Abstract
Administration of mesenchymal stem cells (MSCs) has the potential to ameliorate degenerative disorders and to repair damaged tissues. The homing of transplanted MSCs to injured sites is a critical property of engraftment. Our aim was to identify microRNAs involved in controlling MSC proliferation and migration. MSCs can be isolated from bone marrow and umbilical cord Wharton’s jelly (BM-MSCs and WJ-MSCs, respectively), and WJ-MSCs show poorer motility yet have a better amplification rate compared with BM-MSCs. Small RNA sequencing revealed that miR-146a-5p is significantly overexpressed and has high abundance in WJ-MSCs. Knockdown of miR-146a-5p in WJ-MSCs inhibited their proliferation yet enhanced their migration, whereas overexpression of miR-146a-5p in BM-MSCs did not influence their osteogenic and adipogenic potentials. Chemokine (C-X-C motif) ligand 12 (CXCL12), together with SIKE1, which is an I-kappa-B kinase epsilon (IKKε) suppressor, is a direct target of miR-146a-5p in MSCs. Knockdown of miR-146a-5p resulted in the down-regulation of nuclear factor kappa-B (NF-κB) activity, which is highly activated in WJ-MSCs and is known to activate miR-146a-5p promoter. miR-146a-5p is also downstream of CXCL12, and a negative feedback loop is therefore formed in MSCs. These findings suggest that miR-146a-5p is critical to the uncoupling of motility and proliferation of MSCs. Our miRNome data also provide a roadmap for further understanding MSC biology.
INTRODUCTION
Human mesenchymal stem cells (MSCs) have been identified as multipotent mesoderm-derived stromal cells that have the ability to self-renew and differentiate (1); they have been applied as clinical treatments for bone and other tissue defects (2–4). On activation by tissue damage, MSCs contribute to tissue-repair processes through a multitude of activities, including cell proliferation, migration and differentiation. The mobilization of bone marrow (BM)-derived MSCs from BM to the peripheral blood, and their eventual entry into the injured brain, plays a crucial step in brain plasticity and stroke therapy (5). MSC activities also affect the therapeutic efficacy of engraftment, specifically if only low number of transplanted MSCs migrate to the injured site after infusion, which will limit the therapeutic applications of MSCs (6). The expansion/proliferation rate of MSCs ex vivo also influences cell motility, as MSCs lose their mobility during cultivation (7).
microRNAs (miRNAs) are short non-coding RNAs (∼22 nt) that can repress translations through imperfectly binding to target messenger RNA. After being transcribed and processed by Drosha and Dicer, miRNAs are then loaded into an RNA-induced silencing complex that brings about the regulation of translation (8). To date, relatively few studies have examined miRNA functionality in MSCs: miR-335 has been shown to inhibit cell proliferation, migration and differentiation (9). In addition, miR-138 modulates osteogenesis by MSCs (10). miR-204 has also been found to inhibit osteogenesis but to promote adipogenesis by MSCs (11). We recently found that miR-34a is able to modulate the cellular motility genes of neural precursor cells derived from Wharton’s jelly MSCs (WJ-MSCs) (12).
Till date, hundreds to thousands of miRNAs have been identified in animals and plants, and many more miRNAs are continuously being identified by newly available technologies, including small RNA sequencing (smRNA-Seq). High-throughput sequencing is able to not only reveal the expression profiles of known miRNAs but also identify other non-coding small RNAs and discover new miRNAs that have not been recorded previously in any databases, in particular the miRBase repository. smRNA-Seq has been used to carry out research on various types of stem cells, including embryonic stem cells (13–15), hematopoietic stem cells (16) and neural precursor cells (13). Novel miRNAs have also been identified using smRNA-Seq during neural differentiation of embryonic stem cells (15) and during endothelial differentiation (17). Nevertheless, no smRNA-Seq work has been reported on somatic MSCs.
Because the implanted number and homing of transplanted MSCs to injured sites is one of the critical properties in relation to engraftment, in the present study our aim was to identify miRNAs that are involved in controlling the proliferation and migration phenotypes of MSCs. We hypothesized that miRNAs involved in stem cell motility and proliferation must be present in undifferentiated MSCs, given the variations observed on their mobility. MSCs from different sources have different functionality. MSCs can be obtained from BM as well as other fetal or postnatal tissues, including adipose tissue, umbilical cord blood and the Wharton’s jelly of the umbilical cord (18). WJ-MSCs have been considered as a great alternative source for the harvesting of MSCs (19) and have multilineage differentiation ability that allows them to become osteocytes, chondrocytes, adipocytes, cardiomyocytes and neurons (20). The differentiation, immunomodulation and proliferation abilities, as well as transcriptome profiles, of WJ-MSCs have been compared with other MSCs (21–23). BM-MSCs have better osteogenic and adipogenic abilities, whereas WJ-MSCs have a higher proliferation potential (21). The migration capacity of BM-MSCs was found higher than that of WJ-MSCs (24). In the present study, we applied smRNA-Seq to WJ-MSCs and BM-MSCs for identifying miRNAs that might regulate the activation of repair-related phenotypes of MSCs, as well as various specific target genes associated with these phenotypes.
MATERIALS AND METHODS
Isolation and cultivation of MSCs from BM and the Wharton’s jelly of umbilical cord
All human BM and WJ cells were obtained from unrelated donors with informed consent, and this study follows the guidelines of 1975 Helsinki Declaration. BM-MSCs and WJ-MSCs were isolated and cultured as previously described (21). In brief, all MSCs were cultured in MesenCult (Stem Cells Technology) or MesenPRO RS (Invitrogen) medium at 37°C in a 5% CO2 humidified incubator. Mononuclear cells from BM were separated by centrifugation using a Histopaque-1077 gradient (1077-1; Sigma) and plated in fresh culture medium. WJ cells were isolated from the umbilical cord using collagenase and trypsin within 24 h of collection. In both cases, non-adherent cells were removed after 24–72 h plating. On reaching confluency, the cells were treated with 0.05% trypsin-ethylenediaminetetraacetic acid and subcultured in fresh culture medium.
smRNA-Seq and data analysis
smRNA-Seq was performed on the Illumina Solexa GA IIx sequencing system (Illumina, San Diego, CA, USA) according to manufacturer’s instruction. The results were analyzed by an in-house bioinformatics pipelines (25). In brief, according to the length and sequence, identical adapter-trimmed Fastq sequences were integrated to give a series of non-redundant sequences and aligned to genome [University of California, Santa Cruz (UCSC) hg19] using the Bowtie aligner (26). Sequences that were able to be aligned with the transcribed genome region of known miRNAs (according to miRBase R19, http://www.mirbase.org/) were quantified as absolute expression values for each known miRNA. Expression values of specific miRNAs were calculated as the RPM value (reads per million mapped reads), namely RPM = C/MN × 106, where C is ‘read numbers aligned to a given miRNA chromosomal region’, M is ‘multiple mapping numbers across all miRNA regions of given read (i.e. the number of different miRNA chromosome locations mapped by the same read)’ and N is ‘total read numbers that map to human genome sequence of a single sample’. To minimize the effect of cross-mapping of sequences with uncertained genomic locations, the expression is divided by its number of cross-mapping events. The q-value (false discovery rate-adjusted P-value) was done by edgeR package (27) of the Bioconductor (http://www.bioconductor.org) suite of software for the R statistical programming language (http://www.r-project.org).
All smRNA-Seq data were deposited in NCBI Gene Expression Omnibus database (accession number GSE46989).
Microarray analysis
The total RNA from BM-MSCs transduced with pCDH-146a-5p or pCDH vector, as well as those of WJ-MSCs transfected with short interfering RNAs targeting miR-146a-5p or a green fluorescent protein (GFP) control, was isolated. mRNA profiling was carried out using Affymetrix Human U133 Plus 2.0 chips, and data analysis was performed as described previously (28). Robust Multi-array Average (RMA) log expression units were calculated from Affymetrix GeneChip array data using the ‘affy’ Bioconductor package in the R programming language. Gene expression levels were normalized using default RMA settings. The array results are deposited in the Gene Expression Omnibus database with accession number GSE44186.
Quantification of mRNA and miRNA expression
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR) were performed as described previously (12). Total RNA was extracted using Tri Reagent (Sigma-Aldrich Co., St. Louis, USA) according to the manufacturer’s instructions. For mRNA qPCR, complementary DNA was synthesized from 100–1000 ng of total RNA using RevertAid™ Reverse transcriptase kit (K1622, Fermentas, MA, USA). Amplification of genes was performed using specific primers (Supplementary Table S2), and Maxima™ SYBR Green qPCR Master Mix (K0222, Fermentas), on the StepOne™ sequence detector (Applied Biosystems, USA). The relative expression levels of target genes were normalized against the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). For miRNA qPCR, the expression levels of specific miRNAs were detected using stem-loop RT-qPCR (29). The primers designed and used for each miRNAs are listed in Supplementary Table S2; snRNA U6 was used as an internal control for normalization of miRNA expression.
Dual luciferase reporter assays
The full length of the 3′ untranslated region (3′ UTR) of SIKE1, and a 2-kb fragment from the 3′ UTR of CXCL12 that contains the predicted target site, as well as their various mutant forms that had been mutated within their seed regions, were cloned into the pGL3 basic reporter plasmid (Promega, Charbonnières, France). Next, 293T cells were co-transfected with the firefly luciferase reporter construct (500 ng) and either the miR-146a expression plasmid or the vector control (500 ng), together with an internal Renilla pRL-TK luciferase control plasmid (50 ng). The transformed cells were harvested at 24 h after transfection and their luciferase activity measured as the ratio of firefly luciferase activity to Renilla luciferase activity.
MSC migration, proliferation and differentiation assays
Cell migration
A total of 5 × 104 MSCs in 100 µl of culture medium were seeded into the upper insert with an 8-μm pore-size membrane (Corning, NY, USA), whereas 600 µl fresh culture medium containing 10% serum was added to the lower chamber. After 12 h of incubation, each membrane was fixed with 4% paraformaldehyde, and the MSCs present on the membrane were then stained with Hoechst 33342 solution (Sigma). The various numbers of migrated MSCs were determined by counting the number of nuclei beneath the filter membrane.
Cell proliferation
BM-MSCs and WJ-MSCs had their incubation started at the same passage, and the number of cells was counted every five days after subculture. For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assays, WJ-MSCs were transduced with the appropriate lentivirus expression vector. The day of infection was set as day 0. After various times of incubation, the cells were treated with 1% thiazolyl blue tetrazolium for 1–2 h at 37°C, followed by 0.1% sodium dodecyl sulfate in 2-propanol and thorough mixing. The results were obtained by measuring absorbance at a wavelength of 570 nm using a multiwell scanning spectrophotometer (Thermo, Multiskan Spectrum).
Cell differentiation
MSC differentiation was performed as described previously, with some modifications (21). BM-MSCs were transduced with lentivirus expressing miR-146a-5p or the vector control. One day post-infection, the medium was replaced with osteogenic or adipogenic induction medium.
RNA interference and plasmids of miRNA overexpression and knockdown
miR-146a-5p antagomir (IH-300630-05-0005; Dharmacon, MA, USA) or control oligomers (siRNA against GFP, Sigma, 20 nM) were transfected using a Microporator MP-100 (Invitrogen, USA). miR-146a-5p expression plasmid was purchased from SBI (System Biosciences, CA, USA). The lentiviral expression vector expressing hairpin sequence against miR-146a-5p was also purchased from SBI (System Biosciences, miRZip, MZIP000AA-1).
Flow cytometry and immunoblotting
Flow cytometry and immunoblotting were performed as described previously (21). For flow cytometry, MSCs in phosphate buffered saline were incubated with monoclonal antibodies against CD29 (303004, Biolegend), CD34 (MCA547PE, Serotec), CD44 (312306, Biolegend), CD45 (304006, Biolegend), CD73 (550257, BD), CD90 (MCA90F, Serotec) or HLA-DR (307604, Biolegend). Unstained MSCs were used as a negative control. For immunoblotting, antibodies against phospho-S536 p65 (#3031, Cell Signaling, USA), p65 (06-418, Millipore, USA) or GAPDH (Millipore, USA) were used, and GAPDH acted as the internal control.
Statistical analysis
All results are presented as mean ± standard deviation with statistical significance tested using 2-tailed Student’s t-tests.
RESULTS
Global analysis of miRNA expression in human MSCs by sequencing small RNAs
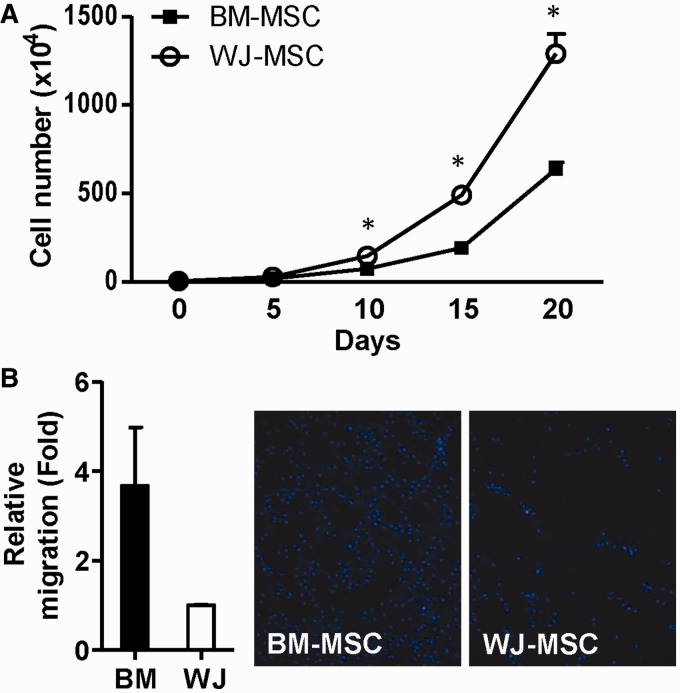
We isolated human MSCs from BM and WJ as described (21). Both types of MSC had a similar morphology and expressed common surface markers, namely CD73, CD29, CD90 and CD44 (Supplementary Figure S1). Hematopoietic markers such as CD34 and CD45 were lacking in both populations (Supplementary Figure S1B). We have previously shown that BM-MSCs show better osteogenic/adipogenic differentiation abilities than WJ-MSCs (21). We further evaluated functional variations in relation to the BM-MSCs and WJ-MSCs by subjecting cells of the same passage to cell proliferation and cell migration assays: WJ-MSCs were found to proliferate faster, whereas BM-MSCs had better cell motility (Figure 1).
Figure 1.
Differential proliferative and migratory abilities between BM-MSCs and WJ-MSCs. (A) Proliferative potentials of two MSCs. The total cell numbers of cultured WJ-MSCs and BM-MSCs were counted every 5 days till day 20 after plating. Results are presented as mean ± SD in two duplicates. *P < 0.05 by t-test. (B) Migratory ability of WJ-MSCs and BM-MSCs. Migrated cells were counted after 12 h incubation. Fold change in migration ability is compared against the WJ-MSCs. Representative images of migrated MSCs after staining with Hoechst 33342 are shown. Results are presented as mean ± SD from two independent experiments.
We hypothesized that stem cell miRNAs are involved in regulating these phenotypes. To explore this, we performed smRNA-Seq to decipher the expression levels of non-coding small RNA, especially miRNAs, expressed by BM-MSCs and WJ-MSCs; these MSCs had been obtained from four and three different donors, respectively, for allowing us to ignore inter-individual variations. To filter out stemness miRNAs that might be involved in MSC motility and/or growth, we also compared the miRNome patterns of MSCs with those of differentiated osteocytes and adipocytes derived from BM-MSCs. The successful differentiation of BM-MSCs was confirmed by the analysis of various markers by qPCR (Supplementary Figure S2). Using the Illumina Solexa platform, each sample smRNA-Seq reaction generated 17–36 million of high-quality reads that corresponded to 87–586 thousand of non-redundant reads (Supplementary Table S1).
Patterns of known miRNAs in undifferentiated and differentiated cells
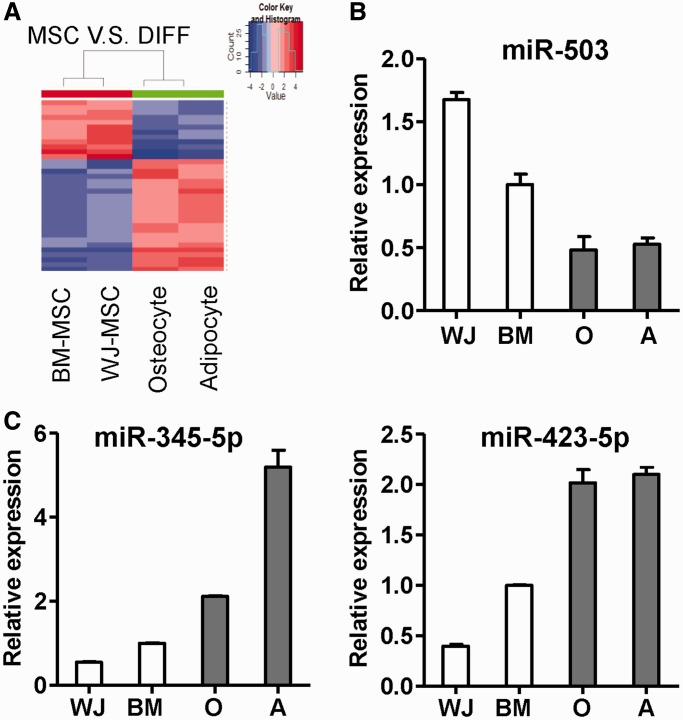
A total of 12 stem cell miRNAs and 23 progeny miRNAs were found to be differentially expressed when undifferentiated and differentiated cells were compared. A heatmap was then created to show details of their differential expression across stem cells and differentiated cells (Figure 2A), and these findings are expanded in Supplementary Figure S3. Examples of the miRNAs that were found to be upregulated in stem cells (miR-503) or in osteocytes/adipocytes (miR-345-5p, miR-423-5p) were validated by RT-qPCR (Figure 2B and C).
Figure 2.
miRNA patterns in undifferentiated MSCs and differentiated cells derived from BM-MSCs. (A) A heatmap shows the differential expression of miRNAs among stem cells and differentiated progeny cells. miRNAs differentially expressed between undifferentiated and differentiated cells (q < 0.01) were shown. (B and C) Validation of differential expressed miRNAs by RT-qPCR. Highly expressed miRNAs in undifferentiated MSCs (B) or differentiated cells (C) are shown. O: osteocytes; A: adipocytes.
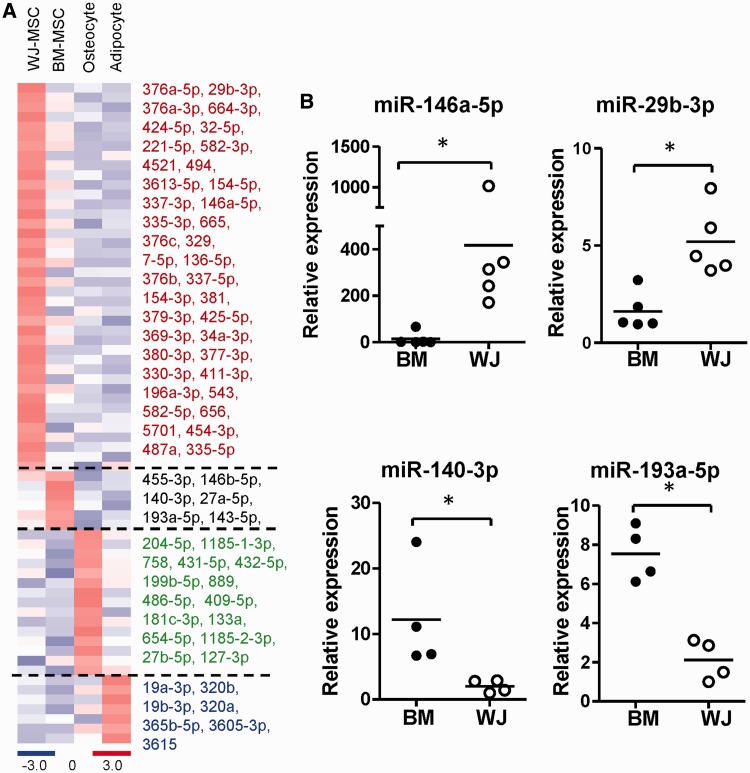
A total of 68 cell type-specific known miRNAs were identified as having fold changes of >2 and RPM >20: 6 miRNAs were found to be unique to BM-MSCs, 40 miRNAs to WJ-MSCs, 15 to osteocytes and 7 to adipocytes. A heatmap and details of their differential expression are presented in Figure 3A. Among all cell type-specific miRNAs in BM-MSC and WJ-MSC, miR-146a-5p was identified as significantly overexpressed at high abundance in WJ-MSCs (Supplementary Figure S4). RT-qPCR verified the unique expression patterns of miR-146a-5p and miR-29b-3p in WJ-MSCs, and miR-140-3p and miR-193-5p in BM-MSCs (Figure 3B). The expression level of miR-146a-5p was not significantly changed during MSC osteogenic or adipogenic differentiation (Supplementary Figure S5).
Figure 3.
Cell type-specific miRNAs. (A) A heatmap shows the specific expression of miRNAs in each cell type. (B) Validation of miRNA levels in different MSCs by RT-qPCR. Each spot represents an independent donor. Data are shown as mean of at least four independent donors. *P < 0.05 by t-test.
Functional module analysis reveals a crucial role for miR-146a-5p in MSC migration and proliferation
The above profiling data imply that miR-146a-5p is a stem cell-specific miRNA that is likely to be involved in MSC motility and/or proliferation, but not in differentiation. We used the Ingenuity Pathway Analysis (IPA) software to evaluate the possible functions of miR-146a-5p in MSCs. A total of 334 BM-MSC-enriched genes (from our previous gene expression array study (21)) were subjected to IPA to pinpoint possible miR-146a-5p functionality in MSCs. Genes regulated by miR-146a-5p (discovered by either wet lab experiments or bioinformatics target prediction) were connected to miR-146a-5p by IPA (Supplementary Figure S6). Genetic network analysis showed that miR-146a-5p targets genes were involved in both cell proliferation and migration, with P-values of 5.69E-3 and 4.40E-3, respectively (Supplementary Figure S6).
miR-146a-5p (previously known as miR-146a) has been associated with nuclear factor kappa-B (NF-κB) activity (Supplementary Figure S6); specifically, expression of miR-146a-5p is regulated by NF-κB through NF-κB binding sites in miR-146a-5p promoter (30), whereas NF-κB is targeted by miR-146a-5p in breast cancer (31); these interactions form a negative feedback loop. miR-146a-5p is also crucial for stem cell properties whereby miR-146a-5p is able to directly induce hematopoietic stem cells to differentiate into macrophages, as well as being able to modulate neural stem cell differentiation (32,33). miR-146a-5p has also been shown to induce the proliferation of vascular smooth muscle cells (34). Furthermore, miR-146a-5p is able to suppress cell invasion in pancreatic cancer (35). However, among MSCs, the various roles of miR-146a-5p in NF-κB regulation, cell proliferation and migration have not been investigated.
miR-146a-5p regulates MSC migration and proliferation, but not differentiation
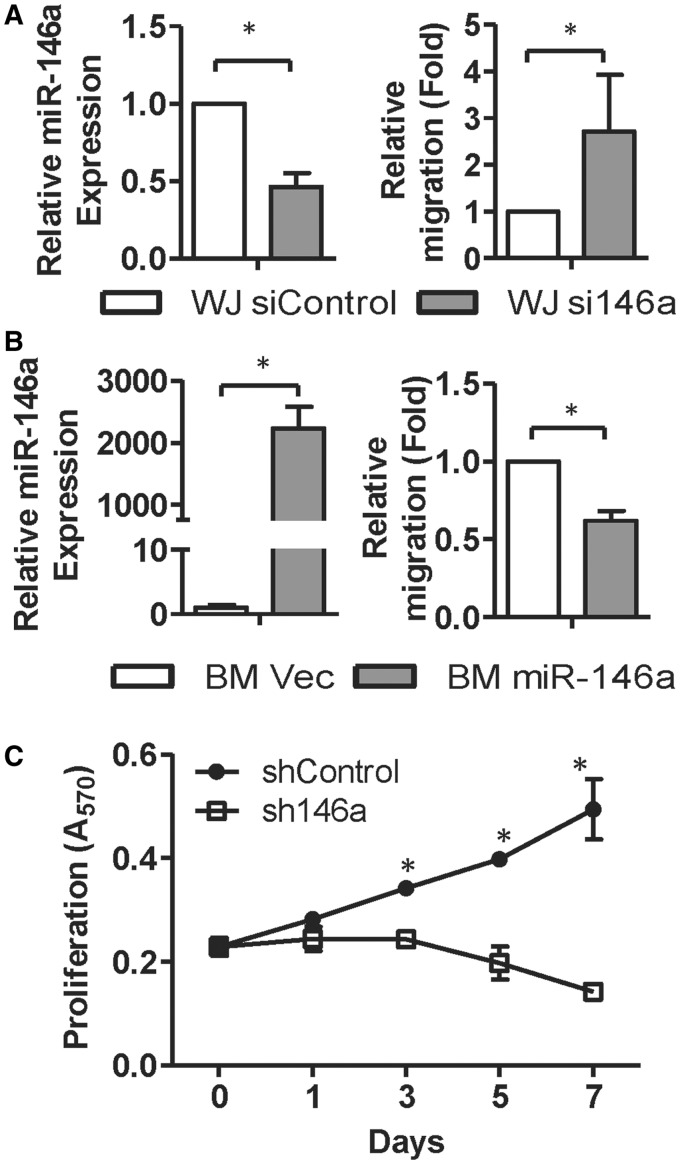
Because miR-146a-5p was found to be more abundant in WJ-MSCs than in BM-MSCs, we analyzed the effects of miR-146a-5p on MSC migration and proliferation. WJ-MSCs transfected with miR-146a-5p antagomirs showed a 50% reduction in expression level of miR-146a-5p, whereas in parallel their cell motility was increased compared with cells transfected with the control oligomer (si-Control in Figure 4A). BM-MSCs transduced with lentiviruses expressing miR-146a-5p at a multiplicity of infection of five showed an increased level of miR-146a-5p, and this was accompanied by decreased cell motility (Figure 4B). These results suggest that miR-146a-5p expression controls MSC motility.
Figure 4.
miR-146a-5p regulates MSC migration and proliferation. (A and B) miR-146a-5p suppresses MSC motility. WJ-MSCs transfected with siRNAs against GFP or miR-146a-5p (siControl or si146a; A), and BM-MSCs transduced with lentiviruses expressing miR-146a-5p or empty lentivirus vector control (B) were used in Transwell assays. miR-146a-5p levels were detected by RT-qPCR (left panels). Results are shown as mean ± SD from four experiments. *P < 0.05 by t-test. (C) miR-146a-5p controls the proliferation of WJ-MSCs. WJ-MSCs were transduced with lentiviruses encoding a short hairpin against miR-146a-5p (sh146a), and cell proliferation rate was measured at the indicated time points using MTT assays. Results are presented as mean ± SD in two duplicates. *P < 0.05 by t-test.
To determine the role of miR-146a-5p in MSC proliferation, MTT assays were conducted on WJ-MSCs at various time points after miR-146a-5p was knocked down. Cell proliferation was reduced in WJ-MSCs transduced with a lentivirus expressing a hairpin sequence targeting miR-146a-5p (sh146a) (Figure 4C). Previous studies have shown that NF-κB plays a crucial role in regulating MSC survival, proliferation and migration: Toll-like receptor ligand is able to activate NF-κB activity, reduce MSC migration ability and increase MSC proliferation, which implies that the NF-κB pathway contributes in MSC functionality (36). We have previously shown that the NF-κB protein level is higher in WJ-MSCs than in BM-MSCs (21). We found that knockdown of miR-146a-5p in WJ-MSCs led to a significant reduction in p65 phosphorylation (Supplementary Figure S7A) when examining NF-κB activity in WJ/sh146a or WJ/shControl cells using immunoblotting with antibodies against the phosphorylated/activated p65 (RelA) subunit of the NF-κB heterodimer. These results suggest that an increase in miR-146a-5p level in MSCs is able to stimulate NF-κB activity.
The various roles of miR-146a-5p in MSC differentiation were also monitored. BM-MSCs transduced with miR-146a-5p lentivirus or vector control were subjected into osteogenic and adipogenic differentiation, and various lineage-specific differentiation markers were measured every week up to week 2 or 3 using RT-qPCR or reverse transcription polymerase chain reaction (RT-PCR). The expression of differentiation markers was found to be unaffected by miR-146a-5p overexpression (Supplementary Figure S7B and C), which implies that miR-146a-5p does not participate in MSC osteogenesis or adipogenesis.
miR-146a-5p targets CXCL12 and SIKE1 in MSCs
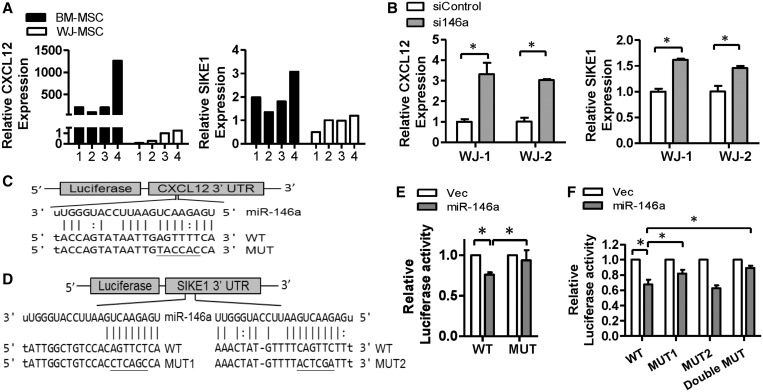
We used a combination of bioinformatics target prediction and gene-expression microarray data to pinpoint potential miR-146a-5p downstream targets to gain mechanistic insights into miR-146a-5p functionality. The transcription profile of BM-MSCs with miR-146a-5p overexpression, as well as that of WJ-MSCs with miR-146a-5p knocked down, was explored by gene-expression microarray. We focused on genes that were downregulated in BM-MSCs when miR-146a-5p was overexpressed, and were upregulated in WJ-MSCs when miR-146a-5p was knocked down (fold change >1.5). A total of 73 genes were identified at this step. Next, because miR-146a-5p has been shown to be downregulated in BM-MSCs, we considered only genes that were more abundant in BM-MSCs. A total of 15 of the original 73 genes fitted this criterion. Among these 15 genes, 7 possessed putative miR-146a-5p binding sites in their 3′ UTRs according to the miRanda and PITA prediction programs. Among these genes, we focused on CXCL12 and SIKE1; this was because CXCL12 is able to induce MSC migration (37), and SIKE1 is a suppressor of IKKε, a NF-κB modulator through a non-canonical pathway (38). We found that the endogenous levels of CXCL12 and SIKE1 were higher in BM-MSCs than in WJ-MSCs (Figure 5A), and that the mRNA expression levels of CXCL12 and SIKE1 were increased in WJ-MSCs that had been transfected with miR-146a-5p antagomirs using cells from independent individuals (Figure 5B).
Figure 5.
CXCL12 and SIKE1 are direct targets of miR-146a-5p. (A) The relative expression levels of CXCL12 and SIKE1 in both MSC types were detected by RT-qPCR (n = 4). (B) CXCL12 and SIKE1 mRNA levels in WJ-MSC transfected with miR-146a-5p antagomirs were analyzed by RT-qPCR. Results are shown as mean ± SD from two duplicates. *P < 0.05 by t-test. (C and D) Putative miR-146a-5p target sites in the CXCL12-3′ UTR (C) and SIKE1-3′ UTR (D). UTR: untranslated region; WT: wild type; MUT: mutant. (E and F) Luciferase reporter assays using the CXCL12-3′ UTR (E) or the SIKE1-3′ UTR (F) in 293T cells transfected with control (Vec) or miR-146a-5p expression vector. Results are presented as mean ± SD of three independent experiments.
The direct repression of CXCL12 and SIKE1 by miR-146a-5p was proved by reporter assays. Specifically, miR-146a-5p was found to repress expression of luciferase in constructs containing the CXCL12 or SIKE1 3′ UTR fused downstream of the luciferase gene, respectively (Figure 5C and D, the wild type (WT) group). Thus, the luciferase assays showed that miR-146a-5p directly interacts with the CXCL12 and SIKE1 3′ UTRs, which results in a modification of gene expression (Figure 5E and F). This repression was reversed by independently mutating the putative miR-146a-5p-binding sites in the CXCL12 and the SIKE1 3′ UTR (Figure 5C and D, the Mut group). In the SIKE1 3′ UTR, only mutation of site1 restored luciferase expression, showing that site1 is the potential binding site for miR-146a-5p (Figure 5F).
miR-146a-5p modulates MSC motility and proliferation via CXCL12 and SIKE1
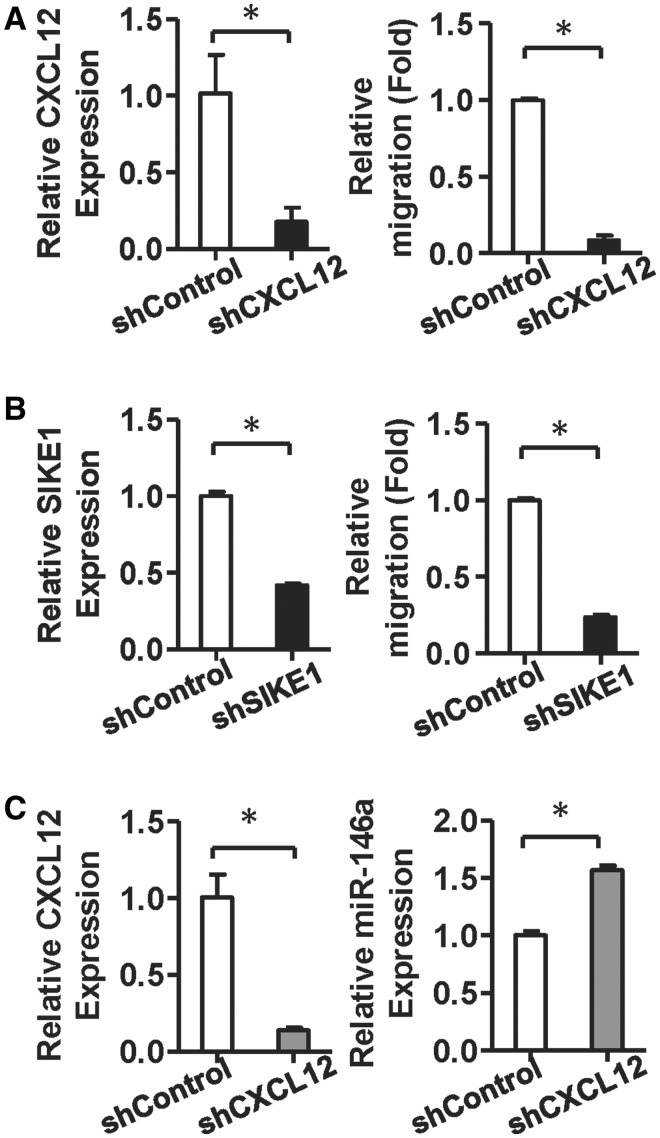
To further dissect the functional impact of CXCL12 and SIKE1 on miR-146a-5p-mediated MSC motility and proliferation, we compared the migration and amplification abilities of MSCs that had either CXCL12 or SIKE1 knockdown. Knockdown of either CXCL12 or SIKE1 was able to significantly decrease the migration ability of WJ-MSCs (Figure 6A and B). The proliferation rate of WJ-MSCs was reduced when the miR-146a-5p level was knocked down. Furthermore, knockdown of SIKE1 in the miR-146a-5p knockdown WJ-MSCs, but not knockdown of CXCL12, was able to restore WJ-MSC proliferation (Supplementary Figure S8A).
Figure 6.
Knocking down CXCL12 and SIKE1 inhibits WJ-MSC migratory ability, and CXCL12 silencing increases miR-146a-5p levels. (A and B) CXCL12 and SIKE1 control MSC motility. A total of 5 × 104 WJ-MSCs were transduced with CXCL12 (A) or SIKE1 (B) shRNA and then assessed by Transwell migration assay. Migrated cells were counted after 12 h. Results are shown as mean ± SD from two duplicates. *P < 0.05 by t-test. (C) miR-146a-5p expression in WJ-MSCs after knockdown of CXCL12. WJ-MSCs transduced with CXCL12 shRNA for 48 h were harvested. All results are presented as mean ± SD from two duplicates experiments. *P < 0.05 by t-test.
We next examined whether the proliferation rate of WJ-MSCs was associated with the phosphorylation level of the NF-κB p65 subunit. p65 phosphorylation was found to be decreased in miR-146a-5p-knockdown WJ-MSCs, but this reduction could be rescued in MSCs when there is miR-146a-5p and SIKE1 double knockdown (Supplementary Figure S8B). Thus, miR-146a-5p enhanced NF-κB activity in WJ-MSCs and, furthermore, NF-κB signaling is able to induce miR-146a-5p expression through binding to the NF-κB binding sites that are present in the miR-146a-5p promoter (30). Knockdown of CXCL12 resulted in an increased expression of miR-146a-5p in WJ-MSCs (Figure 6C), implying that miR-146a-5p positively regulated its expression in MSCs via CXCL12 and NF-κB signaling (Figure 7).
Figure 7.
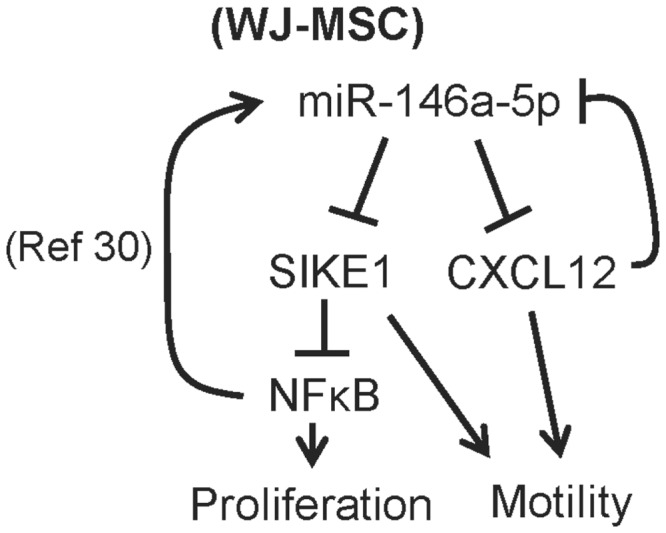
The miR146a-5p gene regulatory pathways involve in WJ-MSC proliferation and motility.
DISCUSSION
Cell-based therapies using multipotent MSCs for organ regeneration have been and are being pursued for a variety of diseases. Clues on molecules controlling MSC biology can be obtained by comparing MSCs with different biological activities. For example, a comparison of MSCs isolated from Murphy Roths Large MRL/MpJ mice was able to demonstrate enhanced regenerative capacity compared with MSCs from C57BL/6 (WT) mice; these results suggest that sFRP2 is a key molecule that affects biogenesis and is important to obtaining a superior MSC regenerative phenotype (39). It is clear that MSCs from different human anatomic locations possess diverse biological activities; these include differences in differentiation, proliferation and migration potentials. miRNA functions are suggested to regulate various biological processes in stem cells (40,41). To thoroughly characterize the miRNA changes in MSC, in this study we used smRNA-Seq to profile miRNA expression patterns in WJ-MSCs, BM-MSCs and their differentiated progenies. Follow-up wet lab studies showed that MSCs use a complex miRNA–mRNA network to modulate their functionalities. In this network, miR-146a-5p and its downstream genes play a crucial role in inhibiting cell migration while promoting cell proliferation.
Previous studies have demonstrated that miRNAs act as important regulators for MSCs. For example, the expression of miR-193 was increased in low-level laser irradiation-treated MSCs, which enhanced MSC proliferation by targeting inhibitor of growth family, member 5 (42). Gain–loss function experiments showed that miR-21 negatively regulates the proliferation of adipose tissue-derived MSCs through regulating signal transducer and activator of transcription 3 (STAT3) expression (43). As to cell differentiation, miR-124 is downregulated during cardiomyocyte differentiation of BM-derived MSCs via targeting STAT3, which is essential in the induction of transdifferentiation (44). On the other hand, miR-138 (10) and miR-22 (45) are involved in MSC osteogenesis, and miR-30 (46) and miR-204 (11) in adipogenesis. In this study, the aforementioned miR-22 is upregulated in both osteogenic and adipogenic differentiated BM-MSCs. miR-22, as well as other miRNAs identified in Supplementary Figure S3, may be involved in common mesenchymal differentiation pathways, rather than being lineage-specific differentiation regulators. miRNAs associated with lineage-specific differentiation were pinpointed in the present study (Figure 3A), and their roles in MSC differentiation awaited characterization. Among these miRNAs, miR-19a-3p and miR-19b-3p, which are encoded by the miR-17-92 cluster, are increased in BM-MSC-derived adipocytes. Overexpression of miR-17-92 could accelerate adipocyte differentiation in mouse preadipocyte 3T3L1 (47). Our results showed that miR-19a-3p and miR-19b-3p may be the critical miRNAs regulating MSC adipogenesis.
CXCL12, also known as stromal cell-derived factor-1, is an α-chemokine that is also a chemoattractant for other stem cells such as hematopoietic progenitor cells. CXCL12 stimulates migration by rearranging the actin cytoskeleton, increasing focal adhesion and matrix metalloproteinases production in MSCs (48,49). CXCL12 is significantly upregulated in MSCs exposed to tumor cell-conditioned medium compared with cells treated with control medium, which suggests that CXCL12 signaling is important to bring about MSC migration toward the tumor microenvironment (48,49). SIKE1 (suppressor of IKKε) is known to interact with IKKε and TANK-binding kinase 1 to bring about inhibition of virus-triggered and toll-like receptor 3 (TLR3)-triggered activation (50). This does not occur via IκB-α degradation, but rather IKKε and TANK-binding kinase 1 regulate NF-κB activity through a non-canonical pathway that directly targets the p65 subunit of the NF-κB complex via phosphorylation of serine 536 (38). In cancer cells, IKKε is involved in regulating constitutive NF-κB activity, but not cytokine-induced NF-κB activity (51). In the context of WJ-MSCs, we proved that miR-146a-5p is a new CXCL12 and SIKE1 inhibitor. Regulating miR-146a-5p expression in MSC may benefit engraftment of transplanted MSC that is homing onto injured tissues.
Injured tissues secrete CXCL12 to recruit MSC for regeneration (52,53). The CXCL12/CXCR4 axis mediates MSC migration through activating PI3K/AKT (54), MAPK/ERK (48) and Jak/Stat (48) signaling pathways. We found that miR-146a-5p is a downstream of CXCL12 (Figure 6C). Inhibiting CXCL12 expression resulted in miR-146a-5p upregulation. How CXCL12/CXCR4 signaling pathways regulate the expression of miR-146a-5p is still unexplored. Promoter of miR-146a gene contains potential binding sites of STAT, c-ETS, PU.1 and NF-κB (55–57). Expression of miR-146a-5p may be regulated by these transcriptional factors in response to CXCL12 stimulation. Transformed B lymphoma cells possess increased AKT phosphorylation and oncomiR miR-21 expression, as well as downregulated miR-146a levels (58), indicating the PI3K/AKT pathway may involve in the activation of miR-146a-5p. Another candidate circuit involved in miR-146a-5p regulation is the NF-κB pathway: there are two NF-κB binding sites within the miR-146a-5p promoter region, and in vitro reporter assays showed that miR-146a-5p is induced via a NF-κB-dependent manner (30). We found that knockdown NFKB1 resulted in downregulated miR-146a-5p in WJ-MSCs (data not shown).
The uncoupled MSC motility and growth in vivo may enable MSCs to migrate to regenerative sites via circulation at a single cell level while avoiding getting stuck in small blood vessels, hence benefitting MSC-mediated tissue repairing. Among MSCs, migrated cells have been shown to consist of more cells in G0/G1 phase and less cells in S and G2/M phase compared with non-migrated cells (59). Activation of NF-κB activity by TLR ligands is able to increase MSC proliferation but decreases MSC basal motility (36). Comparing of migrating and non-migrating fetal BM-MSCs showed that the nuclear receptors, Nur77 and Nurr1, can increase migration of MSC, while decreasing proliferation (60). The relationships between cell cycle and cell motility have been investigated in other stem cells or cancer cells. In HSCs, inhibition of the cell cycle may enhance HSC transplantation efficiency in vivo (61). Transforming growth factor β (TGF-β) and bone morphogenetic protein 4, on the other hand, mediate growth arrest and increase in cellular migration and invasion, which may eventually benefit distant tumor metastasis (62,63). Our results may also be applied to cancer research, as miR-146a-5p is a known migration inhibitor in various types of cancers (35,64).
In summary, this study provides comprehensive miRNome profiles of two different types of MSCs, as well as cells differentiated from BM-MSCs. We envision that our genomic data will help to create a roadmap to a better understanding of MSC stemness and the regulation of MSC differentiation. Because both types of MSCs have potential as sources of stem cells for clinical transplantation, our findings have implications in terms of the clinical applications of MSCs. A manipulation of the level of miR-146a-5p in transplanted MSCs may benefit cell-based therapy in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council [NSC; NSC101-2320-B-010-059-MY3, NSC101-2627-B-010-003 and NSC101-2321-B-010-011]; Tri-Service General Hospital [TSGH-C102-027]; Veterans General Hospitals University System of Taiwan (VGHUST) Joint Research Program, Tsou’s Foundation [102DFA2200016]; National Health Research Institutes [NHRI-EX102-10254SI]; Taipei Veteran General Hospital [Cancer Excellence Center Plan DOH101-TD-C-111-007]; Taipei City Hospital [10201-62-070] and National Yang-Ming University [Ministry of Education, Aim for the Top University Plan]; This work was also supported in part by the UST-UCSD International Center for Excellence in Advanced Bioengineering sponsored by the Taiwan NSC I-RiCE Program [NSC101-2911-I-009-101]. Funding for open access charge: National Science Council [NSC101-2320-B-010-059-MY3].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Wei-Chung Cheng, Cheng-Fong Tsai and Shih-Ting Chang for the help on smRNA-Seq analysis, and technical services of Affymetrix microarray provided by Microarray and Gene Expression Analysis Core and that of smRNA-Seq provided by Sequencing Core of National Research Program for Genome Medicine, National Yang-Ming University, VGH Genome Research Center, as well as the shRNA knockdown plasmids provided by National RNAi Core Facility, Taiwan.
REFERENCES
- 1.Prockop DJ. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 2.Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WW, Gordon PL, Neel M, Sussman M, Orchard P, Marx JC, Pyeritz RE, et al. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat. Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- 3.Kawada H, Fujita J, Kinjo K, Matsuzaki Y, Tsuma M, Miyatake H, Muguruma Y, Tsuboi K, Itabashi Y, Ikeda Y, et al. Nonhematopoietic mesenchymal stem cells can be mobilized and differentiate into cardiomyocytes after myocardial infarction. Blood. 2004;104:3581–3587. doi: 10.1182/blood-2004-04-1488. [DOI] [PubMed] [Google Scholar]
- 4.Battiwalla M, Hematti P. Mesenchymal stem cells in hematopoietic stem cell transplantation. Cytotherapy. 2009;11:503–515. doi: 10.1080/14653240903193806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borlongan CV, Glover LE, Tajiri N, Kaneko Y, Freeman TB. The great migration of bone marrow-derived stem cells toward the ischemic brain: therapeutic implications for stroke and other neurological disorders. Prog. Neurobiol. 2011;95:213–228. doi: 10.1016/j.pneurobio.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Wynn RF, Hart CA, Corradi-Perini C, O'Neill L, Evans CA, Wraith JE, Fairbairn LJ, Bellantuono I. A small proportion of mesenchymal stem cells strongly expresses functionally active CXCR4 receptor capable of promoting migration to bone marrow. Blood. 2004;104:2643–2645. doi: 10.1182/blood-2004-02-0526. [DOI] [PubMed] [Google Scholar]
- 8.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 9.Tome M, Lopez-Romero P, Albo C, Sepulveda JC, Fernandez-Gutierrez B, Dopazo A, Bernad A, Gonzalez MA. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M. MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl Acad. Sci. USA. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–364. doi: 10.1002/stem.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang SJ, Weng SL, Hsieh JY, Wang TY, Chang MD, Wang HW. MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med. Genomics. 2011;4:65. doi: 10.1186/1755-8794-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS One. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skreka K, Schafferer S, Nat IR, Zywicki M, Salti A, Apostolova G, Griehl M, Rederstorff M, Dechant G, Huttenhofer A. Identification of differentially expressed non-coding RNAs in embryonic stem cell neural differentiation. Nucleic Acids Res. 2012;40:6001–6015. doi: 10.1093/nar/gks311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissels U, Wild S, Tomiuk S, Hafner M, Scheel H, Mihailovic A, Choi YH, Tuschl T, Bosio A. Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells. Stem Cells. 2011;29:847–857. doi: 10.1002/stem.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo JK, Kim J, Choi SJ, Noh HM, Kwon YD, Yoo H, Yi HS, Chung HM, Kim JK. Discovery and characterization of novel microRNAs during endothelial differentiation of human embryonic stem cells. Stem Cells Dev. 2012;21:2049–2057. doi: 10.1089/scd.2011.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J. Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 19.Secco M, Zucconi E, Vieira NM, Fogaca LL, Cerqueira A, Carvalho MD, Jazedje T, Okamoto OK, Muotri AR, Zatz M. Multipotent stem cells from umbilical cord: cord is richer than blood! Stem Cells. 2008;26:146–150. doi: 10.1634/stemcells.2007-0381. [DOI] [PubMed] [Google Scholar]
- 20.Wang HS, Hung SC, Peng ST, Huang CC, Wei HM, Guo YJ, Fu YS, Lai MC, Chen CC. Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord. Stem Cells. 2004;22:1330–1337. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh JY, Fu YS, Chang SJ, Tsuang YH, Wang HW. Functional module analysis reveals differential osteogenic and stemness potentials in human mesenchymal stem cells from bone marrow and Wharton's jelly of umbilical cord. Stem Cells Dev. 2010;19:1895–1910. doi: 10.1089/scd.2009.0485. [DOI] [PubMed] [Google Scholar]
- 22.Chen HC, Lee YS, Sieber M, Lu HT, Wei PC, Wang CN, Peng HH, Chao AS, Cheng PJ, Chang SD, et al. MicroRNA and messenger RNA analyses of mesenchymal stem cells derived from teeth and the Wharton jelly of umbilical cord. Stem Cells Dev. 2012;21:911–922. doi: 10.1089/scd.2011.0186. [DOI] [PubMed] [Google Scholar]
- 23.Baksh D, Yao R, Tuan RS. Comparison of proliferative and multilineage differentiation potential of human mesenchymal stem cells derived from umbilical cord and bone marrow. Stem Cells. 2007;25:1384–1392. doi: 10.1634/stemcells.2006-0709. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Zhang XA, Wang H, Wang X, Meng CL, Chan CY, Yew DT, Tsang KS, Li K, Tsai SN, et al. Comparative proteomic analysis of mesenchymal stem cells derived from human bone marrow, umbilical cord, and placenta: implication in the migration. Proteomics. 2009;9:20–30. doi: 10.1002/pmic.200701195. [DOI] [PubMed] [Google Scholar]
- 25.Cheng WC, Chung IF, Huang TS, Chang ST, Sun HJ, Tsai CF, Liang ML, Wong TT, Wang HW. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2013;41:D285–D294. doi: 10.1093/nar/gks1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang TS, Hsieh JY, Wu YH, Jen CH, Tsuang YH, Chiou SH, Partanen J, Anderson H, Jaatinen T, Yu YH, et al. Functional network reconstruction reveals somatic stemness genetic maps and dedifferentiation-like transcriptome reprogramming induced by GATA2. Stem Cells. 2008;26:1186–1201. doi: 10.1634/stemcells.2007-0821. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghani S, Riemke P, Schonheit J, Lenze D, Stumm J, Hoogenkamp M, Lagendijk A, Heinz S, Bonifer C, Bakkers J, et al. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood. 2011;118:2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei J, Bachoo R, Zhang CL. MicroRNA-146a inhibits glioma development by targeting Notch1. Mol. Cell Biol. 2011;31:3584–3592. doi: 10.1128/MCB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, Su M, Han Y, Shi HJ, Wen JK. miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 2011;12:56–62. doi: 10.1038/embor.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Vandenboom TG, II, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pevsner-Fischer M, Morad V, Cohen-Sfady M, Rousso-Noori L, Zanin-Zhorov A, Cohen S, Cohen IR, Zipori D. Toll-like receptors and their ligands control mesenchymal stem cell functions. Blood. 2007;109:1422–1432. doi: 10.1182/blood-2006-06-028704. [DOI] [PubMed] [Google Scholar]
- 37.Son BR, Marquez-Curtis LA, Kucia M, Wysoczynski M, Turner AR, Ratajczak J, Ratajczak MZ, Janowska-Wieczorek A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells. 2006;24:1254–1264. doi: 10.1634/stemcells.2005-0271. [DOI] [PubMed] [Google Scholar]
- 38.Chau TL, Gioia R, Gatot JS, Patrascu F, Carpentier I, Chapelle JP, O'Neill L, Beyaert R, Piette J, Chariot A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 2008;33:171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Alfaro MP, Pagni M, Vincent A, Atkinson J, Hill MF, Cates J, Davidson JM, Rottman J, Lee E, Young PP. The Wnt modulator sFRP2 enhances mesenchymal stem cell engraftment, granulation tissue formation and myocardial repair. Proc. Natl Acad. Sci. USA. 2008;105:18366–18371. doi: 10.1073/pnas.0803437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Leonardo TR, Schultheisz HL, Loring JF, Laurent LC. The functions of microRNAs in pluripotency and reprogramming. Nat. Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Huang W, Wu Y, Hou J, Nie Y, Gu H, Li J, Hu S, Zhang H. MicroRNA-193 pro-proliferation effects for bone mesenchymal stem cells after low-level laser irradiation treatment through inhibitor of growth family, member 5. Stem Cells Dev. 2012;21:2508–2519. doi: 10.1089/scd.2011.0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YJ, Hwang SH, Cho HH, Shin KK, Bae YC, Jung JS. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J. Cell Physiol. 2012;227:183–193. doi: 10.1002/jcp.22716. [DOI] [PubMed] [Google Scholar]
- 44.Cai B, Li J, Wang J, Luo X, Ai J, Liu Y, Wang N, Liang H, Zhang M, Chen N, et al. microRNA-124 regulates cardiomyocyte differentiation of bone marrow-derived mesenchymal stem cells via targeting STAT3 signaling. Stem Cells. 2012;30:1746–1755. doi: 10.1002/stem.1154. [DOI] [PubMed] [Google Scholar]
- 45.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, Li H, Han Q, Zhao RC. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaragosi LE, Wdziekonski B, Brigand KL, Villageois P, Mari B, Waldmann R, Dani C, Barbry P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011;12:R64. doi: 10.1186/gb-2011-12-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl Acad. Sci. USA. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao H, Priebe W, Glod J, Banerjee D. Activation of signal transducers and activators of transcription 3 and focal adhesion kinase by stromal cell-derived factor 1 is required for migration of human mesenchymal stem cells in response to tumor cell-conditioned medium. Stem Cells. 2009;27:857–865. doi: 10.1002/stem.23. [DOI] [PubMed] [Google Scholar]
- 49.Ries C, Egea V, Karow M, Kolb H, Jochum M, Neth P. MMP-2, MT1-MMP, and TIMP-2 are essential for the invasive capacity of human mesenchymal stem cells: differential regulation by inflammatory cytokines. Blood. 2007;109:4055–4063. doi: 10.1182/blood-2006-10-051060. [DOI] [PubMed] [Google Scholar]
- 50.Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adli M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J. Biol. Chem. 2006;281:26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 52.Ponomaryov T, Peled A, Petit I, Taichman RS, Habler L, Sandbank J, Arenzana-Seisdedos F, Magerus A, Caruz A, Fujii N, et al. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J. Clin. Invest. 2000;106:1331–1339. doi: 10.1172/JCI10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kollet O, Shivtiel S, Chen YQ, Suriawinata J, Thung SN, Dabeva MD, Kahn J, Spiegel A, Dar A, Samira S, et al. HGF, SDF-1, and MMP-9 are involved in stress-induced human CD34+ stem cell recruitment to the liver. J. Clin. Invest. 2003;112:160–169. doi: 10.1172/JCI17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Yu X, Lin S, Li X, Zhang S, Song YH. Insulin-like growth factor 1 enhances the migratory capacity of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2007;356:780–784. doi: 10.1016/j.bbrc.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 55.Motsch N, Pfuhl T, Mrazek J, Barth S, Grasser FA. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 56.Curtale G, Citarella F, Carissimi C, Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V, Macino G. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115:265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 57.Jurkin J, Schichl YM, Koeffel R, Bauer T, Richter S, Konradi S, Gesslbauer B, Strobl H. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 58.Rosato P, Anastasiadou E, Garg N, Lenze D, Boccellato F, Vincenti S, Severa M, Coccia EM, Bigi R, Cirone M, et al. Differential regulation of miR-21 and miR-146a by Epstein-Barr virus-encoded EBNA2. Leukemia. 2012;26:2343–2352. doi: 10.1038/leu.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maijenburg MW, Noort WA, Kleijer M, Kompier CJ, Weijer K, van Buul JD, van der Schoot CE, Voermans C. Cell cycle and tissue of origin contribute to the migratory behaviour of human fetal and adult mesenchymal stromal cells. Br. J. Haematol. 2010;148:428–440. doi: 10.1111/j.1365-2141.2009.07960.x. [DOI] [PubMed] [Google Scholar]
- 60.Maijenburg MW, Gilissen C, Melief SM, Kleijer M, Weijer K, Ten Brinke A, Roelofs H, Van Tiel CM, Veltman JA, de Vries CJ, et al. Nuclear receptors Nur77 and Nurr1 modulate mesenchymal stromal cell migration. Stem Cells Dev. 2012;21:228–238. doi: 10.1089/scd.2011.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cashman J, Dykstra B, Clark-Lewis I, Eaves A, Eaves C. Changes in the proliferative activity of human hematopoietic stem cells in NOD/SCID mice and enhancement of their transplantability after in vivo treatment with cell cycle inhibitors. J. Exp. Med. 2002;196:1141–1149. doi: 10.1084/jem.20010916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ungefroren H, Groth S, Sebens S, Lehnert H, Gieseler F, Fandrich F. Differential roles of Smad2 and Smad3 in the regulation of TGF-beta1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: control by Rac1. Mol. Cancer. 2011;10:67. doi: 10.1186/1476-4598-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ketolainen JM, Alarmo EL, Tuominen VJ, Kallioniemi A. Parallel inhibition of cell growth and induction of cell migration and invasion in breast cancer cells by bone morphogenetic protein 4. Breast Cancer Res. Treat. 2010;124:377–386. doi: 10.1007/s10549-010-0808-0. [DOI] [PubMed] [Google Scholar]
- 64.Kogo R, Mimori K, Tanaka F, Komune S, Mori M. Clinical significance of miR-146a in gastric cancer cases. Clin. Cancer Res. 2011;17:4277–4284. doi: 10.1158/1078-0432.CCR-10-2866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.