were processed using each of the three alternative preprocessing algorithms: MAS5.0, fRMA and Barcode (Hubbell et al., 2002; McCall et al., 2010, 2011). Default parameters and subroutines were used for each preprocessing approach. Additionally, the absolute expression values from the standard MAS5.0 were log transformed. As our method aims at classifying single expression profiles, series-based preprocessing techniques (i.e. RMA) were excluded from our study (Irizarry et al., 2003). The Illumina Human Bodymap 2.0 RNA-seq data (GSE30611) was downloaded from GEO and mapped to NCBI’s transcript reference using the Bowtie and Tophat alignment algorithms with default parameters (Langmead et al., 2009; Trapnell et al., 2009). For tissue/cell-type prediction, FPKM transcript expression values were given as input to our hgu133plus2-trained method. Data transformation and significance test for RNA-seq (and cross platform) experiments are explained later in this section.
2.3 Individual tissue and cell-type classifiers
Labeling positive and negative samples correctly is essential for any accurate classifier. Conventional multilabel classification assumes that all labels are mutually exclusive. For example in our study, macrophage samples would be considered negative examples when classifying for leukocytes, ignoring the fact that macrophages are merely a specific type of leukocytes (Fig. 1b). Using the tissue ontology, we thus reconsider the positive and negative samples for each individual tissue and cell-type classifier. For a given tissue term, samples annotated directly to that term or any of its descendant terms (i.e. cell-types) are now considered positive; samples annotated to only its ancestor terms are excluded from training; and the remaining samples annotated to other term in the ontology—including sibling terms—are considered negative. Now, macrophage samples would be considered positive examples for the leukocyte classifier. This re-labeling is based on the very design of the tissue ontology, and consequently expands the number of positive examples and removes ambiguous examples.
Each individual tissue or cell-type is first classified using an independent one-versus-all support vector machine (SVM) classifier using the ontology-aware training standard. SVM maximizes the margin between positive (i.e. yi = 1) and negative (i.e. yi = −1) examples and finds a linear decision boundary without any assumptions of the probability distributions (Burges, 1998). Given l pairs (i.e. samples) of expression data xi and its label yi, we use the L2 linear SVM (with the default cost parameter) implemented in the LIBLINEAR software (Fan et al., 2012):
where C > 0 is the cost parameter, and w the linear decision boundary (i.e. feature weight vector). Bayesian correction (explained later in text) is trained and applied using the SVM outputs ŷ1, … , ŷN of these N cell-type-specific models.
2.4 Bayesian network correction
We use the structure of the tissue ontology as a framework of the Bayesian network (Fig. 1 and Supplementary Fig. S2). We model each term’s SVM output as a random event ŷi and treat it as a noisy observation of a latent binary event yi representing the true label (i.e. cell-type) of a given sample (Fig. 1b). The edges from y to ŷ impose the independence of the noisy random variable ŷi to all other noisy variables ŷj (i ≠ j) given its true label yi. This allows us to calculate the likelihood:
The distribution of positive and negative unthresholded SVM outputs varies across different terms (i.e. cell-types), and so the output values were dynamically binned based on the number of positive examples and their range. These empirical distributions represent the conditional probabilities P(ŷi | yi = 0) and P(ŷi | yi = 1). The conditional probability table for each term was estimated based on a 2-fold cross-validation that never split datasets between folds to mitigate potential batch effects. Laplace smoothing was applied for robustness.
The parent–child conditional probability tables were defined as in the original Bayesian correction method (Barutcuoglu et al., 2006). Intuitively, constant priors of 0.5 were assigned to leaf nodes, and the whole-body root node was assigned a probability of 1. This root assignment allows potential dependencies between every latent variable. This allows us to calculate the prior:
where ch(yi) is child labels of yi.
Finally, we infer the posterior probabilities P(yi|ŷ1, … , ŷN) for each term i using the Lauritzen algorithm as implemented in the SMILE library (Druzdzel, 1999; Lauritzen and Wermuth, 1989). These posterior probabilities for each term (i.e. cell-type) are the estimated probabilities that our method uses to annotate gene expression samples.
2.5 Method training and testing
Genomic experiments are known to suffer from potential laboratory and dataset biases (Leek et al., 2010; Zilliox and Irizarry, 2007). Not controlling for this bias (during evaluation of any method applied to these data) may result in an overestimation of performance and overfitting to dataset-specific biases at the expense of the desired signals. Therefore, for each cell-type, the series/datasets of the manually annotated samples were partitioned into three sets with each set containing roughly the same number of samples. Two partitions were used as the training set and the other as the testing set. Never splitting a single series/dataset between training and test sample sets ensures that our approach does not identify signals specific to particular studies, but rather those reflective of cell-types and tissues.
2.6 Cross-platform prediction
The individual classifiers in URSA were trained on samples only from the most popular Affymetrix Human Genome U133 Plus 2.0 platform (hgu133plus2). URSA has not been explicitly modified or tuned for predicting across other platforms. As input to our method, a gene expression profile from other array-based and sequence-based platforms were quantile transformed to generate a hgu133plus2-like expression profile (Section 2.6.1., later in the text). Additionally, a permutation test (Section 2.6.2., later in the text) was performed to correct for potential biases from gene coverage differences across platforms.
2.6.1 Quantile transformation
The individual cell-type models in URSA have been trained on one microarray platform (hgu133plus2). To detect cell-type-specific information from other gene expression platforms, we must first transform those expression values to a comparable expression space. If we can effectively transform those values, our method—without any modifications or retraining—may be able to measure cell-type-specific signals in these cross-platform experiments. The individual expression values xi may not be comparable across different platform technologies (especially between array-based and sequence-based platforms), but signals based on the relative abundance between genes should be more or less preserved irrespective of the technology used. Therefore, we quantile transform these cross-platform samples to preserved their relative gene abundances (or gene order) and compute hgu133plus2-like expression values based on a hgu133plus2 reference distribution. This reference distribution was constructed by averaging the expression value of each quantile across 1000 random hgu133plus2 arrays.
The most crucial bottleneck for cross-platform annotation is the bias in gene coverage across platforms. The hgu95v2 microarray platform (covering ∼12 000 genes), for example, covers about two-thirds the genes covered by hgu133plus2 (∼18 000 genes). Classification is handicapped by thousands of missing values, and hence, the mean expression value of the reference distribution was used to impute missing gene values (Troyanskaya et al., 2001).
2.6.2 Permutation test
A simple permutation test was performed to select significant predictions. The input data xj consist of real and imputed gene values. Introducing noise to the actual data will blur any real signal and decrease its associated probability value. Thus, we permute only the sample data π1(xj), … , πK(xj) to generate a null distribution of SVM outputs π(ŷi) = (πk(ŷ1), … , πk (ŷN)), where πk(ŷi) = wiT· πk(xj). This null distribution is then used to call out questionable annotations: any tissue annotation P(yi|ŷ1, … , ŷN) with a value lower than even a single random annotation P(yi| πk(ŷ1), … , πk(ŷN)) is considered insignificant and assigned a value of 0.
2.7 Double-blind evaluation of sample annotations
In addition to the evaluation based on our manual sample annotations (Section 2.5., earlier in text), we conducted a rigorous double-blind literature-based study to evaluate the quality of URSA’s novel predictions. To control for any subjective bias, we must also evaluate a random group of predictions in the same literature-based study. First, 120 hgu133plus2 array experiments from GEO that were not in our manual annotation were randomly selected. These experiments were partitioned into three groups. URSA annotations were made for all samples, but only group 1 predictions were retained and group 2 samples were assigned predictions from group 3. This procedure provides random annotations while ensuring the same apparent behavior of predictions as true predicted annotations. We use this conservative background to completely blind the evaluator from distinguishing original from random annotations.
The quality of predicted annotations should be judged based on retrieval of both the most precise tissue term and more general terms consistent with the precise term. For example, an acute lymphocytic leukemia (ALL) sample predicted to ALL but also other non-blood related terms such as urinary bladder and colon is precise but not consistent, whereas the same sample predicted to blood cancer cell or leukocyte in addition to ALL is both precise and consistent. Estimated annotations in group 1 (i.e. original) and group 2 (i.e. random) were evaluated as precise and/or consistent based on associated publications and textual sample descriptions. We also repeated this double-blind study for other microarray platforms: hgu133a, hgu95v2 and hugene1.0st.
3 RESULTS
We address the cell-type prediction challenge as a multilabel classification problem with hierarchical constraints to account for the diverse nature of biological samples. We assess the impact of incorporating the tissue ontology in our method and the method’s robustness across different microarray preprocessing methods. Although our method can be readily retrained to any additional expression technologies given manually curated samples, we find that our method is capable of precisely annotating samples across platforms (including next-generation sequencing-based assays) without any modifications to the original method or its parameters. We finally show that our tissue/cell-type predictions are interpretable based on the biological processes enriched among learned informative genes.
3.1 URSA uses the tissue ontology to accurately predict tissue/cell-type signals
To address the challenge of limited gold standards and high noise levels in the tissue/cell-type classification problem, URSA incorporates the BTO to better predict tissue/cell-type signals in a given gene expression sample. BTO systematically defines parent-to-child relationships between tissue and cell-type terms (Gremse et al., 2011). URSA wields the complexity of this ontology to both systematically label samples to train tissue/cell-type SVM classifiers and also apply BNC to make consistent predictions.
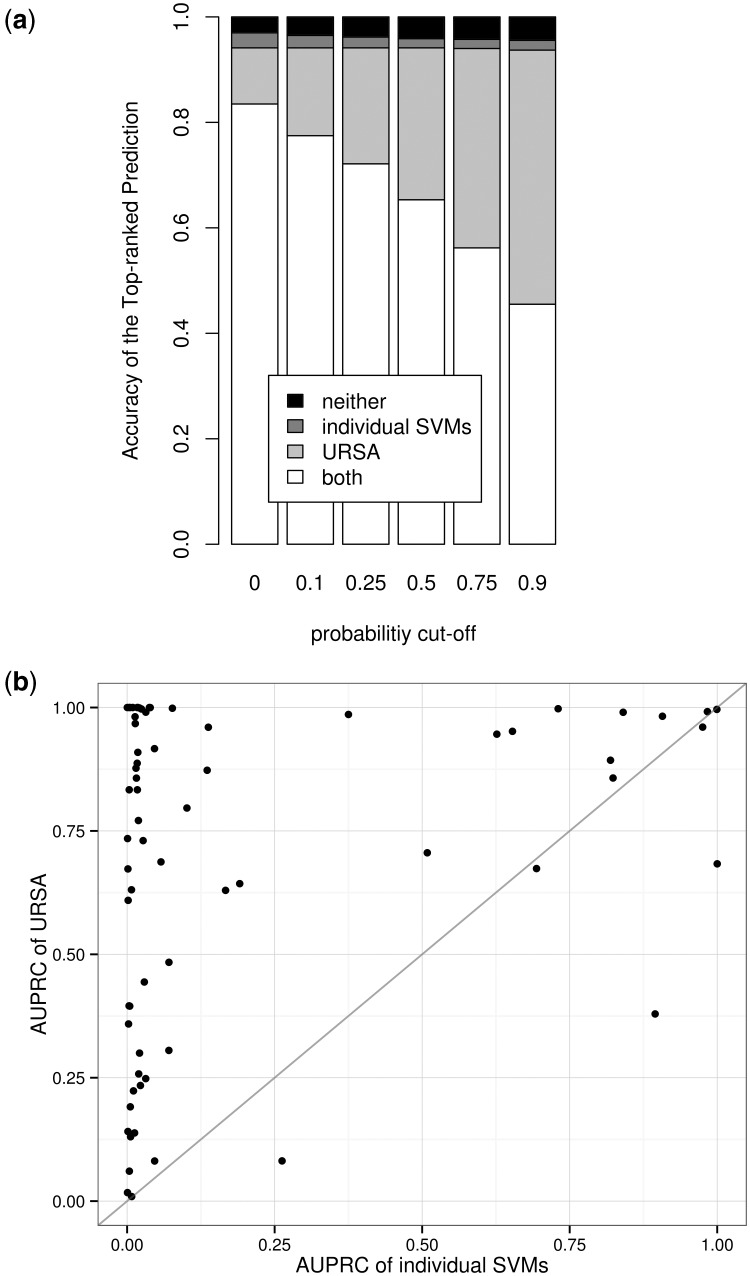
To measure the impact of incorporating the ontology, we compare URSA with individual (i.e. independent) one-versus-all SVM classifiers whose outputs are converted to estimate probability values using logistic regression (Platt, 1999). For these one-versus-all SVMs, whole blood samples are considered as negatives in a leukocyte classifier, for example. Both methods were trained on ∼9000 samples and tested on ∼5000 independent samples (Fig. 2a). The top-predicted term for each sample was evaluated and automatically considered incorrect if the estimated probability value was below a cutoff. Multiple cutoffs from 0 (i.e. no cutoff) to 0.9 (i.e. high-confidence cutoff) were tested (Fig. 2a). This setup simulates the user experience with a predefined cutoff and penalizes correct top predictions with a low probability value.
Fig. 2.
Prediction accuracy improves after integrating the tissue ontology. MAS5 was used for preprocessing. (a) Accuracy of the most probable estimation above a range of probability cutoffs. Estimations below the probability cutoff are discarded. The Bayesian framework corrects many of the mistakes made by individual SVMs and provides meaningful probability values. (b) Scatter plot comparison between URSA and individual SVM classifiers. Each point represents a unique tissue/cell-type with direct sample annotations, and the size of the point represents the number of samples curated to that particular tissue or cell-type. Points above the diagonal correspond to improvements by our method. URSA’s improvements are independent of term size
Across the entire range of probability cutoffs, URSA offers accurate top predictions for more samples in the holdout set. Without a cutoff on the estimated probabilities, both naïve SVM and ontology-aware URSA show considerable accuracy of the top-predicted term over the heterogeneous evaluation set (Fig. 2a, leftmost bar). However, URSA accurately predicts an additional ∼550 samples misclassified by the independent SVMs. Furthermore, URSA conveniently computes a probability value for each predicted tissue/cell-type annotation that provides a natural intuition about the strength of the predicted tissue/cell-type signal present in a given sample. Although probabilities can also be obtained for the individual SVMs, URSA’s Bayesian framework provides a unified probabilistic model that enforces potential dependencies between distant and close tissues. This abstraction consequently computes consistent parameter estimations: e.g. if the probability for leukocyte is high, then the probability for blood should also be high, but not necessarily vice versa. Lending import to the calibrated probability values calculated by BNC, the proportion of URSA’s accurate corrections of SVM’s mis-annotations increases with higher probability cutoffs (Fig. 2a). In case of high confidence predictions (0.9 cutoff), URSA provides accurate annotations for ∼94% of the test samples, 45% (>2200) of which were misclassified by the individual SVMs. More detailed description is provided in the Supplementary Information.
In addition to the overall performance evaluation, it is important to consider how annotation accuracy depends on the number of expression profiles available for training for each tissue term (namely ‘term size’). Term size also serves as an appropriate estimation of the term’s specificity in the tissue ontology, as sample annotations were propagated based on the same ontology. URSA’s ontology-aware Bayesian framework aggregates multiple individual classifiers so that classifiers for large terms (such as blood) could help classify related specific small terms (such as T-cell acute lymphoblastic leukemia cell). Using area under the precision-recall curve (AUPRC), we compare the entire ranking accuracy of URSA and the individual SVM classifiers across tissues/cell-types. URSA provides increased performance for 65 of the 71 tissue terms spanning both large general terms (such as B-lymphocyte > 0.98, breast > 0.89 and lung > 0.95) and small specific terms (such as T-cell acute lymphoblastic leukemia cell = 1, HeLa cell > 0.91 and bronchial epithelial cell > 0.83) (Fig. 2b and Supplementary Table S1). Decreased performance for a few terms could be attributed to the incompleteness of the tissue ontology (e.g. the missing parental relationship between hepatocyte and hepatoma cell). URSA’s improvements over individual SVMs are greater for leaf nodes than non-leaf nodes (Supplementary Fig. S3). The observed inverse relationship and larger improvements for leaf nodes than for non-leaf nodes highlights the need for URSA—especially for the specific terms where individual classifiers often perform poorly due to the lack of training data. Thus, although the number of training samples affects the quality of individual models, our results show that exploiting the known cell-type associations enables URSA to be reasonably immune to this effect. These results are further discussed in the Supplementary Information.
Even without the use of the tissue ontology, independent SVMs perform reasonably well for easy problems such as discriminating blood samples, and so the improvement of our approach is relatively small (AUPRC of 0.9072 for individual SVMs versus AUPRC of 0.9823 for URSA). However, independent SVMs are unable to effectively discriminate more specific cell-type samples such as T-cell acute lymphoblastic leukemia cell samples (SVM AUPRC 0.0034), whereas our ontology-aware approach accurately classifies holdout samples of this specific blood cancer subtype (URSA AUPRC 1.0). This improvement of URSA can be attributed to the effective incorporation of the ontological complexity (Supplementary Fig. S2). Notice this improvement also holds true across a wide range of non-blood cell-types such as prostate gland (0.0317 versus 0.9906), bronchial epithelial cell (0.0174 versus 0.8333) and mesenchymal stem cell (MSC) (0.0017 versus 0.6093) (Fig. 2b and Supplementary Table S1). The fact that these signals were learned in a completely data-driven approach—not from known biomarkers—indicates that our method can provide a data-driven estimation of specific blood (and non-blood) cell-type signals.
3.2 URSA’s performance is robust to expression data preprocessing
Data preprocessing and normalization can have a significant impact on downstream analysis, including prediction of tissues/cell-type signals (Zilliox and Irizarry, 2007). MAS5.0 and fRMA are the two most well-known algorithms for preprocessing single arrays (Hubbell et al., 2002; McCall et al., 2010). Additionally, the barcode preprocessing algorithm was shown to accurately estimate whether a gene is expressed in a given microarray experiment and in specific tissues (McCall et al., 2011; Zilliox and Irizarry, 2007).
We test the robustness of URSA’s ranking accuracy to different preprocessing methods. Our first evaluation (using AUPRC) shows that URSA improves performance over individual SVMs across all three data processing methods: MAS5.0 (Fig. 2b), fRMA and Barcode (Supplementary Fig. S4). Next, we compare URSA with a NN classifier after barcode processing, which is, to our knowledge, the only previous approach shown to predict cell-type (Zilliox and Irizarry, 2007). It is important to note that the overall accuracy of the NN classifier relies on the accuracy of the barcode preprocessing algorithm. URSA correctly annotates ∼95% of the test samples independent of the preprocessing algorithm used, with >650 samples being correctly predicted exclusively using URSA (Supplementary Fig. 5a). Furthermore, our method returns better ranking accuracy for at least 50 of the 71 tissues/cell-types than the NN classifier (Supplementary Figs. S5b and S6). Again, the performance improvements appear to be robust to term size.
These analyses show that URSA can adapt to both generic (e.g. MAS5.0) and specific (e.g. Barcode) preprocessing methods to discover tissue/cell-type-specific information in genome-scale experiments. Moreover, robustness to preprocessing suggests that URSA is modeling biological signals rather than systematic biases or data processing artifacts present in these large compendia. We focus our remaining analyses using the most commonly used MAS5.0, chosen for its simplicity and application to many array platforms.
3.3 URSA is precise for experiments from other expression platforms
URSA is trained using data from the most popular gene expression microarray platform HG-U133 Plus 2.0 (hgu133plus2)—with ∼70 000 samples (from 2500 datasets/series) in GEO. We have shown that URSA performs well for samples from this platform, but there exist many other expression datasets that use other platforms, with new ones emerging continuously. The Affymetrix Human Genome U133A (hgu133a), for example, is arguably the second most common microarray platform, associated with ∼1000 studies in GEO. Other genome-wide array platforms such as HG-U95Av2 (hgu95av2) and HuGene 1.0 ST (hugene1st) have been used for their focused gene coverage. As hundreds of such platforms have been used for human gene expression measurements, re-training classifiers for each platform is impractical. Instead, the challenge is to overcome technical differences across platforms and predict tissue/cell-type signals in a platform-independent manner.
We test URSA’s potential to measure the tissue-specific signatures in profiles from other array-based platforms without re-training its parameters. For this, we quantile-transform input data from cross-platform samples and filter final predictions by using a permutation test (see Section 2). To evaluate these predictions in a manner that best emulates an end-user’s experience, we conduct a double-blind literature study on ‘original’ and ‘random’ annotations (see Section 2). The evaluation shows that the majority of URSA’s predicted annotations are both precise and consistent regardless of the microarray platform (Supplementary Fig. S7). Despite missing expression values for >10 000 genes (due to limited gene coverage), our method is still able to provide high-quality annotations even for hgu95av2 samples. These consistent trends illustrate URSA’s potential to detect cell-type-specific signals across microarray platforms rising above technical biases and even substantial gene coverage differences.
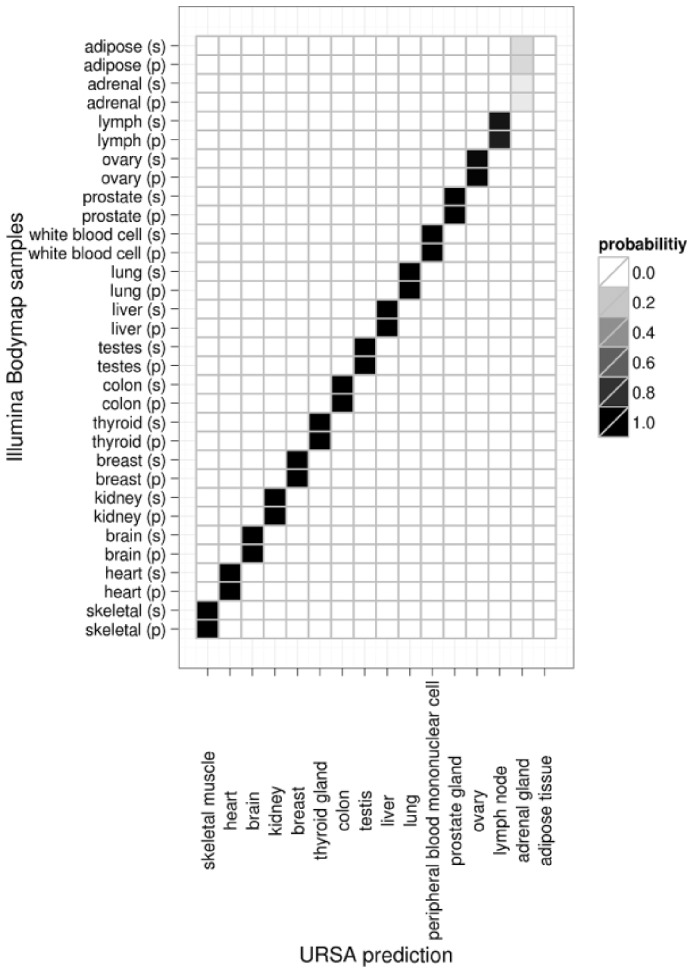
Next-generation sequencing is another rapidly growing technology for transcriptome profiling. A sample annotation method that can be applied to this burgeoning technology is also of great interest, and yet the current number of available tissue/cell-type-specific experiments limits the prospect of effectively training classifiers specifically for RNA-seq experiments. To address this problem, we test URSA’s ability to detect tissue-specific signatures in RNA-seq experiments using the model trained on microarray data. At the outset, this is a challenging task due to the substantial technical differences between microarrays and RNA-seq. We challenge URSA to annotate RNA-seq experiments in the Illumina Bodymap 2.0 reference dataset (GSE30611), which consists of a diverse set of samples from 16 different tissues, generated with both single-end and pair-end sequencing methods. URSA correctly predicts the tissue of origin for all single-end and pair-end samples, except for adrenal gland and adipose tissue samples (Fig. 3). For adrenal gland, URSA ranked adrenal gland as the second most significant tissue signal for adrenal gland samples (and not for any of the other tissue types such as kidney or thyroid gland). Although URSA can eventually be re-trained to better fit growing next-generation sequencing data, its robustness across platforms and technologies demonstrates URSA’s promise to remain applicable and relevant to emerging experimental approaches and data processing methods.
Fig. 3.
Accurate prediction of tissue of origin for RNA-seq samples. Heatmap of URSA’s estimated tissue probabilities of 32 RNA-seq experiments (16 different tissues) in the Illumina Bodymap dataset. The rows are the individual samples, either single-end (s) or pair-end (p), and the columns are the estimated cell-types
3.4 URSA’s tissue and cell-type-specific models are biologically interpretable
With accurate models constructed from >14 000 diverse samples representing over 244 tissue/cell-type terms, URSA’s discriminative features (i.e. genes) could paint a molecular portrait of tissue/cell-type-specific gene expression. To test this hypothesis, we use the PAGE algorithm (Kim and Volsky, 2005) to examine the Gene Ontology (GO) biological processes (Ashburner et al., 2000) represented in each tissue/cell-type model. In effect, the analysis summarizes the gene weights and offers a rich description of the models. Testing all 244 cell-type models, we find that many of the processes enriched among most informative genes in these models appear to be the relevant cell-type-specific GO terms. For example, the top GO term for the B-lymphocyte model is B cell activation (adjusted P < 0.001), whereas the top GO term for the B-cell lymphoma cell model is regulation of inflammatory response (adjusted P < 0.001). GO terms astrocyte differentiation, regulation of synaptic transmission and behavior are enriched in the brain model (adjusted P < 0.001). Complete results with associated z-scores are provided in Supplementary Table S2 and further discussed in the Supplementary Information.
Certain associations are not necessarily obvious. The top GO terms in the MSC model include mesenchymal-specific developmental processes such as skeletal system development, cartilage condensation and muscle organ morphogenesis (Supplementary Table S2). The enrichment of glycosaminoglycan biosynthetic process in the MSC model has some support in that glycosaminoglycans regulate osteoblast differentiation of bone marrow-derived human MSCs and chondrogenesis in mouse MSCs (Kim et al., 2007; Mathews et al., 2012). The top specific GO terms in the embryonic stem cell (ESC) model include calcium-dependent cell–cell adhesion, positive regulation of Wnt receptor signaling pathway and glutamine family amino acid metabolic process. During mouse embryogenesis, inner mass formation and cell surface polarization is regulated by the calcium-dependent cell–cell adhesion system (Shirayoshi et al., 1983). Highly conserved Wnt family proteins play a key role in embryogenesis and oncogenesis, but moreover the positive regulation (i.e. activation) of Wnt signaling maintains the pluripotency in human ESCs (Logan and Nusse, 2004; Peifer and Polakis, 2000; Sato et al., 2004). L-glutamine is needed for the culture and maintenance of human ESCs and is shown to inhibit mouse embryogenesis in high concentrations (Amit and Itskovitz-Eldor, 2006; Kent, 2009; Nakazawa et al., 1997). The enrichment of these non-trivial and specific biological processes demonstrates the expressive (and accurate) interpretation of URSA’s predictions.
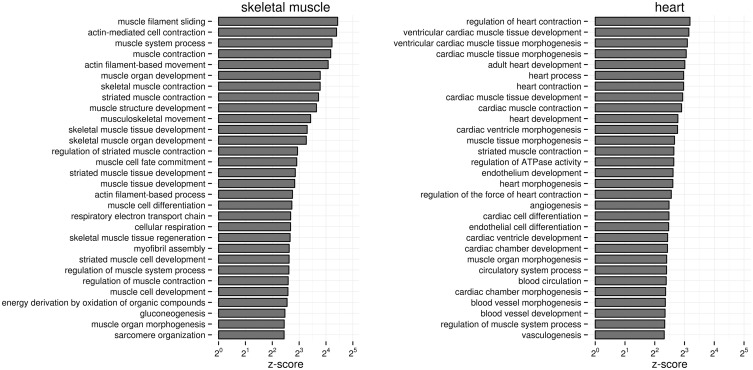
Based on the enriched biological processes (i.e. GO terms), we examine whether the models are specific enough to distinguish even closely related cell-types such as skeletal muscle cells and heart cells (Fig. 4). Skeletal muscle and heart are among the most studied human tissues, and thus are appropriate examples to test the specificity of our models, which are based solely on genome-wide expression experiments. Both skeletal muscle and heart are comprised of muscle cells, and so one might expect that the top GO terms for both tissue models would be general muscle-related GO terms such as actin-mediated cell contraction. Instead, we find that although all top enriched processes for skeletal muscle are general muscle GO terms as expected, the top processes for heart (e.g. ventricular cardiac muscle tissue development and heart contraction) are specific to heart cells (Fig. 4). Thus, without prior knowledge of tissue and cell-type-specific genes, URSA’s models identify genes involved in corresponding cell-type-specific biological processes. This approach could be extended for understanding poorly characterized cell-types including specific cancer subtypes. Our analysis altogether provides biological intuition and credence to the basis for URSA’s tissue and cell-type annotations.
Fig. 4.
Tissue-specific biological processes enriched in URSA’s skeletal muscle and heart models. Barplot of enrichment z-scores of top GO terms in the two models are shown. Both skeletal muscle and heart are primarily populated by muscle cells; yet, the heart tissue model selects genes specifically involved in cardiac muscle processes
4 DISCUSSION
In multicellular organisms, integrative analysis leveraging large gene expression compendia requires accurate annotations of samples to their tissue and cell-type of origin. In this article, we present a scalable computational method URSA that predicts tissue/cell-type signals in expression profiles across platforms and technologies. Key to its performance is the incorporation of the tissue ontology. Much of URSA’s improved performance can be attributed to the construction of more than one hundred additional intermediate (i.e. non-leaf) classifiers, which are then integrated using a Bayesian framework.
URSA can be used to automatically annotate samples in public gene expression repositories where most samples are currently lacking tissue/cell-type-specific information. Researchers can discover specific signals in their own samples via our interactive interface at ursa.princeton.edu. Others interested in integrative studies can download the URSA C++ software and annotate samples on a large scale.
Despite URSA’s current applicability to a wide variety of tissues/cell-types, its predictions can be further improved as the ontology used for integration adds additional terms and associations. For example, immunologists may be interested in the signal of specific T-lymphocytes such as CD4+ T cells, Th17 cells, germinal B cells, and so forth. Unfortunately, the current BRENDA ontology (which was used as a controlled vocabulary and the ontology structure of our method) does not include such terms. Nonetheless, URSA’s ability to delineate tissue/cell-type signals without known biomarker genes makes it naturally extendable to such specific cell-types as the BRENDA ontology is extended with more terms and associations (Gremse et al., 2011). We plan to regularly maintain and update the software with new tissue and cell-type annotations and the latest version of the BRENDA ontology.
Both the strength and the limitation of our method across platforms and technologies depend on the amount of tissue signal in the gene order and the number of missing values. For a given gene expression profile from a different platform, quantile transformation is applied to compute hg133plus2-like expression values. In consequence, our method is robust to different normalization techniques used because only the information of relative gene abundance is transferred. However, specific signals associated with the particular gene expression value may be lost, and properly incorporating such signals may provide greater prediction accuracy. Furthermore, expression values for genes not measured in hg133plus2 could affect the accuracy of our method, although simple mean imputation seems to alleviate that effect.
URSA’s tissue and cell-type-specific models provide a biological interpretation of its predictions. As such, URSA could potentially be used to test and identify possible sample contaminations, resolve cancer samples of unknown primary origin and perhaps provide insight into the molecular basis of poorly characterized clinical subtypes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Bobak Hadidi for processing the RNA-seq data and the Function group at Princeton University for the valuable discussions.
Funding: National Science Foundation (NSF) CAREER award (DBI-0546275); National Institutes of Health (NIH) (R01 GM071966, R01 HG005998 and T32 HG003284); National Institute of General Medical Sciences (NIGMS) Center of Excellence (P50 GM071508). O.G.T. is a Senior Fellow of the Canadian Institute for Advanced Research.
Conflict of interest: none declared.
REFERENCES
- Amit M, Itskovitz-Eldor J. Maintenance of human embryonic stem cells in animal serum- and feeder layer-free culture conditions. Methods Mol. Biol. 2006;331:105–113. doi: 10.1385/1-59745-046-4:105. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–209. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barutcuoglu Z, DeCoro C. Shape Modeling and Applications, 2006. SMI 2006. IEEE International Conference. 2006. Hierarchical shape classification using bayesian aggregation; p. 44. [Google Scholar]
- Barutcuoglu Z, et al. Hierarchical multi-label prediction of gene function. Bioinformatics. 2006;22:830–836. doi: 10.1093/bioinformatics/btk048. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998;2:121–167. [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzdzel MJ. Proceedings of the Sixteenth National Conference on Artificial Intelligence. Menlo Park, CA, USA: American Association for Artificial Intelligence; 1999. SMILE: structural modeling, inference, and learning engine and genie: a development environment for graphical decision-theoretic models; pp. 902–903. [Google Scholar]
- Engreitz JM, et al. ProfileChaser: searching microarray repositories based on genome-wide patterns of differential expression. Bioinformatics. 2011;27:3317–3318. doi: 10.1093/bioinformatics/btr548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R-E, et al. LIBLINEAR: a library for large linear classification. J. Mach. Learn. Res. 2012;9:1871–1874. [Google Scholar]
- Greene CS, Troyanskaya OG. PILGRM: an interactive data-driven discovery platform for expert biologists. Nucleic Acids Res. 2011;39:W368–W374. doi: 10.1093/nar/gkr440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremse M, et al. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–D513. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, et al. Predicting gene function in a hierarchical context with an ensemble of classifiers. Genome Biol. 2008;9(Suppl. 1):S3. doi: 10.1186/gb-2008-9-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser LM, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl Acad. Sci. USA. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs MA, et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Bayesian approach to transforming public gene expression repositories into disease diagnosis databases. Proc. Natl Acad. Sci. USA. 2010;107:6823–6828. doi: 10.1073/pnas.0912043107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell E, et al. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Juric D, et al. Gene expression profiling differentiates germ cell tumors from other cancers and defines subtype-specific signatures. Proc. Natl Acad. Sci. USA. 2005;102:17763–17768. doi: 10.1073/pnas.0509082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent L. Culture and maintenance of human embryonic stem cells. J. Vis. Exp. 2009:e1427. doi: 10.3791/1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, et al. Cytokine-like 1 (Cytl1) regulates the chondrogenesis of mesenchymal cells. J. Biol. Chem. 2007;282:29359–29367. doi: 10.1074/jbc.M700965200. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Volsky DJ. PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics. 2005;6:144. doi: 10.1186/1471-2105-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M, et al. Analysis of biological processes and diseases using text mining approaches. Methods Mol. Biol. 2010;593:341–382. doi: 10.1007/978-1-60327-194-3_16. [DOI] [PubMed] [Google Scholar]
- Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen SL, Wermuth N. Graphical models for associations between variables, some of which are qualitative and some quantitative. Ann. Stat. 1989;17:31–57. [Google Scholar]
- Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. A comparative study of feature selection and multiclass classification methods for tissue classification based on gene expression. Bioinformatics. 2004;20:2429–2437. doi: 10.1093/bioinformatics/bth267. [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Ann. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- Lukk M, et al. A global map of human gene expression. Nat. Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, et al. Glycosaminoglycans enhance osteoblast differentiation of bone marrow derived human mesenchymal stem cells. J. Tissue Eng. Regen. Med. 2012 doi: 10.1002/term.1507. [Epub ahead of print, doi:10.1002/term.1507, April 10, 2012] [DOI] [PubMed] [Google Scholar]
- McCall MN, et al. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, et al. The Gene Expression Barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res. 2011;39:D1011–D1015. doi: 10.1093/nar/gkq1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, et al. Effect of different concentrations of amino acids in human serum and follicular fluid on the development of one-cell mouse embryos in vitro. J. Reprod. Fertil. 1997;111:327–332. doi: 10.1530/jrf.0.1110327. [DOI] [PubMed] [Google Scholar]
- Park CY, et al. Simultaneous genome-wide inference of physical, genetic, regulatory, and functional pathway components. PLoS Comput. Biol. 2010;6:e1001009. doi: 10.1371/journal.pcbi.1001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis–a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Prasad TS, et al. Human protein reference database and human proteinpedia as discovery tools for systems biology. Methods Mol. Biol. 2009;577:67–79. doi: 10.1007/978-1-60761-232-2_6. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc. Natl Acad. Sci. USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat. Rev. Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- Rustici G, et al. ArrayExpress update—trends in database growth and links to data analysis tools. Nucleic Acids Res. 2013;41:D987–D990. doi: 10.1093/nar/gks1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schmid PR, et al. Making sense out of massive data by going beyond differential expression. Proc. Natl Acad. Sci. USA. 2012;109:5594–5599. doi: 10.1073/pnas.1118792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayoshi Y, et al. The calcium-dependent cell-cell adhesion system regulates inner cell mass formation and cell surface polarization in early mouse development. Cell. 1983;35:631–638. doi: 10.1016/0092-8674(83)90095-8. [DOI] [PubMed] [Google Scholar]
- Shyamsundar R, et al. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22. doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Wong AK, et al. IMP: a multi-species functional genomics portal for integration, visualization and prediction of protein functions and networks. Nucleic Acids Res. 2012;40:W484–W490. doi: 10.1093/nar/gks458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat. Methods. 2007;4:911–913. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.