Abstract
5-Hydroxymethylcytosine (5-hmC) was recently identified as a relatively frequent base in eukaryotic genomes. Its physiological function is still unclear, but it is supposed to serve as an intermediate in DNA de novo demethylation. Using X-ray diffraction, we solved five structures of four variants of the d(CGCGAATTCGCG) dodecamer, containing either 5-hmC or 5-methylcytosine (5-mC) at position 3 or at position 9. The observed resolutions were between 1.42 and 1.99 Å. Cytosine modification in all cases influences neither the whole B-DNA double helix structure nor the modified base pair geometry. The additional hydroxyl group of 5-hmC with rotational freedom along the C5-C5A bond is preferentially oriented in the 3′ direction. A comparison of thermodynamic properties of the dodecamers shows no effect of 5-mC modification and a sequence-dependent only slight destabilizing effect of 5-hmC modification. Also taking into account the results of a previous functional study [Münzel et al. (2011) (Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chem. Eur. J., 17, 13782−13788)], we conclude that the 5 position of cytosine is an ideal place to encode epigenetic information. Like this, neither the helical structure nor the thermodynamics are changed, and polymerases cannot distinguish 5-hmC and 5-mC from unmodified cytosine, all these effects are making the former ones non-mutagenic.
INTRODUCTION
The 5-hydroxymethylcytosine (5-hmC) was described in the past few years as a ‘sixth base’ of the genome (1,2), although it was discovered in the animal genome almost 40 years ago (3). 5-hmC has an uneven tissue distribution with preferential occurrence in the brain, especially in parts associated with higher brain functions (4,5). Its physiological function is still unclear; however, it is considered to be an intermediate in demethylation of cytosine mediated by ten-eleven translocation proteins (6,7).
The presence of 5-hmC within eukaryotic genomes was unknown mainly because standard methods for detection of 5-methylcytosine (5-mC) cannot distinguish between this base and 5-hmC (8–10). A newly developed method for single-nucleotide resolution 5-hmC sequencing, based on oxidative conversion of 5-hmC to 5-formyl-cytosine and its further bisulfite conversion to uracil, was recently presented (11). Overall, 5-hmC content might also be determined via selective chemical labeling by β-glucosyltransferase of 5-hmC by a glucose moiety containing an azide group modified, for example, by biotin (12,13). Another approach uses electric signal detection on a DNA solution flown through nanopores (14). A better understanding of the principles of recognition and binding of 5-hmC, however, requires its atomic structure in biological context. The interaction of 5-hmC-modified DNA with the SET and RING-associated domain of the Uhrf1 protein that belongs to the big family of 5-mC binding proteins was studied by crystal structure and molecular dynamics analyses (15). The authors showed that the 5-hmC base is flipped out of the DNA double helix and that the 5-hmC hydroxyl group participates in hydrogen bonding, which helps to stabilize the interaction. Expansion of studies on 5-hmC was enabled by the development of suitable phosphoramidites (16). These authors found that both 5-hmC and 5-mC were not causing any insertion of wrong nucleotides even by low-fidelity polymerases, and therefore concluded that position 5 of cytosine is the ideal place to store epigenetic information.
As the presence of 5-hmC might have important biological consequences, we were interested whether and how the 5-hmC for cytosine substitution influences the structure and thermodynamics of B-DNA. To address this question, we chose the well-known Drew–Dickerson dodecamer (17,18). It is a self-complementary oligonucleotide, having the sequence d(CGCGAATTCGCG) (19), which was used as a model for B-DNA and as a starting point for many consecutive studies. Thermodynamic properties of non-modified Drew–Dickerson dodecamer were thoroughly studied (17,20). The influence of cytosine modification on the B-DNA stability was established (21,22). It was proposed that cytosine 5-methylation stabilizes the DNA duplex, mostly by entropic contribution, whereas 5-hydroxymethylation reverses this stabilization, and the stability of the 5-hmC-containing duplex is somewhere between the non-modified and 5-mC-modified sequences. Influence of 5-hmC modification on the geometry of a modified base pair within a 27-bp DNA double helix was earlier studied by Wanunu et al. (14).
We have determined by X-ray diffraction the high-resolution structures of the Dickerson dodecamer containing either 5-hmC or 5-mC instead of either the second or third cytosine and compared all sequences with the non-modified one.
MATERIALS AND METHODS
Synthetic oligonucleotides were purchased from VBC Biotech (Austria). Oligonucleotides were synthesized at a 10 µM scale, purified by high-performance liquid chromatography (RP C18 column, dimethoxytrityl (DMT)-on with cleavage of the last DMT directly on the column) and lyophilized by the provider. Lyophilized oligonucleotides were dissolved in water and annealed by heating at 90°C for 5 min, followed by fast cooling to room temperature. Precise concentration of oligonucleotides was determined from ultraviolet (UV) absorption at 260 nm using the average nucleoside molar absorption coefficient ε = 9500 M−1·cm−1 calculated according to Gray (23) for all oligonucleotides. UV absorption measurements for concentration determination were performed on UNICAM 5625 UV/Vis spectrophotometer (UK) in water at 70°C in 0.1 cm quartz cells (Hellma, Germany). Water solutions of oligonucleotides were then split into aliquots of defined mass and lyophilized for optimal storage and transport. For X-ray diffraction experiments, the aliquots were then dissolved in water to a 10 mg/ml concentration and for circular dichroism (CD) and UV spectroscopic measurements to a 0.01 M nucleoside concentration, respectively.
CD measurements were conducted on a J-815 dichrograph (Jasco, Japan) in 0.1 cm quartz cells at 25°C. Spectra were collected as an average of four accumulations between 330 and 200 nm with a data pitch of 0.5 nm and digital integration time of 0.25 s at 200 nm·min−1 scan speed. Before any measurements were made, the DNA samples were heated directly in cells to 70°C and precise concentration was determined, similarly as described above. CD signals are expressed as the difference in the molar absorption Δε of the right- and left-handed circularly polarized light. The molarity was related to nucleosides. Experimental conditions were changed directly in the cells by adding a 0.4 M solution of sodium phosphate buffer, pH 7.4, and 3 M sodium chloride, and the final nucleoside concentration was corrected for the volume increase.
UV absorption spectra and melting experiments were conducted on a Varian Cary 4000 UV/Vis spectrophotometer (USA) in 0.1 cm quartz cells under the same conditions as with the CD experiments. Whole spectra were taken within two temperature ramps ranging from 98 to 8°C and 8 to 98°C, respectively, at 1°C step with a 2 min delay at each temperature step. Including measurement time, the average temperature decrease/increase rate was ∼0.25°C·min−1. Each spectrum was measured between 330 and 220 nm with a data pitch of 1 nm at a scan rate of 600 nm·min−1. Melting curves are monitored by absorbance at 260 nm. Thermal difference spectra (TDS) were calculated as the difference between UV absorption spectrum measured at 98°C and at 8°C in conditions similar to the CD experiments (24).
Crystallization
Initially, crystallization condition optimization was performed using sparse matrix crystallization screens Nucleix (Qiagen), Natrix2 (Hampton Res.) and the one described by Doudna et al. (25) with the help of a crystallization robot at the Institute of Biochemistry, UZH, Switzerland. Crystals were grown in 288-well plates by the vapor diffusion method using the sitting drop technique with a drop volume of 300 nl at 20°C in the crystallization farm. In some cases, further optimization of the initial crystallization conditions was necessary. Then, the hanging drop technique at 25°C was used in which 1 µl of 10 mg/ml DNA was mixed with 1 µl of modified reservoir solution and equilibrated against 800 µl of this solution. In all cases, crystals grew within 3–6 days. Precise crystallization conditions and solution composition of particular oligonucleotides are summarized in Table 1. Crystals were then transferred to a drop of particular reservoir solution, mounted to either a loop or foil, depending on crystal size, and flash frozen by quickly dropping into liquid N2 without any particular cryoprotection procedure.
Table 1.
Crystallization techniques and precise crystallization conditions.
| Condition | DDm3 | DDm9 | DDh3 | DDh9 | DDh3b |
|---|---|---|---|---|---|
| Technique/drop size | hanging drop/2 µl | hanging drop/2 µl | sitting drop/0.3 µl | hanging drop/2 µl | Sitting drop/0.3 µl |
| Temperature | 298 K | 298 K | 293 K | 298 K | 293 K |
| pH | 6.0 | 7.0 | 7.0 | 7.0 | 7.0 |
| Buffer | 40 mM Na cacodylate | 40 mM Na cacodylate | 40 mM Na cacodylate | 40 mM Na cacodylate | 40 mM Na cacodylate |
| Salts | 20 mM MgCl2, 80 mM NaCl | 80 mM NaCl | 80 mM NaCl | 12 mM NaCl, 80 mM KCl | 20 mM MgCl2, 80 mM NaCl, 12 mM KCl |
| Additives | 12 mM spermine·4HCl | 12 mM spermine·4HCl | 12 mM spermine·4HCl | 12 mM spermine·4HCl | 12 mM spermine·4HCl |
| 2-Methyl-2,4-pentanediol (MPD) | 35% (V/V) | 27% (V/V) | 30% (V/V) | 37% (V/V) | 35% (V/V) |
X-ray diffraction
Data were collected at 100 K at the X06DA beamline of the Swiss Light Source (SLS) at the Paul–Scherrer Institute using radiation of a wavelength of 1.00067 Å with the help of a Pilatus-2MF (Dectris) detector. Initial indexing and scaling of recorded diffraction images, together with further reflection merging, were done by the XDS software package (see Tables 1 and 2) (26). All crystals were annealed on the diffractometer by blocking the stream of cold nitrogen for 3 s (27).
Table 2.
Data collection and refinement statistics
| Parameter | DDm3 | DDm9 | DDh3 | DDh9 | DDh3b |
|---|---|---|---|---|---|
| Crystal data | |||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Unit cell | |||||
| A [Å] | 25.495 | 24.744 | 24.513 | 25.637 | 25.175 |
| B [Å] | 40.400 | 41.885 | 41.602 | 41.767 | 40.590 |
| C [Å] | 65.500 | 66.062 | 66.044 | 63.997 | 65.399 |
| Z | 4 | 4 | 4 | 4 | 4 |
| Data collection | |||||
| Resolution range [Å] | 1.41–40.40 | 1.72–41.92 | 1.83–41.60 | 1.66–41.78 | 1.99–40.59 |
| Outer shell [Å] | 1.41–1.50 | 1.72–1.82 | 1.83–1.94 | 1.66–1.76 | 1.99–2.11 |
| Reflections | |||||
| Merged | 24635 | 7618 | 6201 | 8542 | 4775 |
| Test set | 1235 | 458 | 435 | 427 | 334 |
| Completeness | 98.2 | 97.9 | 97.5 | 99.4 | 96.5 |
| In the outer shell | 97.1 | 96.1 | 96.6 | 99.0 | 94.6 |
| Redundancy | 3.61 | 3.65 | 3.68 | 3.61 | 3.55 |
| Rmerge [%] | 2.7 | 2.4 | 3.4 | 2.5 | 3.8 |
| In the outer shell | 16.3 | 21.3 | 21.6 | 20.2 | 21.7 |
| I/σ | 30.59 | 28.99 | 24.06 | 28.66 | 21.23 |
| In the outer shell | 8.12 | 6.94 | 6.65 | 6.93 | 5.81 |
| Structure refinement | |||||
| Resolution range [Å] | 1.412–40.40 | 1.721–41.92 | 1.831–41.60 | 1.662–41.78 | 1.991–40.59 |
| R-work | 0.177 | 0.220 | 0.233 | 0.237 | 0.252 |
| R-free | 0.215 | 0.287 | 0.306 | 0.276 | 0.328 |
| RMS deviation | |||||
| Bond lengths [A] | 0.0183 | 0.0085 | 0.0070 | 0.0087 | 0.0070 |
| Angle distances [A] | 0.0388 | 0.0300 | 0.0270 | 0.0293 | 0.0730 |
| Mean temperature factor | 22.46 | 34.75 | 30.06 | 33.11 | 36.29 |
| Number of ions | 1Mg2+·6H2O | 2K+ | 1Mg2+·6H2O, 1K+ | ||
| Number of water molecules | 111 | 88 | 40 | 68 | 29 |
Structure modeling and refinement
The structures were solved by molecular replacement (28) using the PHASER program (29) in the CCP4 package (30) in the space group P212121. As a starting model, we used the 0.95 Å resolution structure of the Drew–Dickerson dodecamer, PDB code 1DPN, with 2′-deoxy-2′-fluoroarabino-thymine substituted for thymine at position 7 (31). Before phasing, the thymine modification and all solvent molecules including ions were removed from the PDB file. Further refinement to yield the final structures was done with the help of the SHELXL program (32). All models of data with a resolution higher than 1.8 Å, i.e. DDm3, DDm9 and DDh9, were refined anisotropically with enhanced rigid bond restraints, the RIGU instruction in SHELXL (33). A limit value of 1.8 Å is recommended by the authors of the SHELXL and the new instruction set (33). We tested the anisotropic refinement with RIGU restraints for all models; however, for DDh3 with a resolution of 1.82 Å, no improvement was observed; thus, DDh3 and DDh3b were refined isotropically. Identified cations in solvent were refined anisotropically also by using RIGU restraints. Final R-values together with unit cell dimensions and other parameters are summarized in Table 2.
Structure parameter calculations
Root mean square deviations (RMSD) between particular structures and for individual residues between these structures were calculated using the YASARA modeling software (34). All PDB files with structures were adjusted first by removing all solvent molecules, ions, and special atoms of modified bases, and then atom numbering and labels were unified. Base pair parameters were calculated using the 3DNA software package (35).
All density functional theory (DFT) calculations were performed with the Gaussian 03 program package (36) using the hybrid functional PBE1PBE (37–39) in conjunction with the standard 6-31+G(d) basis set (40). This functional uses 25% exchange and 75% correlation weighting, and is known in the literature as PBE0. The relaxed potential energy surface scans were carried out with geometry optimization at each point. Eleven optimizations were performed for each scan as the chosen torsional angle was stepped 11 times by 30°. Solvent effects were taken into account using the polarizable continuum model (41) with water as solvent (the calculations were performed in the presence of a solvent by placing the solute in a cavity within the solvent reaction field) for all optimized geometries.
Solvation energies were calculated with the help of the ‘solvate’ module of the SEQMOL-Kd software (version 3.5.7c, biochemlabsolutions.com, 2007). For this calculation, all water molecules and cations of the PDB files were removed, the nomenclature of the atoms was harmonized and the solvation energies were calculated.
RESULTS AND DISCUSSION
Overall folding revealed by spectroscopic methods
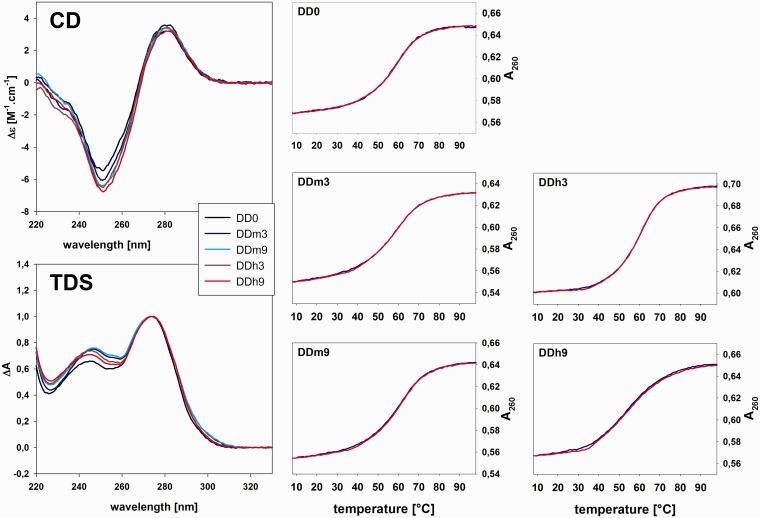
The CD spectrum of the non-modified oligonucleotide d(CGCGAATTCGCG) (Figure 1) corresponds to a B-DNA spectrum (42) and is similar to the one presented earlier for this sequence (20). Typical B-DNA is characterized by a spectrum with a positive peak at ∼280 nm and a negative one at 250 nm having comparable amplitudes, whereas the spectrum of d(CGCGAATTCGCG) displays a more pronounced negative band. The spectrum is similar to those of alternating purine–pyrimidine sequences like poly d(GC) (42). The shift toward negative CD values is usually caused by an elevated ionic strength of the solvent and is attributed to the increased winding of the B-DNA double helix (43). The spectra of all modified sequences (Figure 1) behave similarly to the non-modified sequence; however, the deepening effect is slightly more pronounced for the modified sequences. As the CD spectra of nucleic acids reflect relative base positions, base stacking and also overall helix conformation of the molecule (44,45), this indicates that the cytosine modifications do not cause changes in the overall folding, but do have some impact on double helix parameters. The structural modifications are also visible in the thermal difference spectra (Figure 1): these are similar for all studied structures with two positive peaks at ∼246 and 274 nm, respectively, and absence of any negative peak at ∼300 nm. The spectra resemble mostly that of a GC duplex (24), which is in line with the primary structure of the studied oligonucleotides: d(CGCGAATTCGCG) with a dominating content of G:C pairs.
Figure 1.
CD and UV spectroscopic characteristics of the oligonucleotides in 10 mM sodium phosphate buffer, pH 7.0, with 0.1 mM EDTA and 50 mM sodium chloride. Top left: CD spectra. Bottom left: normalized thermal difference spectra. Right: UV melting curves.
Overall molecule structure
We obtained five crystal structures with four different sequences: two with C to 5-mC substitutions, DDm3 and DDm9, and three with C to 5-hmC substitutions, DDh3, DDh3b and DDh9 (Table 1). For each sequence we obtained several crystals, usually at different conditions, and for publication we selected the crystals providing the highest resolution diffraction data. Thus, the presented crystals were obtained at different conditions, including pH value, MPD concentration and especially cation composition. DDm9 (PDB entry 4GLG) and DDh3 (PDB entry 4GLC) were crystallized in similar conditions, with sodium as the dominant cation. It is difficult to distinguish the sodium cation from a water molecule in electron density maps during structure refinement at these resolutions. As we did not observe strong enough evidence for placing a sodium ion at any of these positions, all were assigned as water molecules. DDh9 (PDB entry 4GLH) was crystallized furthermore in the presence of potassium. We identified two potassium cations within the structure (see below). DDm3 (PDB entry 4GJU) was crystallized in the presence of sodium and magnesium. We identified one fully hydrated magnesium cation in the structure. DDh3b (PDB entry 4HLI) was crystallized in the simultaneous presence of sodium, potassium and magnesium, and we could unambiguously identify one potassium and one fully hydrated magnesium. All sequences crystallized in the same space group, P212121, and provided similar cells (Table 2) as the originally described dodecamer structure (19).
First, we compared several published PDB models of the Dickerson dodecamer to get an idea about their overall structural differences, expressed as RMSDs of individual nucleotides. As a reference model, 1BNA, the original structure published by Drew and Dickerson, was taken (19). The test set included 1DPN (31), which we used as a starting model for molecular replacement, 355D (46), 436D and 460D (47), 1FQ2 (48) and 1JGR (49). The RMSD for nucleotides C3–G10 was usually <0.6 Å (Supplementary Figure S1). Both terminal di-nucleotides showed increased RMSD reaching 1.0 Å, which indicates increased flexibility of the duplex ends even in a crystal.
We compared our models both to reference 1BNA and to each other. DDm3 is the closest to the reference with C3–C10 RMSD <0.4 Å and slightly increased for terminal nucleotides (Supplementary Figure S2). This may be because all reference models including 1BNA were crystallized in the presence of magnesium ions, similarly as DDm3, whereas our other sequences crystallized in the absence of magnesium.
DDh3 compared with DDm9 showed RMSD at ∼0.3 Å for most nucleotides (Supplementary Figure S3), excluding the B-strand terminal 5′-end cytosine, with RMSD 0.77 Å, and A-strand C3 and G4 with RMSD 0.47 Å. The latter two nucleotides correspond to these either directly modified, i.e. 5-hmC at position 3 for DDh3, or pairing with modified nucleotide, i.e. G4 for DDm9 pairing with 5-mC of B-strand at position 9. Thus, this increased RMSD relative to other nucleotides is probably due to the modification. These two sequences were crystallized in similar conditions (see Materials and Methods). The low RMSD values indicate that the crystallization conditions may influence the oligonucleotide structure more than the modifications.
DDh9 has higher overall RMSD values for most nucleotides compared with the other sequences presented here. This may be because of the presence of potassium as the major monovalent cation in solution instead of sodium. A composition of the spine in the minor groove was supposed to influence the bending of the dodecamer. Substitution of water or sodium by potassium, which has different size and coordination properties, would probably cause changes in dodecamer double helix parameters. DDh9 has, except for terminal dinucleotides, high RMSD at positions G4 of A-strand and C9 of B-strand (Supplementary Figure S2). The C9 position contains the 5-hmC, and position 4 is its pairing partner. This indicates that this base pair structure is strongly altered. It is not clear why this occurs for only one modification-containing base pair and not also for the G4/B:C9/A pair.
Comparing DDh3 structures obtained in two different conditions, with and without magnesium (DDh3b; Supplementary Figure S4), revealed that their RMSD values are, on average, higher than values obtained when DDh3b was compared with DDm3. The magnesium is located at the same position as in DDm3. This again indicates that the influence of the co-crystallized magnesium is higher than the influence of the cytosine modification.
Structure and environment of modified cytosine bases
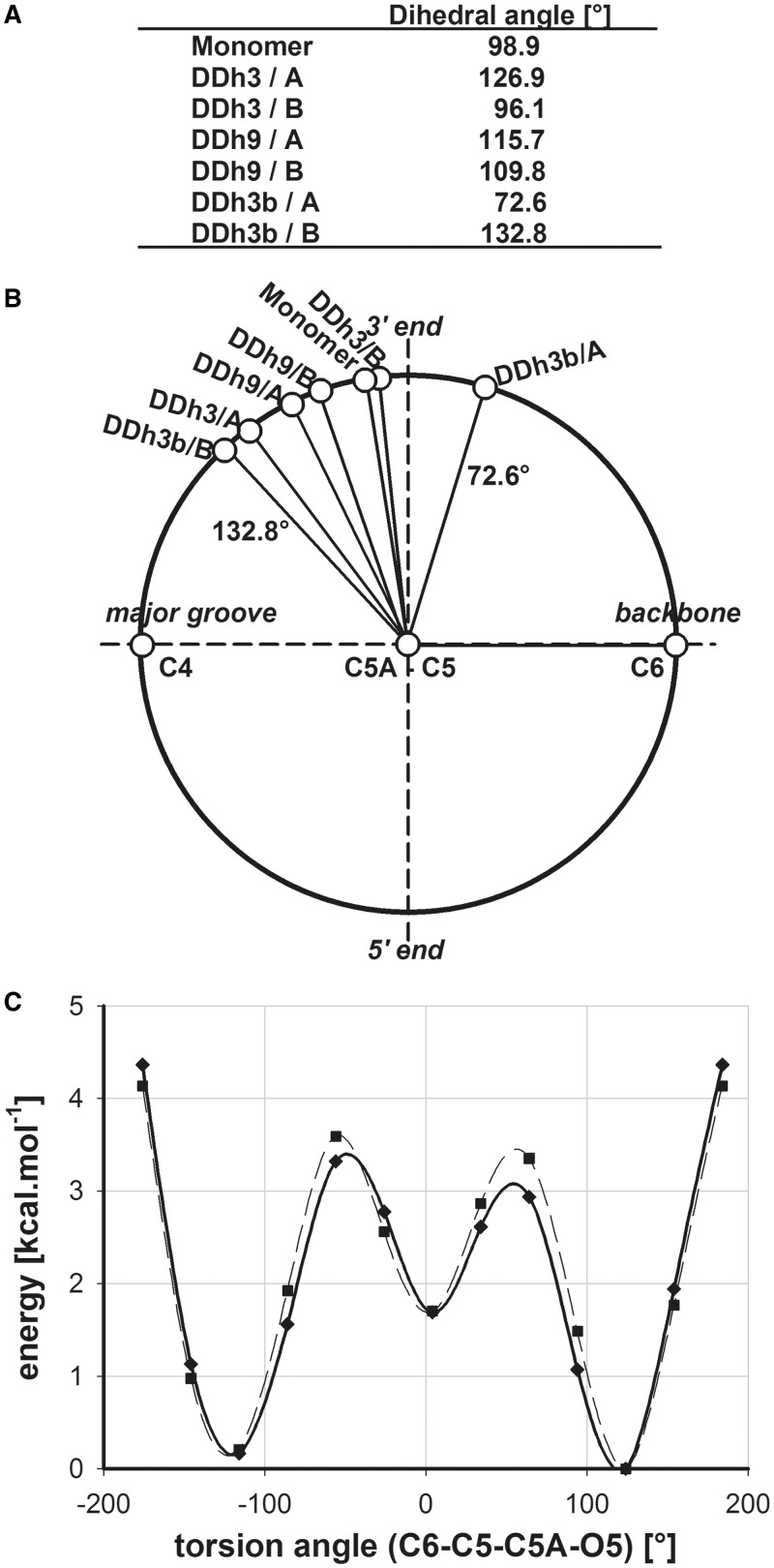
Analysis of C6-C5-C5A-O5 torsion angles for all four 5-hmC residues of DDh3, DDh3b and DDh9 showed that they are always between 72 and 133° (Figure 2A and B). Compared with the 360° range theoretically possible, it corresponds to only 17% of the potential values. Observed values are also in line with the angle determined from the monomer 5-hydroxymethyl 2′-deoxycytidine (50). Its C6-C5-C5A-O5 torsion angle is 98.9°, which is within the range determined for 5-hmC incorporated in DNA. However, in the case of the monomer, the O5 atom of the hydroxyl group participates in a hydrogen bond network with its neighboring 5-hmC monomers within the crystal. The agreement between the torsion angle values of the B-DNA double helix and the nucleoside monomer indicates that this torsion angle is most likely determined at the mononucleoside level. The observed torsion angle values ensure that the C5A-O5 bond is always pointing in the 3′-direction and away from the phosphate backbone of the strand. To further evaluate the possible torsion angles, DFT calculations were performed. Starting from the X-ray structure of the monomer 5-hydroxymethyl 2′-deoxycytidine (50), the C6-C5-C5A-O5 torsion angle was systematically changed in 30° steps and the energy of each conformation calculated. The same procedure was repeated with an explicit water molecule present, as some of us have shown this to be of essential importance in the determination of lowest energy conformations (51). As can be seen in Figure 2C, the energy landscape of the rotating hydroxyl group is rather shallow. Therefore, it is possible that the C5A-O5 orientation may be easily changed according to some external trigger.
Figure 2.
Values (A) and schematic representation (B) of dihedral angle defined by C6-C5-C5A-O5 atoms for 5-hydroxymethyl-2′-deoxycytidine monomer (50) and for this base incorporated in DDh3, DDh3b or DDh9. (C) Calculated energy of 5-hmC as a function of torsional angle C6-C5-C5A-O5 without (solid line) and with (dashed line) an additional water molecule.
Now we will address the question how the cytosine modification influences intra- and inter-base pair parameters. Wanunu et al. reported about a 27-bp oligonucleotide molecular dynamic-based fluctuation of altered base parameters for 5-mC and 5-hmC base pairs (14). On these relative fluctuations, compared with a non-modified sequence, they claimed that 5-mC modification causes increased rigidity of the double helix, whereas 5-hmC causes DNA to be more flexible.
Using the 3DNA software package (35), we calculated all intra- and inter-base pair parameters for the whole sequence (Supplementary Figure S5A–L). Although we observed some differences between sequences in different parameters, we were not able to conclude any general rules of cytosine modification on the DNA helix parameters. As was shown by the RMSD values above, it is possible that base pair parameters are more influenced by crystallization conditions than by specific cytosine modification.
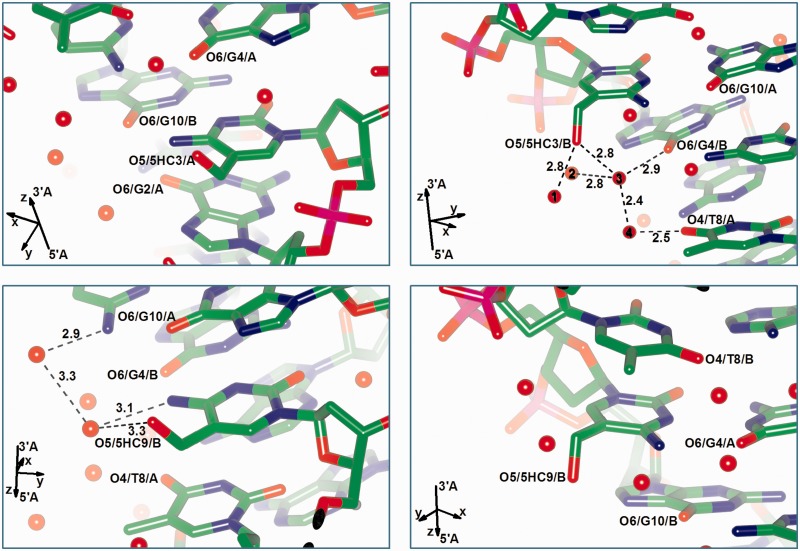
One of the main properties that is affected when substituting 5-mC by 5-hmC is the hydrophobicity, as we replace a hydrophobic methyl group with the more hydrophilic hydroxymethyl group. We wanted to know what is the hydration pattern of the hydroxymethyl group in the context of an oligonucleotide. A search range of 2.4–3.3 Å for the distance between O5 of 5-hmC and the oxygen of a hydrogen bond bound water molecule was set. Within this range, we did not identify any hydrogen bond for O5 of strand A of DDh3 and of strand B of DDh9 (Figure 3). However, for O5 in strand B of DDh3, we identified a hydrogen bond network: two water molecules (Figure 3, top right, labeled as 1 and 3) are bound directly to the O5 atom. One of them (labeled 3) is further bonded to two other water molecules (labeled 2 and 4) and also to the O6 atom of G4 in strand B. Water 4 is then bound to the O4 atom of T8 in strand A. Superposing all four presented models, we were able to identify common water positions: position 1 is unique for 5-hmC in strand B of DDh3, whereas positions 2, 3 and 4 are common for at least three sequences. As expected, there is no water in close contact with any of the methyl groups of the 5-mC nucleobases (Supplementary Figure S6).
Figure 3.
Structures of individual 5-hmCs. Selected hydrogen bonds together with the distances are represented by dashed lines. Top left: DDh3/A strand. Top right: DDh3/B strand. Bottom left: DDh9/A strand. Bottom right: DDh9/B strand. See text for further explanations.
The solvation energy for all four oligonucleotides was calculated with help of the program SEQMOL-Kd (Table 3) (52). It is obvious that the two oliogonucleotides with the 5-hydroxymethylcytosine DDh3 and DDh9 have higher solvation energies than the ones with the 5-methylcytosine DDm3 and DDm9. The energy difference is slightly bigger for the DDm3/DDh3 pair than for the DDm9/DDh9 pair, which could be explained by a better solvation of the hydroxy group in the position 3 compared with the position 9.
Table 3.
Solvation energies of DDm3, DDh3, DDm9 and DDh9 calculated by SEQMOL-Kd
| Sequence | Energy [kcal/mol] | Difference (DDhX−DDmX) [kcal/mol] |
|---|---|---|
| DDm3 | −49.8 | −2.2 |
| DDh3 | −52.0 | |
| DDm9 | −48.6 | −1.6 |
| DDh9 | −50.2 | |
Cation/water network in minor groove
All presented structures contain a spine of water molecules in the minor groove as described previously in the literature. The rules for assigning the electron density spots to either water molecule or cation were the same as commonly used: electron density spot size, number and length of coordination distances to DNA atoms, occupancy/thermal factors and valency.
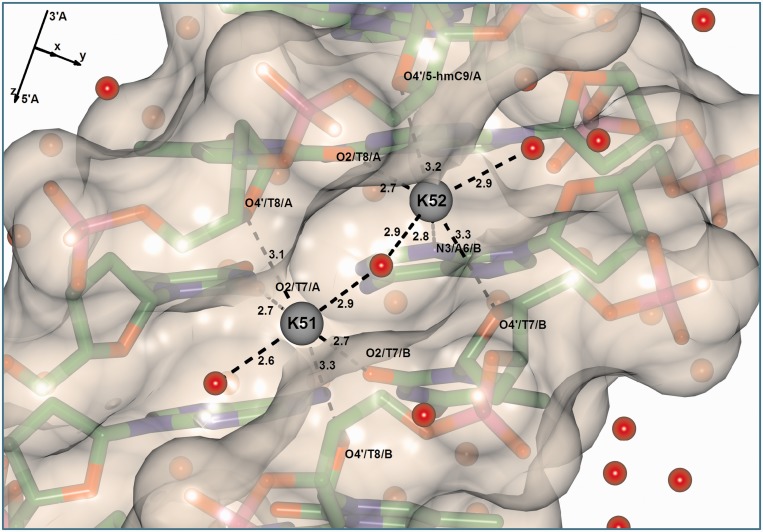
Structure DDh9 was crystallized in the presence of potassium (Table 2). We identified two potassium ion positions in the minor groove (Figure 4). The first one, labeled as K51, is located between the A6/A:T7/B and T7/A:A6/B base pairs. It is coordinated to four oxygen atoms of thymines, O2/T7/A, O4’/T8/A, O2/T7/B and O4’/T8/B, and two water molecules, positioned in the outer layer of the minor groove spine. This potassium is characterized by 80% occupancy, a thermal factor of 30.9 Å2 and a potassium-specific valence of 0.79, which was calculated similarly as in (47), according to the approach described in (53) with potassium–oxygen bond parameters from (54). This site corresponds to a P3 position described earlier (55). The presence of potassium in an ApT:TpA base pair step in the minor groove is known, as it was shown several times for modified Drew–Dickerson dodecamer structures (56,57) and even for the nucleosome (58). The second potassium, labeled as K52, is located between T7/A:A6/B and T8/A:A5/B base pairs and is coordinated to O2/T8/A, O4’/5-hmC9/A, N3/A6/B, O4’/T7/B and two water molecules. This site corresponds to a P2 position described earlier (55). All potassium ions possess complete coordination geometry. The water/cation networks have also been analyzed with the help of the program Nucplot (Supplementary Figure S7) (59).
Figure 4.
Structure and Conolly surface of the minor groove of DDh9. Two K+ cations are represented with gray circles. Coordination bonds of K+ cations together with the distances are represented by dashed lines.
Structure stability and thermodynamic parameters
To compare our sequences with others studying 5-hmC-modified DNA, published earlier (22), we also calculated the thermodynamic parameters. As experimental background, we used UV melting experiments. Thermodynamic parameters were calculated using van’t Hoff calculations according to a 2A → C model (60), from melting curves expressed as absorbance at 260 nm at different temperatures (Figure 1). We assumed a simple two-state melting for the 12-bp duplex at 55 µM strand concentration in ∼65 mM salt. The absence of hysteresis during the renaturation/denaturation cycle indicates thermodynamic equilibrium, which means that the obtained absorbance at a particular temperature is directly related to the ratio of folded versus unfolded state.
Obtained and calculated thermodynamic parameters are shown in Supplementary Table S1. The values of the non-modified sequence (DD0) are in line with the previously reported ones also determined by UV spectroscopy after having taken into account the different ionic strength (20). Tm values observed by nuclear magnetic resonance are >10°C higher, but they were observed at an ∼10 times higher DNA concentration (17). Except for DDh9, the thermodynamic parameters and also the folded fraction curves (Supplementary Figure S8) are within a range of 1.3°C and, therefore, similar for all sequences. Introduction of 5-methylcytosine instead of cytosine either did not change Tm at all (DDm3) or slightly increased Tm by 1.3°C (DDm9). Substitution with 5-hydroxymethylcytosine, on the other hand, either led to a minimal increase (DDh3) or decrease by 2.5°C (DDh9) of the melting temperatures. These data highlight the importance of sequence as well as position of the modification. Our results essentially indicate that 5-hmC and 5-mC are both modifications of the DNA that do not lead to great changes in melting behavior in line with previous reports (16,21,22). Our structural and thermodynamic results confirm the conclusion of Münzel et al. (16) that position 5 of cytosine is an ideal place to store epigenetic information.
Other features
All reference structures, except for the unclear case of 1BNA, were crystallized in the presence of magnesium. In all these cases, at least one magnesium cation was localized in the structures. In our case, sequences DDm3 and DDh3 were crystallized in the presence of magnesium. For DDm3 at a resolution of 1.41 Å, we were able to identify one magnesium site; the magnesium displays the typical octahedral coordination sphere with six bound water molecules. This position corresponds to a site Mg1 described earlier (47). The other reported magnesium positions in Drew–Dickerson dodecamer are present only at higher Mg2+ concentration, whereas we used only 20 mM Mg2+.
DDh3 was measured also in conditions containing 20 mM Mg2+; however, the observed resolution was only 2.0 Å (see DDh3b). Magnesium in this case is positioned at the same place as in DDm3. Also the RMSD between DDm3 and DDh3b is much smaller than between DDh3 and DDh3b. This indicates that the influence of crystallization conditions is much higher than the influence of cytosine modifications (see end of the subchapter “Overall molecule structure”).
CONCLUSIONS
We determined five high-resolution X-ray structures of the palindromic dodecamer d(CGCGAATTCGCG) modified at a single position either as an 5-hmC or a 5-mC at either the third or the ninth position. These modifications do not influence the overall B-DNA helix structure, the thermodynamic behavior or the altered base pair structure and orientation. The O5 atom of the hydroxyl group of 5-hydroxymethylcytosine points in the 3′ direction, away from the phosphate backbone. This conformation was also found as an energy minimum calculated by DFT methods for the 5-hydroxymethyl 2′-deoxycytidine monomer. The presence of the hydroxyl group substantially increases the overall solvation energy. This high quality set of similar DNA structures allows differentiating between the influences of cations present in the crystallization buffers and the modified nucleobases. Also taking into account the results of a previous functional study (16), we come to the conclusion that the 5 position of dC is an ideal place to encode epigenetic information. Like this, neither the helical structure nor the thermodynamics are changed, and polymerases cannot distinguish 5-hmC and 5-mC from unmodified cytosine, all these effects are making the former ones non-mutagenic.
ACCESSION NUMBERS
4GLG, 4GLC, 4GLH, 4GJU and 4HLI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [61].
FUNDING
The CRUS organization [Sciex-NMSch fellowship [11.137] (to D.R.); European Regional Development Fund [“CEITEC–Central European Institute of Technology’’ [CZ.1.05/1.1.00/02.0068] and [P205/12/0466 by GA CR] (to M.V.); University of Zurich (to B.S. and O.B.). Funding for open access charge: PD Stiftung University Zurich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the anonymous reviewers for many helpful comments.
REFERENCES
- 1.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. Presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew. Chem. Int. Ed. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 5.Kriukiene E, Liutkeviciute Z, Klimasauskas S. 5-hydroxymethylcytosine - the elusive epigenetic mark in mammalian DNA. Chem. Soc. Rev. 2012;41:6916–6930. doi: 10.1039/c2cs35104h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. TET proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Münzel M, Lercher L, Müller M, Carell T. Chemical discrimination between dC and (5Me)dC via their hydroxylamine adducts. Nucleic Acids Res. 2010;38:e192. doi: 10.1093/nar/gkq724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 12.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J. Am. Chem. Soc. 2010;133:486–492. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Münzel M, Lischke U, Stathis D, Pfaffeneder T, Gnerlich FA, Deiml CA, Koch SC, Karaghiosoff K, Carell T. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chem. Eur. J. 2011;17:13782–13788. doi: 10.1002/chem.201102782. [DOI] [PubMed] [Google Scholar]
- 17.Patel DJ, Kozlowski SA, Marky LA, Broka C, Rice JA, Itakura K, Breslauer KJ. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982;21:428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- 18.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Crystal-structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 19.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCAATTCGCG): evidence for hairpin formation. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez López CM, Lloyd AJ, Leonard K, Wilkinson MJ. Differential effect of three base modifications on DNA thermostability revealed by high resolution melting. Anal. Chem. 2012;84:7336–7342. doi: 10.1021/ac301459x. [DOI] [PubMed] [Google Scholar]
- 22.Thalhammer A, Hansen AS, El-Sagheer AH, Brown T, Schofield CJ. Hydroxylation of methylated CpG dinucleotides reverses stabilisation of DNA duplexes by cytosine 5-methylation. Chem. Commun. 2011;47:5325–5327. doi: 10.1039/c0cc05671e. [DOI] [PubMed] [Google Scholar]
- 23.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 24.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doudna JA, Grosshans C, Gooding A, Kundrot CE. Crystallization of ribozymes and small RNA motifs by a sparse-matrix approach. Proc. Natl Acad. Sci. USA. 1993;90:7829–7833. doi: 10.1073/pnas.90.16.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 28.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 29.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egli M, Tereshko V, Teplova M, Minasov G, Joachimiak A, Sanishvili R, Weeks CM, Miller R, Maier MA, An H, et al. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers. 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 33.Thorn A, Dittrich B, Sheldrick GM. Enhanced rigid-bond restraints. Acta Crystallogr. A. 2012;68:448–451. [Google Scholar]
- 34.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA - a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 35.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. Gaussian 03, Revision D.01. Wallingford, CT: Gaussian Inc.; 2004. [Google Scholar]
- 37.Adamo C, Barone V. Toward reliable density functional methods without adjustable parameters: the PBE0 model. J. Chem. Phys. 1999;110:6158–6170. [Google Scholar]
- 38.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996;77:3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 39.Perdew JP, Burke K, Ernzerhof M. Generalized gradient approximation made simple (vol 77, pg 3865, 1996) Phys. Rev. Lett. 1997;78:1396. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 40.Ditchfield R, Hehre WJ, Pople JA. Self-consistent molecular-orbital methods. 9. An extended Gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 1971;54:724–728. [Google Scholar]
- 41.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 42.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson BB, Dahl KS, Tinoco I, Ivanov VI, Zhurkin VB. Correlations between deoxyribonucleic-acid structural parameters and calculated circular-dichroism spectra. Biochemistry. 1981;20:73–78. doi: 10.1021/bi00504a013. [DOI] [PubMed] [Google Scholar]
- 44.Gray DM, Ratliff RL, Vaughan MR. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 45.Johnson WC. New York: VCH Publishers; 1994. CD of nucleic acids. [Google Scholar]
- 46.Shui XQ, McFail-Isom L, Hu GG, Williams LD. The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry. 1998;37:8341–8355. doi: 10.1021/bi973073c. [DOI] [PubMed] [Google Scholar]
- 47.Tereshko V, Minasov G, Egli M. The Dickerson–Drew B-DNA dodecamer revisited at atomic resolution. J. Am. Chem. Soc. 1999;121:470–471. [Google Scholar]
- 48.Sines CC, McFail-Isom L, Howerton SB, VanDerveer D, Williams LD. Cations mediate B-DNA conformational heterogeneity. J. Am. Chem. Soc. 2000;122:11048–11056. [Google Scholar]
- 49.Howerton SB, Sines CC, VanDerveer D, Williams LD. Locating monovalent cations in the grooves of B-DNA. Biochemistry. 2001;40:10023–10031. doi: 10.1021/bi010391+. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Kumar SVP, Stuart AL, Delbaere LTJ, Gupta SV. Structure and conformation of 5-hydroxymethyl-2′-deoxycitidine, C10H15N3O5. Acta Crystallogr. C. 1994;50:1837–1839. [Google Scholar]
- 51.Zobi F, Blacque O, Schmalle HW, Spingler B, Alberto R. Head-to-head (HH) and head-to-tail (HT) conformers of cis-bis guanine ligands bound to the [Re(CO)3]+ core. Inorg. Chem. 2004;43:2087–2096. doi: 10.1021/ic035012a. [DOI] [PubMed] [Google Scholar]
- 52.Fernández-Recio J. Prediction of protein binding sites and hot spots. WIRES Comput. Mol. Sci. 2011;1:680–698. [Google Scholar]
- 53.Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- 54.Brown ID, Wu KK. Empirical parameters for calculating cation-oxygen bond valences. Acta Crystallogr. B. 1976;32:1957–1959. [Google Scholar]
- 55.Shui XQ, Sines CC, McFail-Isom L, VanDerveer D, Williams LD. Structure of the potassium form of CGCGAATTCGCG: DNA deformation by electrostatic collapse around inorganic cations. Biochemistry. 1998;37:16877–16887. doi: 10.1021/bi982063o. [DOI] [PubMed] [Google Scholar]
- 56.Juan ECM, Kondo J, Kurihara T, Ito T, Ueno Y, Matsuda A, Takénaka A. Crystal structures of DNA:DNA and DNA:RNA duplexes containing 5-(N-aminohexyl)carbamoyl-modified uracils reveal the basis for properties as antigene and antisense molecules. Nucleic Acids Res. 2007;35:1969–1977. doi: 10.1093/nar/gkl821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsunoda M, Karino N, Ueno Y, Matsuda A, Takenaka A. Crystallization and preliminary X-ray analysis of a DNA dodecamer containing 2′-deoxy-5-formyluridine; what is the role of magnesium cation in crystallization of Dickerson-type DNA dodecamers? Acta Crystallogr. D Biol. Crystallogr. 2001;57:345–348. doi: 10.1107/s0907444900017583. [DOI] [PubMed] [Google Scholar]
- 58.Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: a program to generate schematic diagrams of protein–nucleic acid interactions. Nucleic Acids Res. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 61.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. 2011;D67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.