Abstract
Mediator is a prominent multisubunit coactivator that functions as a bridge between gene-specific activators and the basal RNA polymerase (Pol) II initiation machinery. Here, we study the poorly documented role of Mediator in basal, or activator-independent, transcription in vivo. We show that Mediator is still present at the promoter when the Pol II machinery is recruited in the absence of an activator, in this case through a direct fusion between a basal transcription factor and a heterologous DNA binding protein bound to the promoter. Moreover, transcription resulting from activator-independent recruitment of the Pol II machinery is impaired by inactivation of the essential Mediator subunit Med17 due to the loss of Pol II from the promoter. Our results strongly support that Mediator is an integral component of the minimal machinery essential in vivo for stable Pol II association with the promoter.
INTRODUCTION
Mediator is a large multisubunit complex, conserved throughout eukaryotes, that plays an essential role in transcription of protein-encoding genes. It was first discovered in yeast due to its ability to support activator-driven transcription on naked DNA templates in a cell-free system reconstituted with purified RNA polymerase II (Pol II) and general transcription factors (GTFs; including TATA-binding protein (TBP), Transcription Factor (TF) IIB, TFIIF, TFIIE and TFIIH) (1). Mediator was shown to interact with numerous yeast and mammalian gene-specific transcription activators as well as with Pol II and GTFs, supporting a role as a bridge between gene-specific regulators bound at enhancers and the basal Pol II machinery assembled at the core promoter (2). Electron microscopy analyses combined with biochemical and genetics data have led to a model of topological organization in which the 25 subunits forming the yeast Mediator complex are distributed into four distinct modules named as head, middle, tail and CDK8 (3). The head, middle and tail modules constitute the core Mediator. In the presence of Pol II, the core Mediator assumes an elongated shape and makes multiple contacts with Pol II through the head and middle modules (4). The CDK8 module, which comprises a cyclin–kinase pair, associates reversibly and under specific conditions with the core complex and is mainly involved in negative regulations (5,6).
A widespread model assumes that Mediator is recruited to promoters by gene-specific activators bound to enhancer elements before binding of Pol II and GTFs (2). This model is supported by various studies, notably in yeast, showing spatial, temporal and physical separation between Mediator and Pol II recruitment at some promoters (7–9). Mediator tail module is viewed as a major target of gene-specific activators because of the numerous interactions evidenced between subunits of this module and various activators both in yeast and in mammals; however, a number of interactions involving subunits of other modules were also reported (10,11). A primary key role of Mediator once recruited by activators would be to facilitate the recruitment of GTFs and Pol II to form a pre-initiation complex (PIC). Studies using electron microscopy and single-particle-reconstruction techniques have shown that human Mediator undergoes conformational shifts on binding of activators to its tail module, supporting an allosteric model in which activators would trigger a wave of remodeling within Mediator ultimately stabilizing its association with Pol II and impacting assembly and activity of the PIC (12–14). Considering the biochemical stability of the ternary complex between Mediator, Pol II and TFIIF (13,14), an alternative possibility could be that Mediator is recruited as a larger, holoenzyme-like complex containing also Pol II. The two models are not exclusive in view of the results by Esnault et al. (15), suggesting multiple pathways for recruitment of Pol II and formation of the PIC in vivo.
In addition to its well-documented role in supporting activated transcription, Mediator was also shown to stimulate basal, or activator-free, transcription in both yeast and mammalian in vitro systems (16–19). This observation, together with the fact that inactivation of Med17 (also known as Srb4) impairs transcription of the majority of protein-encoding genes in yeast (6), has been taken into account as evidence that Mediator functions as a GTF, comparable in importance with Pol II and other basal factors for transcription initiation (19). However, there is no evidence to date that Mediator can bind promoter DNA and function independently of activator in vivo. Genome-wide localization studies using chromatin immunoprecipitation coupled with microarrays were carried out in yeast, but the results were conflicting and it has been unclear whether Mediator is always present at active promoters (20–23). Interpretation of these studies is also complicated by the strong possibility that formaldehyde does not crosslink Mediator at enhancer and core promoter sequences with the same efficiency. Therefore, the question of knowing whether Mediator is an intrinsic component of the basal machinery required for transcription initiation by Pol II in vivo remains unresolved. To address this issue, we have carried out ‘activator bypass’ experiments in yeast using molecular tools designed for recruiting the transcription machinery to a test promoter in the absence of classical activator, in this case through artificial tethering of a GTF (24,25). We have used these tools to determine whether Mediator is recruited alongside the Pol II machinery and is still required for transcription in this context. Our results show that Mediator is indeed found at the promoter when the Pol II transcription machinery is recruited through tethering of TFIIB or TBP. Moreover, we show that inactivation of the head module impairs activator-independent transcription driven by artificial recruitment of TFIIB or TBP. Altogether, our results provide evidence that Mediator is an integral part of the basal Pol II transcription machinery in vivo and functions as a general transcription initiation factor.
MATERIALS AND METHODS
Yeast strains and media
Genotypes are given in Supplementary Table S1. Strains expressing the chromosomal SUA7 allele under the control of the GAL1 promoter were generated by integrating the KanMX6-PGAL1 cassette from Longtine et al. (26) upstream of the coding sequence of SUA7 using standard procedures. Y805 was generated from W303-1A. Y807, Y809 and Y811 were generated from CL18, CL7 and DY3168, respectively, described in Leroy et al. (27). Y822 and Y823 were generated from Y400 and Y402, which contain a null allele of MED17(SRB4) on the chromosome and plasmids RY2844 (SRB4+, CEN, LEU2) or RY2882 (srb4-138, CEN, LEU2) in the W303-1A genetic background (6). Y892 was generated from Y14, which corresponds to W303-1A with an amino terminal HA-tagged TBP at the chromosome (28). Y893 and Y894 were generated from Y80 and Y84, which contain a TAP-KlTRP1 cassette inserted before the stop codon of Med5 or Med14, respectively (9). Y909 and Y911 were generated from two isogenic strains containing either the wild-type RPB1 allele or the rpb1-1 allele at the chromosome, described in Nonet et al. (29). Y959 and Y960 were generated from two isogenic strains containing either the wild-type KIN28 allele or the kin28ts3 allele at the chromosome, described in Valay et al. (30). Y963 was generated from a met4Δ::TRP1 strain, in which nucleotides 180–1848 of MET4 open reading frame (ORF) were replaced by TRP1. This strain was crossed with Y84 and appropriate segregants combining met4Δ::TRP1 and MED14-TAP-klTRP1 were selected by tetrad analysis and confirmed by polymerase chain reaction (PCR).
YNB medium contains 0.7% yeast nitrogen base, 0.5% ammonium sulfate and 2% glucose. Complete Supplement Mixture (CSM) medium contains, in addition, a CSM amino acid drop-out mixture.
Plasmids
pRS313-TFIIB-RFX (HIS3, CEN) was generated from pRS314-TFIIB-RFX (TRP1, CEN) (a gift from Michel Strubin) by subcloning an Xba1-Xho1 fragment containing the promoter region of TBP followed by the ORFs of TFIIB and the human regulatory factor X (RFX) protein. A short linker coding for the nuclear localization signal (NLS) of SV40 and the HA epitope from influenza virus is found in between TFIIB and RFX. YCp91-LexA (CEN, TRP1) contains the entire LexA coding sequence (residues 1–202) under the control of the ADH1 promoter, followed by SV40 NLS, an HA epitope and the CYC8 terminator (24). YCp91-LexA-TBP (CEN, TRP1) contains the SPT15 coding sequence and terminator in place of the HA epitope (24). pRS315-LexA was generated by subcloning into pRS315 a BamH1-Sac1 fragment from YCp91-LexA containing ADH1 promoter, SV40 NLS, HA epitope and CYC8 terminator. pRS315-LexA-TBP was generated by subcloning into pRS315 a BamH1 fragment from YCp91-LexA-TBP containing ADH1 promoter, SV40 NLS and SPT15 coding sequence and terminator. YCplac33-xMET17-GFP (URA3, CEN) was constructed by cloning into YCplac33-GFP (a gift from Dieter Kressler) a PCR fragment spanning nucleotides −1 to −400 of MET17 and containing at position −200 the sequence CAGTTGCCTAGCAACTACATATGGTCACC (refer-red to as X box) including the RFX binding site found in the polyomavirus enhancer (underlined). The xhis3 fragment in YCplac33-xhis3-GFP originates from a LexAop-his3 construct containing a LexA operator inserted into a derivative of his3 lacking the Tc element and a functional Gcn4 binding site (31). It was generated by replacing the LexA operator by the X box using PCR. YCplac33-xPHO5-GFP and YCplac33-xGAL1-GFP (URA3, CEN) were constructed by cloning into YCplac33-GFP PCR fragments spanning nucleotides −1 to −392 of PHO5 or nucleotides −1 to −500 of GAL1, and containing the X box inserted at position −167 and −200, respectively. YCplac33-LexAop-MET17-GFP was constructed by cloning into YCplac33-GFP a fragment, engineered by PCR, containing the sequence CTACTGTATGTACATACAGTAGTTTGTT (LexA operator underlined) inserted at −200 in a MET17 fragment spanning from −1 to −400 (relative to ATG).
Chromatin immunoprecipitation (ChIP)
Cell fixation, chromatin preparation and immunoprecipitation were performed essentially as described previously (27). In Figures 2 and 7A, formaldehyde fixation was performed at 37°C to maximize crosslinking efficiency, and cells were shifted to 37°C for 60 min before adding formaldehyde. Pol II was immunoprecipitated using the rabbit polyclonal antibody y-80 (Santa Cruz Biotechnology) directed against amino acids 1–80 of the Rpb1 subunit. Typically, 10 µl of antibody was incubated overnight at 4°C with crosslinked chromatin extracted from 10 ml of culture at OD650 = 1. TAP-tagged proteins were immunoprecipitated by incubating the same amount of crosslinked chromatin with 30 µl of rabbit Immunoglobulin G (IgG)-agarose (Sigma) for 4 h at 4°C. DNA was quantified by real-time PCR using the LightCycler 480 instrument (Roche), and SYBR Premix Ex TaqTM (Takara). Sequences of primers are given in Supplementary Table S2. A typical run included duplicates of each IP and input DNA, and serial dilutions of one input DNA to create a standard curve and determine the efficiency of the amplification. Data were analysed with the LightCycler 480 software using the ‘second derivative maximum’ method for quantification. The level of occupancy at a specific DNA locus is calculated as the percentage of DNA present in the immunoprecipitate relative to the total input. Relative occupancy was obtained by normalizing to occupancy at the IME2 locus.
Figure 2.
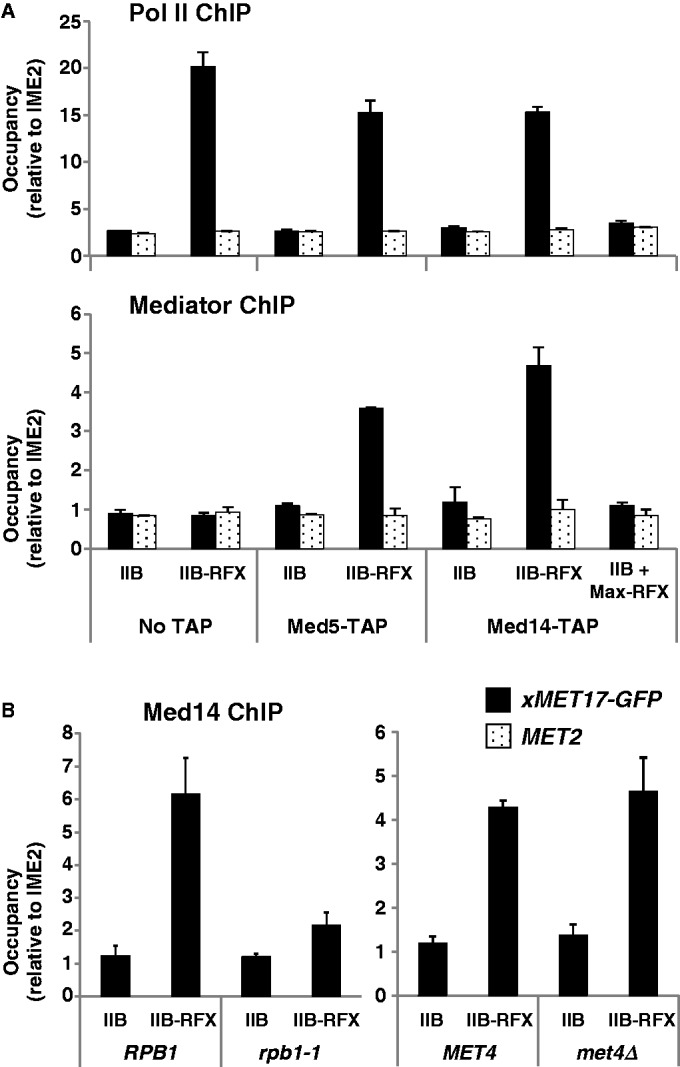
Transcriptional activation of xMET17 by TFIIB-RFX leads to Mediator recruitment. (A) Untagged (No TAP) and TAP-tagged cells (Y892, Y893 and Y894) containing YCp33-xMET17-GFP and either pRS313-IIB or pRS313-IIB-RFX, or pRS313-IIB plus pRS315-Max-RFX, were grown in glucose-containing CSM medium supplemented with 0.5 mM methionine. Mediator and Pol II occupancy at xMET17-GFP and MET2 was measured by ChIP. DNA was quantitated by qPCR using primers specific for the ORF (Pol II ChIP) or for the promoter (Mediator ChIP). Occupancy levels were normalized using the ORF of the transcriptionally inactive gene IME2. Error bars indicate standard deviations from three independent experiments. (B) Left graph: A Med14 TAP-tagged rpb1-1 mutant and the isogenic wild-type strain (Y909 and Y911) containing YCp33-xMET17-GFP and pRS313-IIB or pRS313-IIB-RFX, were grown to early log phase at 25°C in CSM medium supplemented with 0.5 mM methionine, and were shifted to 37°C for 45 min before formaldehyde fixation. Mediator occupancy was measured by ChIP as in (A). Right graph: A met4-disrupted, Med14 TAP-tagged strain (met4Δ) and the isogenic wild-type strain (Y894 and Y963) containing YCp33-xMET17-GFP, and pRS314-IIB or pRS314-IIB-RFX, were grown and submitted to ChIP as above.
Figure 7.
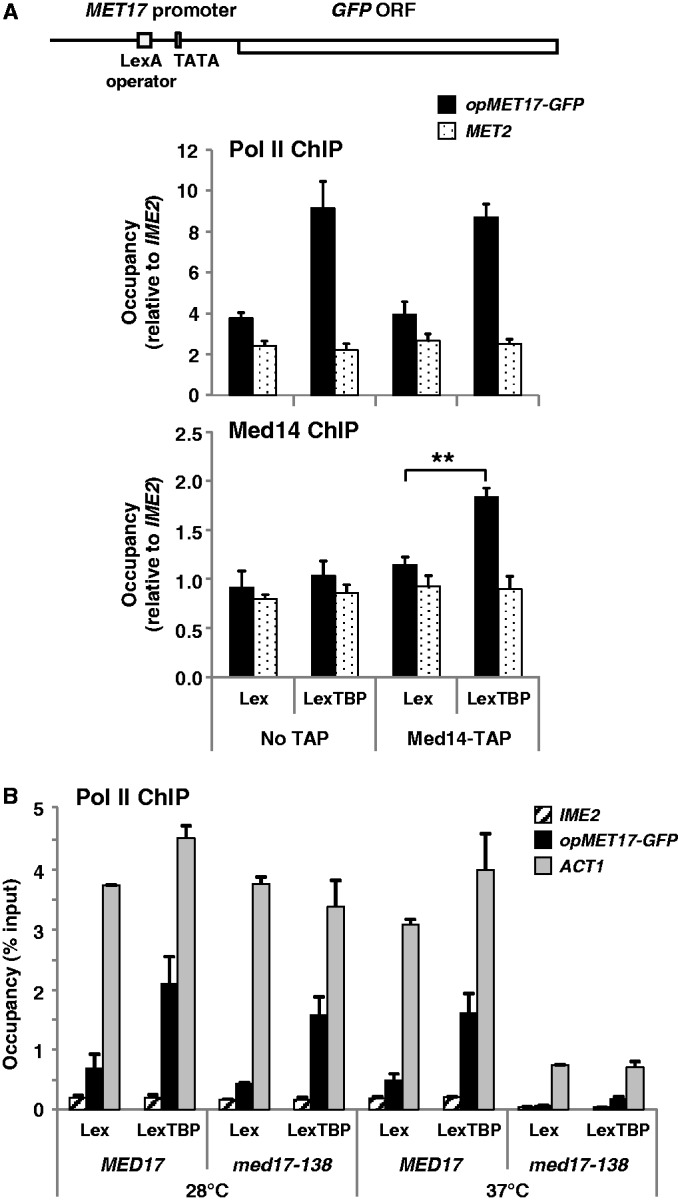
Transcriptional activation by LexA-TBP from a MET17 derivative containing the LexA operator requires Mediator. (A) Untagged (No TAP) and Med14 TAP-tagged cells (Y14 and Y84) containing YCp33-opMET17-GFP (see schematic representation) and either pRS313-LexA (Lex) or pRS313-LexA-TBP (LexTBP) were grown as shown in Figure 2. Pol II and Med14-TAP occupancy at opMET17-GFP and MET2 was measured by ChIP. DNA was analysed by qPCR using primers for the ORF (Pol II ChIP) or the promoter (Mediator ChIP). Occupancy levels were normalized using IME2 ORF. Error bars indicate standard deviations from three (Pol II ChIP) or four (Mediator ChIP) independent experiments. Asterisks indicate P < 0.005 in a Student’s t test. (B) The med17(srb4)-138 mutant and an isogenic wild-type strain (Y400 and Y402) containing YCp33-opMET17-GFP and either YCp91-LexA (Lex) or YCp91-LexA-TBP (LexTBP) were grown at 28°C to early log phase in CSM medium supplemented with 0.5 mM methionine, and were shifted to 37°C. Pol II occupancy was measured by ChIP before and 60 min after the shift. DNA was analysed by qPCR using primers for the 5′-end of GFP ORF, ACT1 ORF and IME2 ORF. Error bars indicate standard deviations from two independent experiments.
RNA analysis
Total RNA was extracted with hot acidic phenol following a protocol derived from Schmitt et al. (32). Briefly, cells collected from 10 ml of culture at OD650 = 0.5–1 were resuspended in 400 µl of cold AE buffer (50 mM sodium acetate, pH 5.3; 10 mM ethylenediaminetetraacetic acid, pH 8; 10% sodium dodecyl sulphate) and the suspension was mixed with 400 µl of cold phenol saturated with 0.1 M citrate buffer pH 4.3 (Sigma). The mixture was incubated for 8 min at 65°C with agitation, quickly frozen in liquid nitrogen, incubated again at 65°C with agitation for 4 min and centrifuged at room temperature for 10 min, 12 000 rpm. The aqueous phase was extracted once with 1 vol acidic phenol/chloroform (1:1), once with 1 vol chloroform and was precipitated with 0.1 vol 3 M LiCl and 2.5 vol absolute ethanol. After centrifugation, the RNA pellet was washed with absolute ethanol, air-dried and resuspended in water.
Reverse transcription (RT)-quantitative PCR was conducted following a two-step procedure using the RevertAid H Minus M-MuLV reverse transcriptase (Thermo Scientific) and random hexamers for priming, and the SYBR Green I Master mix and LightCycler 480 system (Roche) for real-time PCR. Sequence of primers is given in Supplementary Table S2.
Primer extension was performed as follows. Ten microliters containing 10 µg of RNA, 1 pmol of 32P-5′-labelled gene-specific oligonucleotides and 20 nmol of each dNTP was heated for 15 min at 65°C and cooled down on ice. Ten microliters containing 200 units of SuperScript II Reverse Transcriptase (Invitrogen) in 2× first-strand buffer and 0.02 M DTT was added, and the mixture was incubated at 42°C for 30 min. Reaction was stopped by adding 10 µl containing 0.9 M LiCl, 40 µg of glycogen and 10 µg of RNase. After 5 min at room temperature, the DNA was ethanol-precipitated, centrifuged, air-dried, dissolved in 6 µl of formamide-containing loading buffer and electrophoresed in a 6% denaturing polyacrylamide gel. Sequence of primers is given in Supplementary Table S3.
Protein analysis
Protein extracts were prepared using mechanical breakage with glass beads as described in Dunn and Wobbe (33). One microgram of total protein was separated by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Green fluorescent protein (GFP) was visualized using mouse anti-GFP monoclonal antibodies (Roche) and the SuperSignal West Pico chemiluminescence detection system (Thermo Scientific).
RESULTS
Experimental system
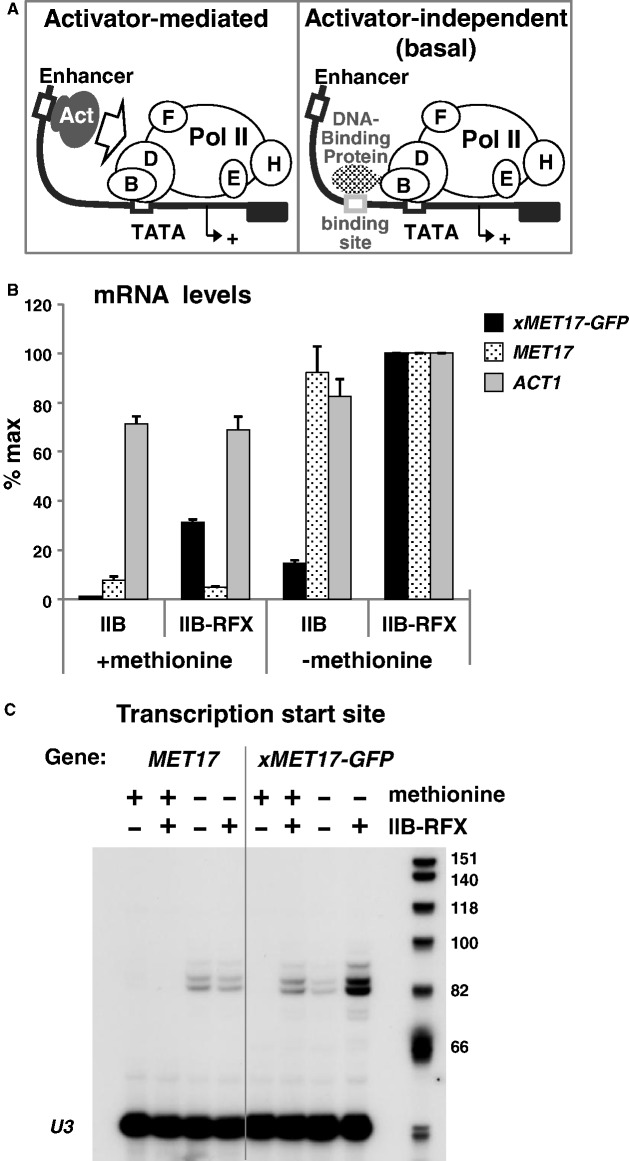
To assess the role of Mediator in basal transcription in vivo, we took advantage of molecular tools allowing artificial recruitment of the Pol II machinery to a test promoter in the absence of classical activator protein in yeast (see Figure 1A). We first used the system developed by Strubin et al., in which TFIIB is fused to RFX, a human sequence-specific DNA binding protein with no activation potential in yeast (34,35). The RFX binding site was introduced at 73 bp upstream of the unique TATA element present in the promoter of the methionine biosynthetic gene MET17. Transcription of MET17 is induced by the activator Met4 in the absence of methionine; however, under conditions of excess of methionine, Met4 activity is downregulated through a mechanism involving degradation by the ubiquitin–proteasome system, and as a result, MET17 transcription is shut down (36,37). MET17 is also induced by cadmium, which inactivates the ubiquitin-ligase targeting Met4 even in the presence of excess of methionine (38). A plasmid bearing this test promoter (named as xMET17) fused to the coding sequence of the GFP was introduced, with a plasmid bearing TFIIB or TFIIB-RFX, into a strain containing the chromosomal TFIIB allele under control of the glucose-repressed GAL1 promoter to shut down expression of the endogenous TFIIB. As expected, the xMET17 promoter remained inactive in cells expressing TFIIB in the presence of 0.5 mM methionine, and attaching TFIIB to RFX led to a strong increase in xMET17-GFP transcription (Figure 1B). We confirmed through primer extension analysis that transcription driven by TFIIB-RFX was initiated at the same positions as transcription driven by Met4 (Figure 1C). Moreover, inactivation of Met4 rendered cells auxotroph for blocked transcriptional induction of the endogenous MET17 genes by cadmium but had no effect on activation of xMET17-GFP by TFIIB-RFX (Supplementary Figure S1). Therefore, artificial tethering of TFIIB to MET17 results in efficient and accurate transcription initiation in the absence of the activator.
Figure 1.
TFIIB-RFX supports accurate transcription initiation from a MET17 promoter containing an upstream RFX binding site. (A) Experimental strategy. See text for details. (B) Strain Y892 (see Supplementary Table S1) containing YCp33-xMET17-GFP, and either pRS314-TFIIB (IIB) or pRS314-TFIIB-RFX (IIB-RFX), were grown in glucose-containing CSM medium supplemented with 0.5 mM methionine (+methionine), or in glucose-containing YNB medium with no methionine (−methionine). RNA levels for xMET17-GFP, MET17 and ACT1 were quantified by RT-qPCR and normalized to 25S rRNA levels. Values are expressed as a percentage of the maximum value. Error bars represent standard deviations from three independent experiments. (C) Same RNA preparations as in (B) were subjected to primer extension analysis with primers for MET17 and xMET17-GFP starting in both cases at exactly the same distance from MET17 TATA box (175 bp). U3 snoRNA was used as a control.
Tethering TFIIB to the xMET17 promoter leads to recruitment of Mediator
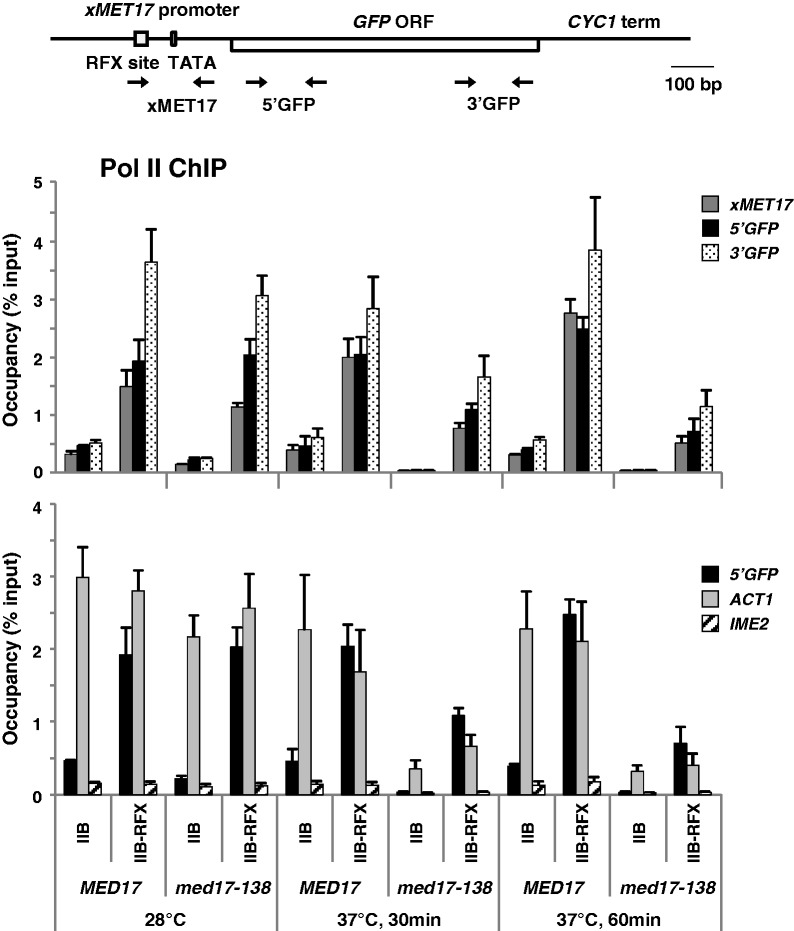
To determine whether Mediator is present at the xMET17 promoter when transcription is driven by TFIIB-RFX, we carried out chromatin immunoprecipitation (ChIP) experiments using cells containing TAP-tagged versions of Med5 and Med14, two Mediator subunits belonging to the middle and tail modules, respectively. The results for Med14-TAP and Med5-TAP showed a 3- to 4-fold increase in occupancy at the xMET17 promoter in the strains expressing TFIIB-RFX compared with the strains expressing TFIIB (Figure 2A, bottom). In contrast, no increase of occupancy was observed in the untagged strains or at the promoter of MET2, another Met4-regulated gene not activated in the presence of methionine. As expected, Pol II occupancy at xMET17 was similar in the tagged and untagged strains expressing TFIIB-RFX (Figure 2A, top). To ascertain that Mediator recruitment occurred in association with the Pol II machinery and not through interaction with the RFX moiety, we also used Med14-TAP cells expressing RFX fused to the helix-loop-helix-leucine zipper dimerization motif present in the human Max oncogene (25). Expression of Max-RFX did not lead to any increase in Med14 occupancy at xMET17 (Figure 2A, bottom), indicating that the RFX moiety is not able to recruit Mediator. In parallel, we checked Mediator occupancy at xMET17 in cells containing the rpb1-1 temperature-sensitive mutation within Pol II, which causes a genome-wide transcriptional arrest at 37°C (6). The results showed that Med14 occupancy was 3-fold lower in the rpb1-1 mutant compared with the wild-type cells after 45 min at 37°C (Figure 2B, left graph). Finally, to completely rule out any participation of Met4, Med14-TAP occupancy was also assayed in the met4Δ strain containing a null allele of the MET4 gene. The results showed no effect of MET4 deletion on Med14-TAP association with xMET17 (Figure 2B, right graph). We concluded from all these results that Mediator can be recruited to xMET17 in cells containing TFIIB-RFX by virtue of its ability to interact with Pol II and its GTFs.
Transcriptional activation by TFIIB-RFX is not affected in mutants of Mediator tail module
Mediator tail module is viewed as the main interface through which transcriptional activators come into contact with the Mediator. Accordingly, we showed in our previous report that transcriptional activation of Met4-dependent genes, including MET17, is severely affected by deletion of the non-essential tail subunits Med2 and Med3, or by a C-terminal truncation in the essential tail subunit Med14 (also known as Rgr1) (27). Therefore, we asked whether the same mutations would also affect transcriptional activation of xMET17-GFP by TFIIB-RFX. The results showed that the med2Δ, med3Δ and med14-100 mutations did not lead to any significant defects in Pol II occupancy at xMET17 (Figure 3A), and they affected neither xMET17-GFP transcript levels (Figure 3B) nor GFP protein levels (Figure 3C). As already reported (39), transcription of ACT1 was not affected in the mutants, whereas transcription of TPI1 was decreased by 2-fold. These results are consistent with the observation that inactivation of Med2 or Med3 affects only a limited subset of genes (40), which suggests no essential role in assembly and activity of the Pol II machinery.
Figure 3.
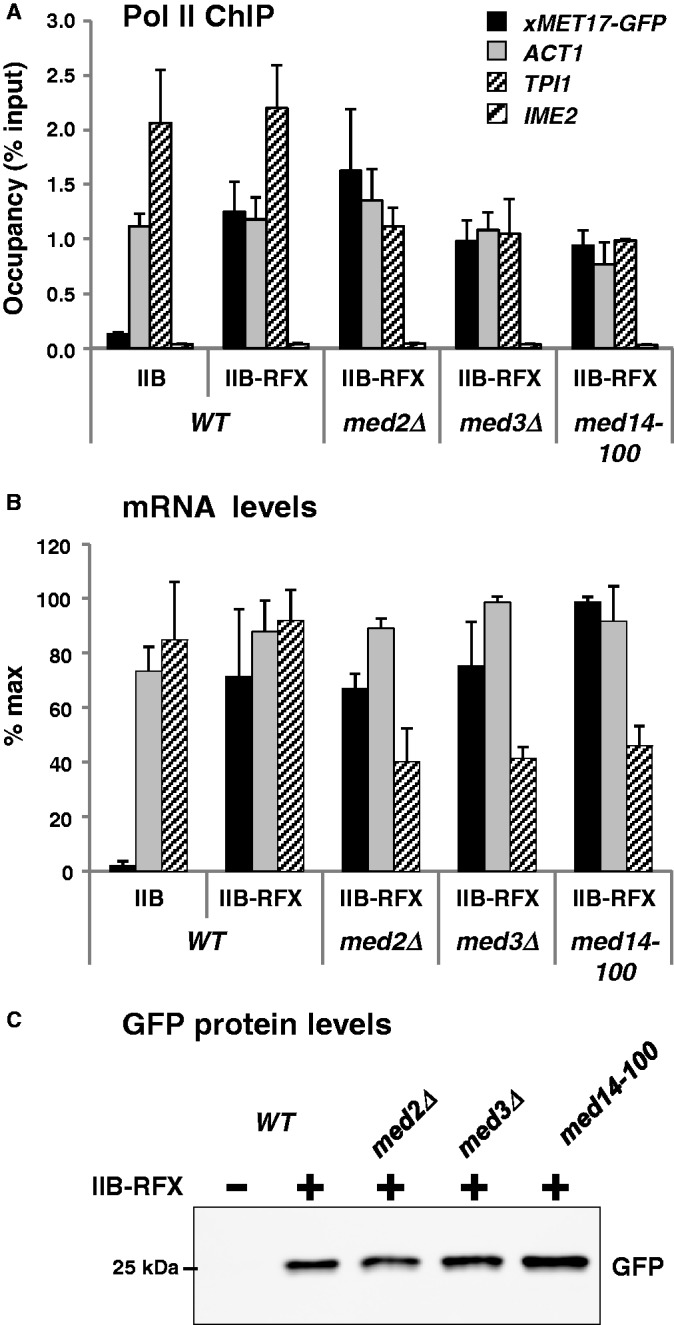
Mutation of Mediator tail module has no effect on transcriptional activation of xMET17 by TFIIB-RFX. The med2Δ, med3 Δ and med14-100 mutants (Y807, Y809 and Y811) and the isogenic wild-type strain (Y805) containing YCp33-xMET17-GFP, and either pRS314-IIB or pRS314-IIB-RFX, were grown at 28°C in glucose-containing CSM medium supplemented with 0.5 mM methionine. (A) Pol II occupancy at xMET17-GFP, ACT1, TPI1 and IME2 was measured by ChIP. DNA was analysed by qPCR using primers for the ORFs of GFP (5′ORF), ACT1, TPI1 and IME2. Error bars indicate standard deviations from three independent experiments. (B) RNA levels quantified as in Figure 1. (C) Western blot on whole cell extracts using anti-GFP antibody.
Med17 is required for transcriptional activation by TFIIB-RFX
We next used a mutation in the Med17 subunit located at the Mediator head module, knowing that this module makes multiple contacts with Pol II in the structural models (4,41). Med17 has recently been shown to interact directly with Pol II in vivo (42). We used the well-characterized med17-138 temperature-sensitive mutant, which ceases transcription of all mRNA in a manner similar to the rpb1-1 mutant at 37°C (6). Mutant and isogenic wild-type cells expressing TFIIB-RFX showed similar levels of Pol II occupancy at xMET17-GFP at 28°C (Figure 4). In contrast, occupancy was decreased 3- to 4-fold in the mutant compared with the wild-type cells 60 min after transfer to 37°C. Importantly, a similar decrease was observed with primers centered on the TATA box of MET17, and primers positioned at the beginning or at the end of the ORF of GFP (Figure 4, top graph), which indicates a defect in Pol II recruitment to the promoter and not a defect in promoter clearance or elongation. Consistently, the decrease in Pol II association was accompanied by a similar decrease in mRNA levels (Supplementary Figure S2).
Figure 4.
Med17 inactivation affects transcriptional activation by TFIIB-RFX from the xMET17 promoter. The med17-138 mutant and an isogenic wild-type strain (Y822 and Y823) containing YCp33-xMET17-GFP, and either pRS314-IIB or pRS314-IIB-RFX, were grown at 28°C to early log phase in glucose-containing CSM medium supplemented with 0.5 mM methionine, and were shifted at 37°C. Pol II occupancy was analysed by ChIP. Samples were fixed with formaldehyde before, and 30 or 60 min after the temperature shift. DNA was analysed by qPCR using primers for the promoter or the ORF of xMET17-GFP (see top schematic diagram), and primers for the ORF of ACT1 and IME2. Error bars indicate standard deviations from four independent experiments.
For comparison, ChIP experiments were also performed with strains carrying a temperature-sensitive mutation in the kinase subunit of TFIIH. We used the kin28-ts3 mutation already known to cause a global shutdown of mRNA synthesis at 37°C (6,30). The mutant and the wild-type cells expressing TFIIB-RFX showed similar levels of Pol II occupancy at xMET17-GFP at the permissive temperature (25°C; Supplementary Figure S3). However, occupancy was decreased by 3- to 4-fold in the mutant cells compared with the wild-type cells at 37°C. Therefore, inactivation of Med17 and Kin28 has the same effect on transcriptional activation of xMET17-RFX by TFIIB-RFX.
To validate further our findings, we used three additional test promoters containing the RFX binding site inserted within HIS3, PHO5 or GAL1 promoters (Figure 5). HIS3 is regulated by the activator of the general amino acid control Gcn4. The HIS3 promoter possesses two TATA elements: a TR element that contains a canonical TATA sequence and is responsible for transcription from the +13 initiation site, and a more upstream TC element that lacks a conventional TATA sequence and is responsible for transcription from the +1 site (43). The his3 derivative used here is devoid of the TC element and carries two mutations that completely inactivate the binding site for Gcn4 (44) (see diagram in Figure 5A). PHO5 and GAL1 are two tightly regulated promoters: PHO5 is not expressed under high-phosphate conditions because of exclusion of the activator Pho4 from the nucleus (45); GAL1 is not expressed in the absence of galactose because of inhibition of the activator Gal4 by the repressor Gal80, which binds Gal4 activation domain and prevents recruitment of the Pol II machinery, including Mediator (46). PHO5 and GAL1 both contain a unique conventional TATA element (see diagram in Figure 5B and C). The results in Figure 5 and Supplementary Figure S4 show that attaching TFIIB to RFX led to an increase in Pol II recruitment (Figure 5A and B) and transcription (Figure 5C and Supplementary Figure S4) for all three genes (note that only RNA levels, not Pol II recruitment, were assessed for xGAL1-GFP because transcriptional activation was too weak to give ChIP signals over background). Moreover, as in the case of xMET17-GFP, inactivation of Med17 resulted in a decrease in transcription activation of the three promoters by TFIIB-RFX (Figure 5 and Supplementary Figure S4). For instance, Pol II occupancy at xhis3-GFP and xPHO5-GFP was decreased by 4- and 6-fold, respectively, in the med17-138 mutant compared with the wild-type cells after 60 min at 37°C (Figure 5A and B), and mRNA levels for xGAL1-GFP were decreased >10-fold (Figure 5C).
Figure 5.
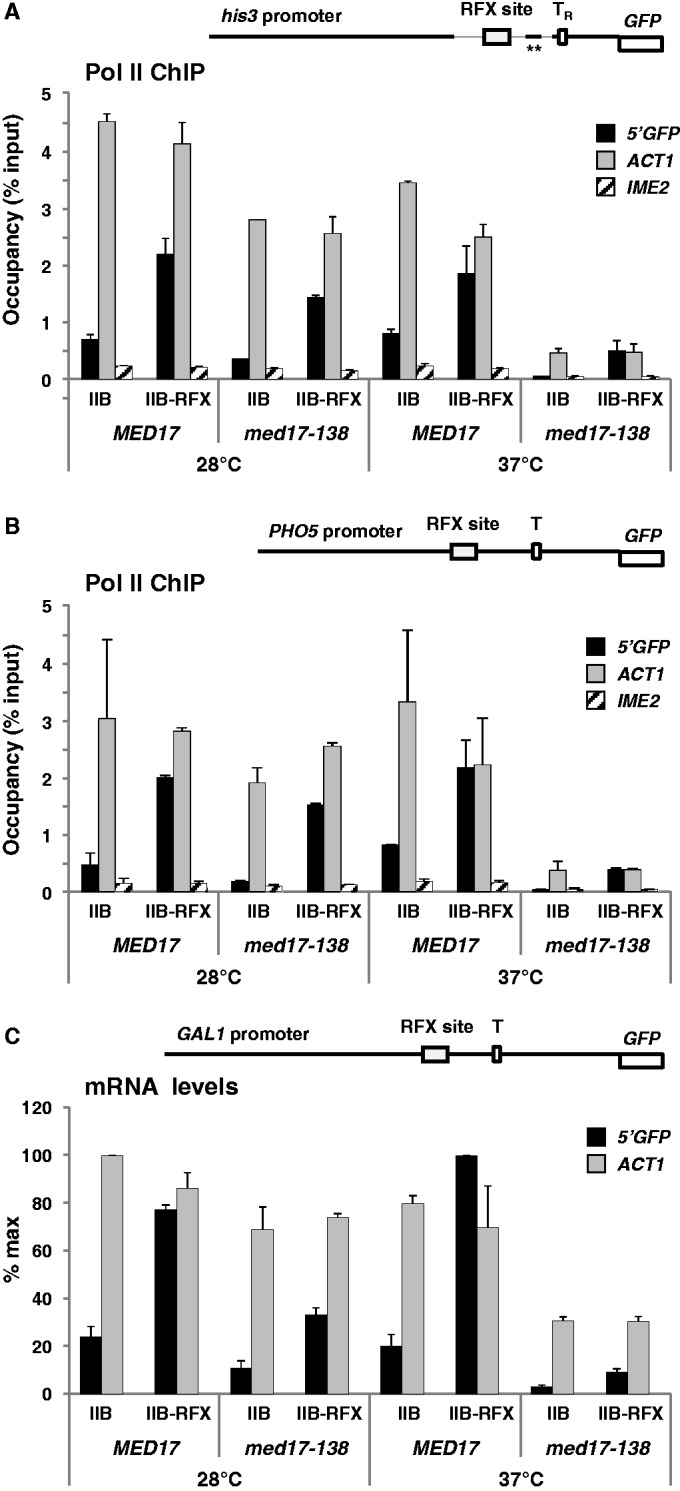
Transcriptional activation by TFIIB-RFX from HIS3, PHO5 and GAL1 promoter derivatives containing the RFX binding site is affected in the med17-138 mutant. (A) Top diagram: schematic representation of the xhis3-GFP test gene containing an RFX binding site 72 bp upstream the canonical TATA element (TR) of HIS3. The thick lines corresponds from left to right to positions −750 to −464, −125 to −108 and −78 to −1 of HIS3, and the asterisks indicate mutations in the Gcn4 binding site. TR is at −70. Graph: Pol II occupancy at GFP, ACT1 and IME2 ORF measured by ChIP. The med17-138 mutant and an isogenic wild-type strain (Y822 and Y823) containing YCp33-xhis3-GFP, and either pRS314-IIB or pRS314-IIB-RFX, were grown in glucose-containing CSM medium at 28°C to early log phase, and were shifted to 37°C. Samples were processed for ChIP analysis as in Figure 4. Error bars represent standard deviations from two independent experiments. (B) Upper diagram: schematic representation of the xPHO5-GFP gene containing an RFX binding site 80 bp upstream PHO5 TATA element. The thick line corresponds to positions −392 to −1 of PHO5. The TATA element is at −101. Graph: Pol II occupancy at GFP, ACT1 and IME2 ORF measured by ChIP. The med17-138 mutant and an isogenic wild-type strain (Y822 and Y823) containing YCp33-xPHO5-GFP, and either pRS314-IIB or pRS314-IIB-RFX, were grown and processed for ChIP analysis as in (A). (C) Top diagram: schematic representation of the xGAL1-GFP gene containing an RFX binding site 67 bp upstream of the GAL1 TATA element. The thick line corresponds to positions −500 to −1 of GAL1. The TATA element is at −147. Graph: RNA levels for xGAL1-GFP and ACT1 quantified by RT-qPCR as shown in Figure 1. Error bars represent standard deviations from two independent experiments.
Primer extension analysis was also performed to assess whether Med17 inactivation had an effect on transcription start site (TSS) selection. We analysed TSS positions at xMET17 and xhis3, which both possess multiple TSSs (Figure 6). The results showed a parallel decrease in transcription initiated at all sites in both cases, and no obvious change in positions (Figure 6). Therefore, inactivation of Med17 leads to a decrease in transcriptional activation by TFIIB-RFX, but does not change the position of the TSSs.
Figure 6.
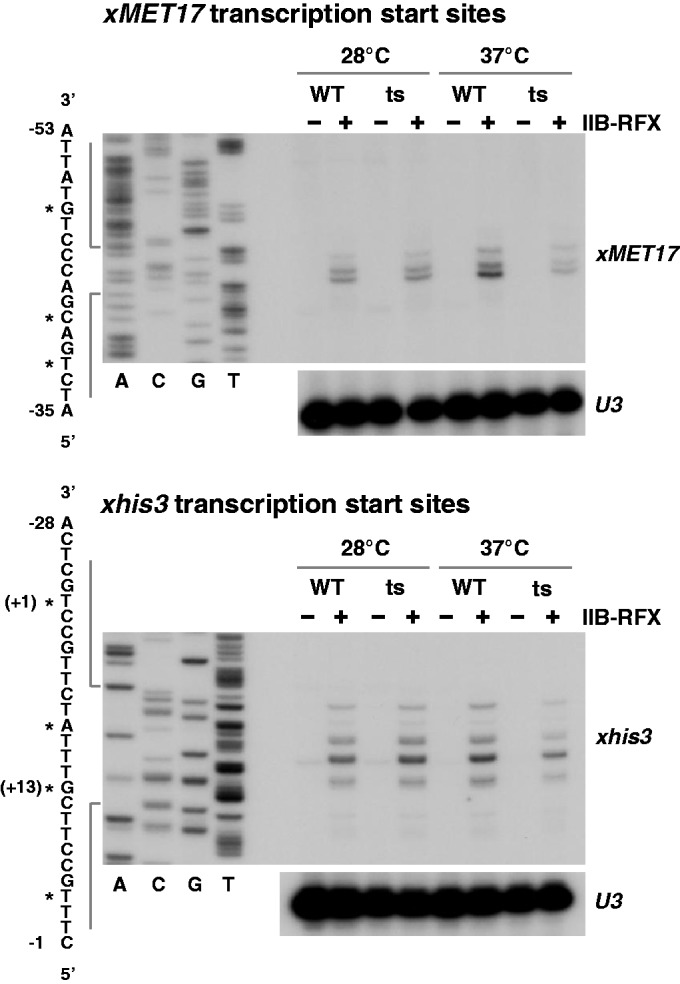
Med17 inactivation has no effect on start site selection at xMET17 and xhis3 promoters. Primer extension analysis was performed using the same RNA preparations as shown in Figure 4 (xMET17 start sites) and Figure 5 (xhis3 start sites) with a primer starting at position +74 of GFP. The sequencing DNA ladder was generated using the same primer and a plasmid bearing xMET17-GFP or xhis3-GFP. The asterisks indicate the major TSSs. Positions are given relative to the start codon. U3 snoRNA was used as a control.
Transcription activation by artificial tethering of TBP also depends on Mediator head module
Finally, we asked whether Mediator is also required when the Pol II machinery is recruited to DNA by tethering TBP instead of TFIIB. To this end, we used a LexA-TBP hybrid containing TBP fused to the full-length bacterial repressor LexA (24), and a LexAop-MET17-GFP test reporter containing one LexA operator inserted 69 bp upstream of the TATA box of MET17 (see schematic in Figure 7A). ChIP experiments were carried out using Med14 TAP-tagged strains. The results showed a modest (1.8-fold) but significant increase in Med14 occupancy at LexAop-MET17 in the TAP-tagged strain expressing LexA-TBP compared with the TAP-tagged strain expressing LexA alone (Figure 7A, bottom graph). This low occupancy is consistent with the fact that LexA-TBP led to lower levels of Pol II recruitment than TFIIB-RFX. ChIP experiments were also carried out using the med17-138 mutant and the isogenic wild-type strain. As shown in Figure 7B, mutant and isogenic wild-type cells expressing LexA-TBP showed similar levels of Pol II occupancy at LexAopMET17-GFP at 28°C. In contrast, occupancy was decreased 9-fold in the mutant compared with the wild type cells after 60 min at 37°C. Therefore, transcription activation by tethering TBP to the promoter is also accompanied by recruitment of Mediator and is also sensitive to inactivation of Med17.
DISCUSSION
Mediator recruitment can occur independently of activator
Conventional and genome-wide ChIP analyses in yeast have clearly demonstrated that Mediator is crosslinked by formaldehyde preferentially to enhancers rather than to core promoters where Pol II binds; moreover, based on time-course experiments and mutant studies, it was also shown that Mediator association with promoters is independent of Pol II, GTFs and core promoter sequences (7–9,20,21,47). Altogether, these studies support the view that the recruitment of Mediator to promoters occurred through activators bound to enhancers. We show here that Mediator can also be recruited to promoters in vivo independently of the presence of an activator, solely via its ability to interact with Pol II and the GTFs. Mediator has been known for a long time to form a biochemically stable complex with Pol II (16,48), and physical contacts with GTFs such as TBP and TFIIH were also shown (15,49). The demonstration of a direct interaction between individual subunits of Pol II and Mediator in vivo has also been reported recently (42) but, to our knowledge, this study provides the first demonstration that the recruitment of Mediator to a promoter can occur in vivo in the absence of interaction with an activator. This scarcity of data is certainly a consequence of the transient nature of the interaction between Mediator and Pol II at promoters, which occurs only during transcription initiation and must be broken to allow Pol II escape (50), whereas the interaction between Mediator and activators is more stable and persists after Pol II has left the promoter (9,51).
Mediator head is required for activator-independent transcription in vivo
Our results with med17-138 demonstrate a role of Mediator in assembly of the basal Pol II initiation machinery at promoters even in the absence of signals transmitted by activators. Importantly, this conclusion was reached using two GTFs tethered to four different test promoters through two different DNA binding proteins. Strikingly, inactivation of Med17 has a more pronounced effect on Pol II recruitment to opMET17 via LexA-TBP than on Pol II recruitment to xMET17 via TFIIB-RFX (compare Figures 4 and 7). This result suggests that tethering of TFIIB to the promoter can, at least partly, compensate for the loss of Med17, thus pointing toward a role of Mediator in stabilizing TFIIB within the PIC in vivo. This view is in line with a previous in vitro study showing that the requirement of human Mediator for basal transcription in nuclear extracts can be alleviated by increasing the TFIIB amount (52). Part of the residual Pol II recruitment observed in the med17-138 mutant at a restrictive temperature is also certainly owing to incomplete inactivation of Med17. Indeed, it was observed that the MED6-101 dominant mutation could suppress the temperature-sensitive phenotype of med17-138 but not the lethality associated with the med17-null allele, indicating that the Med17-138 protein retains some function even at a non-permissive temperature (53). This explains why Pol II occupancy remains higher at ACT1 than at IME2 in med17-138 after 60 min at a non-permissive temperature (Figures 4 and 7).
In contrast with Med17, we have found that inactivation of subunits of the tail module does not affect activation of the xMET17 promoter by TFIIB-RFX, even though this module is required for activation of MET genes by Met4 (27). In agreement with the idea that the tail module serves mainly as an interface for activators, our results provide in vivo confirmation to the finding that Mediator head alone is sufficient to stimulate basal transcription in vitro (41,54).
Mediator association with Pol II in vivo does not require activator-induced conformational change
Structural studies showed that the yeast Mediator complex adopts two different conformations when free or when associated with Pol II: the free complex presents a compact conformation with the middle and tail modules folded on each other, whereas the complex associated with Pol II presents an extended conformation, with Pol II binding in a pocket generated when Mediator changes conformation (55). It is not known whether this change in conformation occurs in the context of the living cell. However, if it does occur, the question remains whether it occurs spontaneously or is triggered by contact with activators. Our results argue that Mediator can be recruited to promoters and exert its function on assembly and activity of the PIC in the absence of an activator. Therefore, if any change in conformation is required, it can happen without the presence of an activator. This conclusion is in line with recent cryo-EM analyses of the human Mediator-Pol II-TFIIF assembly, which showed that Pol II binds Mediator at the same general location in the presence or the absence of an activator (13,14).
Mediator is often presented as a relay conveying regulatory signals from gene-specific activators bound at distal regulatory elements to the basal Pol II machinery located at the core promoter. The present study shows that the yeast Mediator functions not only through targeted recruitment by gene-specific factors, but also in an untargeted manner, strongly supporting a model in which Mediator is an integral part of the basic Pol II initiation machinery and can exert a function independently of regulatory signals conveyed by activators. Considering the functional and structural conservation of Mediator (2,56), this model should apply to all eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique, and grants from Agence Nationale pour la Recherche [ANR-06-JCJC-0045 and ANR-08-BLAN-0229]; and Association pour la Recherche sur le Cancer [subvention 4901]. Funding for open access charge: Centre National de la Recherche Scientifique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michel Strubin, Dieter Kressler, Lisete Fernandes, and Gérard Faye for plasmids and strains. We are grateful to Laetitia Cormier for constructing the strains Y400 and Y402, Christophe Leroy for constructing the strain Y933 and Lisete Fernandes for her comments on the manuscript.
REFERENCES
- 1.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high resolution protein interaction map of the yeast mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 5.Borggrefe T, Davis R, Erdjument-Bromage H, Tempst P, Kornberg RD. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- 6.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 7.Bhoite LT, Yu Y, Stillman DJ. The Swi5 activator recruits the mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 2001;15:2457–2469. doi: 10.1101/gad.921601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 9.Kuras L, Borggrefe T, Kornberg RD. Association of the mediator complex with enhancers of active genes. Proc. Natl Acad. Sci. USA. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borggrefe T, Yue X. Interactions between subunits of the mediator complex with gene-specific transcription factors. Semin. Cell Dev. Biol. 2011;22:759–768. doi: 10.1016/j.semcdb.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Ansari SA, Morse RH. Mechanisms of mediator complex action in transcriptional activation. Cell Mol. Life Sci. 2013;70:2743–56. doi: 10.1007/s00018-013-1265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taatjes DJ, Näär AM, Andel F, 3rd, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 13.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. Molecular architecture of the human Mediator-RNA polymerase II-TFIIF assembly. PLoS Biol. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernecky C, Taatjes DJ. Activator-mediator binding stabilizes RNA polymerase II orientation within the human mediator-RNA polymerase II-TFIIF assembly. J. Mol. Biol. 2012;417:387–394. doi: 10.1016/j.jmb.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 17.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell. Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J. Biol. Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 20.Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Fan X, Chou DM, Struhl K. Activator-specific recruitment of mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Wirén M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 25.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 26.Longtine MS, McKenzie A, IIIrd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Leroy C, Cormier L, Kuras L. Independent recruitment of mediator and SAGA by the activator Met4. Mol. Cell. Biol. 2006;26:3149–3163. doi: 10.1128/MCB.26.8.3149-3163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 29.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD.J. Mol. Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 31.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn B, Wobbe CR. Preparation of protein extracts from yeast. Curr. Protoc. Mol. Biol. 2001 doi: 10.1002/0471142727.mb1313s23. Chapter 13, Unit13.13. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Couto E, Klages N, Strubin M. Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl Acad. Sci. USA. 1997;94:8036–8041. doi: 10.1073/pnas.94.15.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imoberdorf RM, Topalidou I, Strubin M. A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol. Cell. Biol. 2006;26:1610–1616. doi: 10.1128/MCB.26.5.1610-1616.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuras L, Cherest H, Surdin-Kerjan Y, Thomas D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 1996;15:2519–2529. [PMC free article] [PubMed] [Google Scholar]
- 37.Kuras L, Rouillon A, Lee T, Barbey R, Tyers M, Thomas D. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell. 2002;10:69–80. doi: 10.1016/s1097-2765(02)00561-0. [DOI] [PubMed] [Google Scholar]
- 38.Barbey R, Baudouin-Cornu P, Lee TA, Rouillon A, Zarzov P, Tyers M, Thomas D. Inducible dissociation of SCF(Met30) ubiquitin ligase mediates a rapid transcriptional response to cadmium. EMBO J. 2005;24:521–532. doi: 10.1038/sj.emboj.7600556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers LC, Gustafsson CM, Hayashibara KC, Brown PO, Kornberg RD. Mediator protein mutations that selectively abolish activated transcription. Proc. Natl Acad. Sci. USA. 1999;96:67–72. doi: 10.1073/pnas.96.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn TTJP, van Leenen D, Holstege FCP. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 41.Cai G, Chaban YL, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias FJ. Interaction of the mediator head module with RNA polymerase II. Structure. 2012;20:899–910. doi: 10.1016/j.str.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 43.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Tzamarias D, Ellenberger T, Harrison SC, Struhl K. Adaptability at the protein-DNA interface is an important aspect of sequence recognition by bZIP proteins. Proc. Natl Acad. Sci. USA. 1993;90:4513–4517. doi: 10.1073/pnas.90.10.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 46.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell. 2001;7:1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 48.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 49.Larivière L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 50.Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proc. Natl Acad. Sci. USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 52.Baek HJ, Kang YK, Roeder RG. Human mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 53.Lee TI, Wyrick JJ, Koh SS, Jennings EG, Gadbois EL, Young RA. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.