Abstract
Long double-stranded RNA may undergo hyper-editing by adenosine deaminases that act on RNA (ADARs), where up to 50% of adenosine residues may be converted to inosine. However, although numerous RNAs may undergo hyper-editing, the role for inosine-containing hyper-edited double-stranded RNA in cells is poorly understood. Nevertheless, editing plays a critical role in mammalian cells, as highlighted by the analysis of ADAR-null mutants. In particular, the long form of ADAR1 (ADAR1p150) is essential for viability. Moreover, a number of studies have implicated ADAR1p150 in various stress pathways. We have previously shown that ADAR1p150 localized to cytoplasmic stress granules in HeLa cells following either oxidative or interferon-induced stress. Here, we show that the Z-DNA-binding domain (ZαADAR1) exclusively found in ADAR1p150 is required for its localization to stress granules. Moreover, we show that fusion of ZαADAR1 to either green fluorescent protein (GFP) or polypyrimidine binding protein 4 (PTB4) also results in their localization to stress granules. We additionally show that the Zα domain from other Z-DNA-binding proteins (ZBP1, E3L) is likewise sufficient for localization to stress granules. Finally, we show that Z-RNA or Z-DNA binding is important for stress granule localization. We have thus identified a novel role for Z-DNA-binding domains in mammalian cells.
INTRODUCTION
Adenosine deaminases that act on RNA (ADARs) catalyze the conversion of adenosine (A) to inosine (I) within double-stranded RNA (dsRNA). Three ADARs have been described in mammalian cells (ADARs 1–3), although only ADAR1 and ADAR2 appear to be catalytically active (1,2).
Although a single gene encodes ADAR1, two different isoforms are generated by use of alternative promoters and alternative splicing (3,4). The short form of ADAR1 (ADAR1 p110; ADAR1p110) is constitutively expressed and localizes mainly to the nucleus, whereas the long form of ADAR1 (ADAR1p150; ADAR1p150) is interferon-inducible and shuttles between the nucleus and cytoplasm. However, the localization of ADAR1p150 is predominantly cytoplasmic (3–7). With the exception of the N-terminal region, where ADAR1p110 is truncated by 295 amino acids relative to ADAR1p150, the two proteins are otherwise identical (1,3,8). Both proteins have in common three dsRNA-binding domains (dsRBDs) and a C-terminal deaminase domain. The proteins differ in that ADAR1p150 has two Z-DNA/Z-RNA-binding domains (ZBDs; Zα and Zβ) within the N-terminal region, whereas only the Zβ domain is present in the N-terminally truncated ADAR1p110 (9). This difference will have functional consequences, as only the Zα domain has the ability to bind Z-DNA/Z-RNA (10). Recent in vitro studies have shown that binding of ADAR1p150 to a Z-RNA-forming motif enhances editing within adjacent A-form dsRNA sequences. The Zα domain may thus be used to target ADAR1p150 to sequences with the potential to form Z-RNA for selective editing (11).
Z-DNA or Z-RNA comprises an alternative, higher energy conformation of DNA or RNA, respectively, that preferentially occurs within sequences containing alternating purine and pyrimidine residues to give a characteristic zigzag helix (12). Although transition to the higher-order structure is energetically unfavorable, proteins containing Z-DNA-binding domains can specifically induce and stabilize both Z-DNA and Z-RNA. Moreover, structural analyses have revealed that recognition and binding of both Z-DNA and Z-RNA by Zα domains is conformation-specific rather than sequence-specific (10,13,14). As the left-handed Z-conformations of Z-DNA and Z-RNA are similar, the Zα domain may thus represent an unusual class of domains that specifically bind to both duplex DNA and RNA (15).
In addition to ADAR1p150, Z-DNA/Z-RNA-binding domains have been identified in only a limited number of proteins. These include the Z-DNA-binding protein 1 [ZBP1; also known as DLM-1 or DAI (DNA-dependent activator of IFN-regulatory factors)] (14,16), the poxvirus virulence factor E3L (17,18), a protein kinase (PKZ) that was uniquely identified in fish (19) and a protein (ORF112) from the family Alloherpesviridae, which includes important pathogens of fish and amphibian species (20). Intriguingly, all of these proteins play a role in immune pathways. ADAR1p150, ZBP1 and PKZ are upregulated by interferon and are implicated in host defense mechanisms (3,4,16,19,21,22). In contrast, the Zα domain of the E3L protein is required for viral pathogenicity (22). It is possible that the Zα domain of E3L competes with cellular Z-DNA-binding proteins for binding of specific target sequences, and thus subverts the host interferon response (23). The Zα domain of ORF112 similarly has the potential to antagonize PKZ function in fish by binding its cellular targets (20).
We have recently shown that ADAR1p150 localizes to cytoplasmic stress granules (24). Stress granules are cytoplasmic foci that provide eukaryotic cells a means of survival during stress (25–27). Stress granules typically form as the result of translational inhibition and therefore comprise translationally arrested mRNA as well as stalled initiation complexes and small ribosomal proteins (28). Importantly, sequestration of a set of translationally arrested mRNAs allows selective synthesis of proteins required for cell survival. When the stress conditions are removed, mRNAs sequestered within stress granules may reassemble on polysomes and resume translation (25). Alternatively, mRNAs may be degraded within cytoplasmic-processing bodies (P-bodies), which are dynamically linked to stress granules (29).
Here, we present experimental data showing that the Zα domain of ADAR1p150 is necessary and sufficient for localization of ADAR1p150 to stress granules. Fusion of the Zα domain to either green fluorescent protein (GFP) or polypyrimidine tract binding protein 4 (PTB4) resulted in their localization to stress granules. Moreover, we demonstrate that the Zα domain from ZBP1 and E3L also direct localization to stress granules. Finally, we provide evidence that binding of the Zα domain to Z-RNA/Z-DNA is important for localization of ADAR1p150 to stress granules.
MATERIALS AND METHODS
Transfections
HeLa cells were maintained in DMEM with GlutaMAX™-I (Gibco) supplemented with 10% (v/v) fetal bovine serum. HeLa cells (2 × 105 cells per well (6 well plate)) were used for transfections. Plasmids were transfected into HeLa cells using Lipofectamine-2000 (LF-2000; Invitrogen), and cells visualized after 24 h. Poly(IC) (10 ng; Sigma) was typically used to transfect HeLa cells using LF-2000, and cells were visualized after 7 h. Mock transfections used LF-2000 alone. For arsenite treatment, cells were incubated in media supplemented with 0.5 mM sodium arsenite (Sigma) for 30 min, then allowed to recover for 30 min in the absence of arsenite (28). Stress granules were identified in HeLa cells with reference to the stress granule marker TIAR (red). As the localization of TIAR in untreated cells is predominantly nuclear, stress granules were readily identifiable as distinct cytoplasmic foci. GFP-tagged or Flag-tagged proteins (green) were then analyzed for co-localization with TIAR in stress granules. To determine the efficiency of localization of ADAR1p150 (or other constructs) to stress granules, at least 100 cells positive for the stress granule marker TIAR were analyzed for co-localization of ADAR1p150 (n ≥ 3; error bars are mean ± SD). The t-tests (two-tailed, unequal variance) were used. Cell lysates were prepared using NET buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% [v/v] NP40, 1 mM EDTA), and protein concentrations were determined by Bradford assays.
Expression constructs
GFP-ADAR1 p150 (GFP-Adar1p150) was as described previously (6). Other constructs were prepared using standard cloning procedures (30). For GFP-ADAR1p110, the ADAR1p110 sequence [amino acids (aa) 296–1226; numbered as for ADAR1p150] was amplified by PCR using primers containing Xho1 and HindIII restriction sites, and inserted into pEGFP-C1 (Clontech). The same strategy was used to clone the following ADAR1 domains into pEGFP-C1: Z (aa 1–361), ZΔ (aa 1–295), ZR (aa 1–807), R (aa 486–807), RD (aa 486–1226), D (aa 812–1226), and ZαADAR1 (aa 123–209). For Flag-ADAR1p150, full-length ADAR1 (aa 1–1226) was amplified by PCR using primers containing HindIII and XbaI restriction sites, and inserted into p3xFLAG-CMV™-7.1 (Sigma). The same strategy was used to clone Flag-ADAR1p110 (aa 296–1226) and Flag-ZΔ (aa 1–295). For Flag-ΔN-NES-ADAR1, the oligonucleotide sequence 5′- GGCAGAGAGGTGTTGATTGCCTTTCCTCACATTTCCAGGAACTGAGTATCTACGAA was cloned into the SacII and BglII sites within Flag-ADAR1p150. Mutations within Flag-ADAR1p150 (K169A, E171A, Y177A, E912A) were introduced by site-directed mutagenesis (Stratagene). For GFP-ZαZBP1, the Zα domain from ZBP1 (aa 4–85) was amplified by PCR using primers containing Xho1 and HindIII restriction sites, and inserted into pEGFP-C1 (Clontech). ZBP1-GFP was as described previously (23). For Flag-PTB4-ZαADAR1 and Flag-ZαADAR1-PTB4, the ZαADAR1 domain (aa 123–209) was initially amplified by PCR using primers with EcoRI and BglII restriction sites, and inserted into p3xFLAG-CMV™-7.1. The inclusion or exclusion of a stop codon following ZαADAR1 gave rise to Flag-ZαADAR1-stop or Flag-ZαADAR1, respectively. Sequences corresponding to PTB4 (aa 1–557) were subsequently amplified by PCR using primers containing HindIII and EcoRI restriction sites, and inserted into Flag-ZαADAR1-stop to give Flag-PTB4-ZαADAR1. Alternatively, the sequence corresponding to PTB4 was amplified by PCR using primers containing BglII and SalI restriction sites, and inserted into Flag-ZαADAR1 to give Flag-ZαADAR1-PTB4. Flag-PTB4 was as described previously (31,32). For GFP-E3L, the E3L gene from vaccinia virus (Western Reserve strain) (aa 1–190) was amplified from genomic DNA by PCR using primers with XhoI and HindIII restriction sites, and inserted into pEGFP-C1 (Clontech). For GFP-ZαE3L, the Zα domain from E3L (aa 1–83) was amplified by PCR using primers containing Xho1 and HindIII restriction sites, and inserted into pEGFP-C1 (Clontech). All constructs were confirmed by sequencing.
Immunofluorescence
HeLa cells were plated on 13 mm glass cover slips in six-well plates, and grown for 24 h (cells were 50–70% confluent). Cells were washed in phosphate buffered saline (PBS), incubated 20 min in 4% (v/v) paraformaldehyde and then rewashed in PBS. Cells were permeabilized in 0.5% Triton-X (Fisher Scientific) for 3 min, washed with PBS, then incubated in blocking buffer (1% (w/v) BSA in PBS) for 1 h. Cells were subsequently incubated for 1 h with primary antibodies diluted in blocking buffer, washed extensively with PBS and then incubated with secondary antibodies for 1 h. Primary antibodies used were TIAR (Sc-1749; Santa Cruz Biotech, Inc.) and Flag (F3165; Sigma). Secondary antibodies were labeled with either Rhodamine Red-X RRX (red) or DyLight 488 or Alexa Fluor 488 (green) (Jackson ImmunoResearch). Cells were washed three times with PBS, dried and then mounted in ProLong® Gold antifade reagent with DAPI (Invitrogen). Cells were viewed and photographed using a Zeiss Axioimager M1 microscope (Carl Zeiss MicroImaging). Images were compiled using Adobe Photoshop.
Immunoblots
Proteins (50 μg) were separated by SDS–PAGE, transferred to PVDF and detected using enhanced chemiluminescence (ECL). Antibodies were GFP (sc-9996; Santa Cruz Biotech, Inc.); Flag (F3165; Sigma) and actin (A2066; Sigma). Secondary antibodies were horseradish peroxidase coupled (Jackson ImmunoResearch).
RESULTS
We have previously shown that ADAR1p150 localizes to cytoplasmic stress granules in HeLa cells during either arsenite-induced oxidative stress or following transfection of long dsRNA (24). We now aimed to further characterize the localization of ADAR1p150 in stress granules.
Localization of ADAR1p150 to stress granules does not require deaminase activity
We previously considered mechanisms by which ADAR1p150 may be recruited to cytoplasmic stress granules (24). One possible explanation was that ADAR1p150 localized to stress granules by virtue of association with IU-dsRNA following editing of cytoplasmic RNA targets. This idea was based on previous data that showed that IU-dsRNA bound to a ‘stress-granule-like complex’ that may precede stress granule assembly (33). We thus asked whether a functional deaminase domain of ADAR1p150 was required for its localization to stress granules.
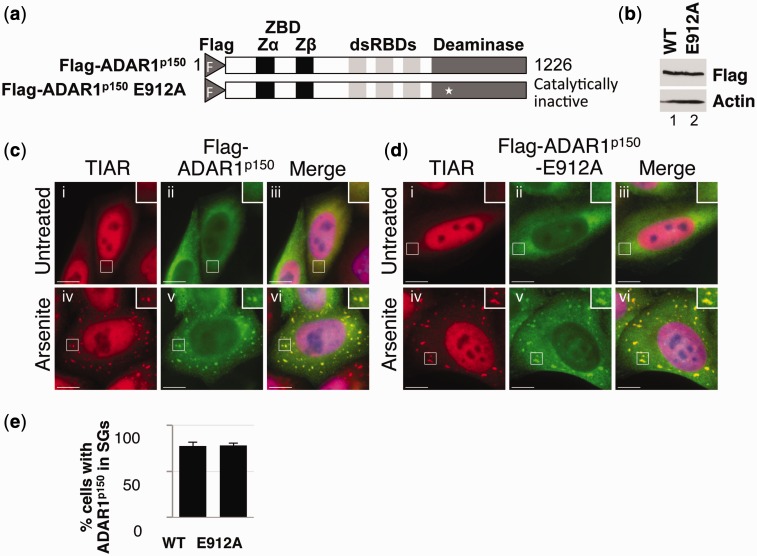
Expression constructs were prepared which encoded Flag-tagged full-length ADAR1p150 (Flag-ADAR1p150) or a Flag-tagged catalytically inactive mutant of ADAR1p150 (Flag-ADAR1p150 E912A), as described previously (34) (Figure 1a). HeLa cells were then transfected with Flag-ADAR1p150 and Flag-ADAR1p150 E912A, and after 24 h, the cells were cultured in the absence or presence of arsenite. Immunoblotting using a Flag antibody confirmed that expression was similar for each construct (Figure 1b). HeLa cells were subsequently fixed and stained with antibodies specific for TIA-1 related protein (TIAR), a stress granule marker (red) or Flag (green) and visualized using fluorescent microscopy. DAPI (blue) was used to stain the nucleus in this and all subsequent experiments. In the absence of arsenite (Untreated), TIAR localized to both the nucleus and cytoplasm, although was predominantly nuclear (Figure 1c and d; i and iii). In contrast, Flag-ADAR1p150 and Flag-ADAR1p150 E912A were largely cytoplasmic (Figure 1c and d; ii and iii). This is consistent with previous observations (6,24). Following arsenite treatment (Arsenite), TIAR, Flag-ADAR1p150 and Flag-ADAR1p150 E912A co-localized to stress granules (Figure 1c and d; iv–vi). The relative efficiency of localization of Flag-ADAR1p150 and Flag-ADAR1p150 E912A to stress granules was subsequently determined, which showed that Flag-ADAR1p150 and Flag-ADAR1p150 E912A localized to cytoplasmic stress granules with comparable efficiencies (Figure 1e). Together, these data showed that a functional deaminase domain was not required for localization of ADAR1p150 to stress granules.
Figure 1.
Deaminase activity is not required for stress granule localization. (a) A schematic diagram of Flag-ADAR1p150 and Flag-ADAR1p150 E912A. The white star indicates the position of the point mutation in Flag-ADAR1p150 E912A. The Flag epitope tag (F), Z-DNA-binding domains (ZBD; Zα and Zβ), dsRBDs and deaminase domain are indicated. (b) HeLa cells were transfected with expression vectors for wild-type Flag-ADAR1p150 (WT) and Flag-ADAR1p150 E912A (E912A), and lysates prepared after 24 h. Immunoblotting with a Flag antibody was used to analyze expression. Actin was a loading control. (c and d) HeLa cells transiently transfected with expression vectors for Flag-ADAR1p150 (c) and Flag-ADAR1p150 E912A (d). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) or Flag-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (e) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for wild-type Flag-ADAR1p150 (WT) or Flag-ADAR1p150 E912A (E912A) was then determined. Error bars are mean ± SD (n = 3).
ADAR1p150 constructs that contained the Zα domain localized to stress granules
Localization of ADAR1p150 to stress granules occurred independently of a functional deaminase domain (Figure 1). We next undertook experiments that aimed to understand how ADAR1p150 was localized to stress granules. Initially, we asked which domains of ADAR1p150 were responsible for its localization to stress granules.
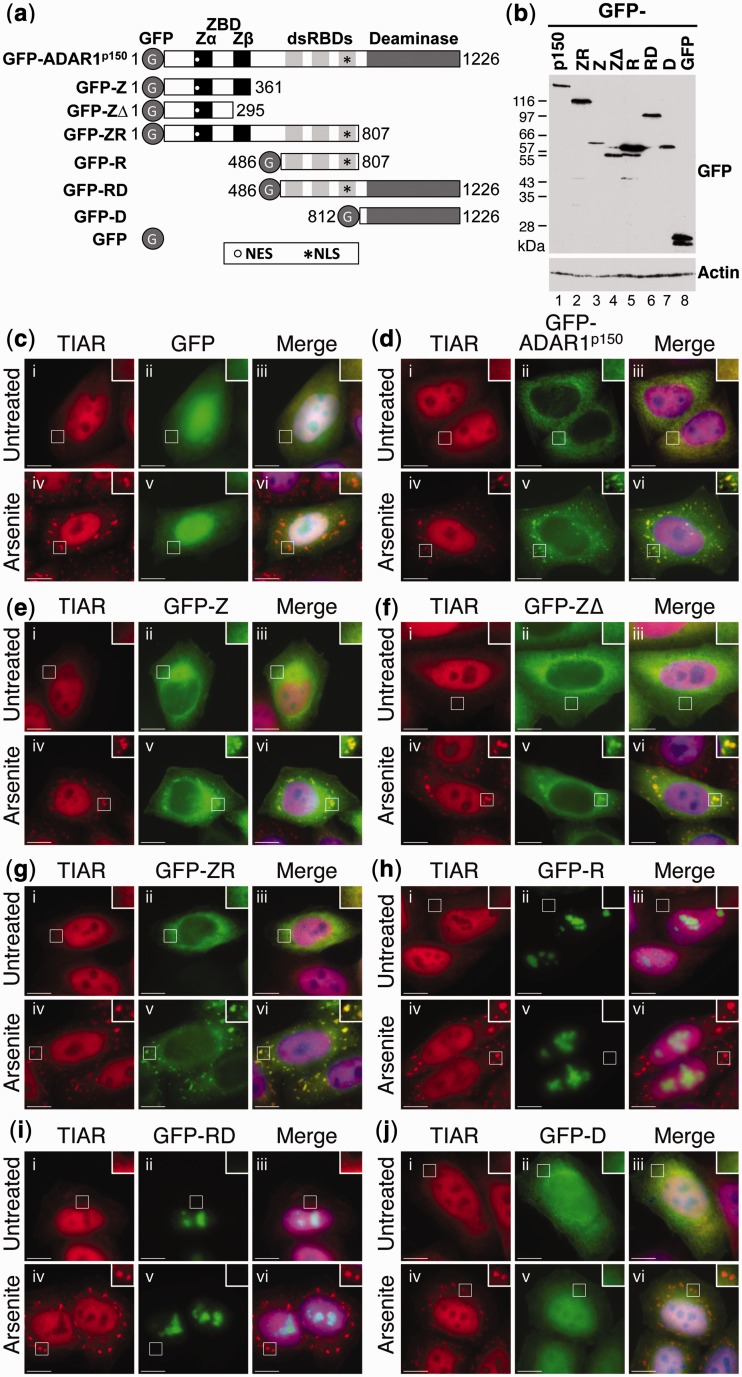
Expression constructs were prepared where GFP was fused to various domains of ADAR1p150 (Figure 2a), where the domain boundaries were in accordance with those used in previous studies (21,35). HeLa cells were subsequently transfected with the various ADAR1 expression constructs (GFP-ADAR1p150, GFP-Z, GFP-ZΔ, GFP-ZR, GFP-R, GFP-D) or with a control construct expressing GFP alone (GFP), and after 24 h, the cells were cultured in the absence (Untreated) or presence of arsenite (Arsenite). Immunoblotting using a GFP antibody confirmed that expression was similar for each construct (Figure 2b). HeLa cells were subsequently fixed and processed for visualization of TIAR (red) and the GFP-tagged proteins (green) using fluorescent microscopy. Although the GFP-tagged proteins could be visualized directly, specific antibodies were used to detect TIAR. In the absence of arsenite treatment, GFP and GFP-D co-localized with TIAR in both nuclear and cytoplasmic compartments, although they were substantially enriched in the nucleus (Figure 2c and j, respectively; i–iii). In contrast, GFP-ADAR1p150, GFP-Z, GFP-ZΔ and GFP-ZR were largely cytoplasmic in untreated cells (Figure 2d–g; ii and iii). Finally, GFP-R and GFP-RD appeared to localize exclusively to the nucleus, where they were found predominantly in the nucleolus (Figure 2h; i, ii and iii). The observed distribution of the ADAR1 domains was consistent with the position of nuclear export and import signals previously identified (Figure 2a). A nuclear export signal (NES) is present within the Zα domain of ADAR1p150, whreas a nuclear localization signal (NLS) is located within the third dsRBD of ADAR1 (5,7). Thus, although GFP-ADAR1p150, GFP-Z, GFP-ZΔ and GFP-ZR contained a NES and were localized to the cytoplasm, its absence in GFP-R and GFP-RD resulted in nuclear localization. Following treatment of HeLa cells with arsenite, GFP-ADAR1p150, GFP-Z, GFP-ZΔ and GFP-ZR co-localized with TIAR in stress granules (Figures 2d–g; iv–vi). In contrast, GFP-R, GFP-RD and GFP-D were not detected in stress granules following arsenite treatment (Figures 2h–j; iv–vi). Equivalent observations were made using Flag-tagged ADAR1 domains (data not shown). Analysis of these data together revealed that the only domain common to all of the ADAR1 constructs that localized to stress granules was the Zα domain. We therefore speculated that the N-terminal Zα domain was responsible for localization of ADAR1p150 to stress granules.
Figure 2.
The Zα domain of ADAR1p150 is required for localization to stress granules. (a) A schematic diagram of the GFP-ADAR1p150 constructs; the amino acids of ADAR1p150 included in each construct are shown. The position of the NES and NLS in each construct is shown as a white circle and a black asterisk, respectively. GFP (G), the Z-DNA-binding domains (ZBD; Zα and Zβ), dsRBDs and deaminase domain are indicated. (b) HeLa cells were transfected with expression vectors for GFP-ADAR1p150 (p150), GFP-ZR, GFP-Z, GFP-ZΔ, GFP-R, GFP-RD, GFP-D or GFP, and lysates were prepared after 24 h. Immunoblotting with a GFP antibody was used to analyze expression. Actin was a loading control. (c–j) HeLa cells were transiently transfected with expression vectors for GFP (c), GFP-ADAR1p150 (d), GFP-Z (e), GFP-ZΔ (f), GFP-ZR (g), GFP-R (h), GFP-RD (i) and GFP-D (j). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) or GFP-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm.
ADAR1p110 does not localize to stress granules
As described earlier, ADAR1p110 is truncated at the N-terminus by 295 amino acids, relative to full-length ADAR1p150. Importantly, this truncation effectively removes the Zα domain, although the Zβ domain remains intact (Figure 3a). We thus investigated whether ADAR1p110 localized to stress granules in the absence of the Zα domain.
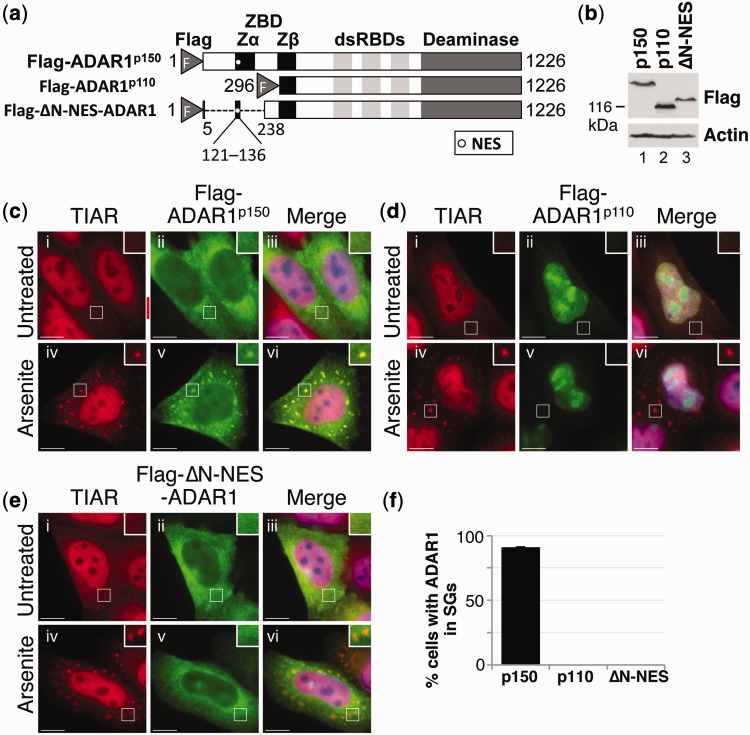
Figure 3.
ADAR1p110 does not localize to stress granules. (a) A schematic diagram of Flag-ADAR1p150, Flag-ADAR1p110 and Flag-ΔN-NES-ADAR1. The position of the NES is shown as a white circle. The Flag epitope tag (F), Z-DNA-binding domains (ZBD; Zα and Zβ), dsRBDs and deaminase domain are indicated. (b) HeLa cells were transfected with the expression constructs Flag-ADAR1p150 (p150), Flag-ADAR1p110 (p110) or Flag-ΔN-NES-ADAR1 (ΔN-NES), and lysates are prepared after 24 h. Immunoblotting with a Flag antibody was used to analyze expression. Actin was a loading control. (c–e) HeLa cells were transiently transfected with expression vectors for Flag-ADAR1p150 (c), Flag-ADAR1p110 (d) and Flag-ΔN-NES-ADAR1 (e). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) or Flag-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (f) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for Flag-ADAR1p150 (p150), Flag-ADAR1p110 (p110) or Flag-ΔN-NES-ADAR1 (ΔN-NES) was then determined. Error bars are mean ± SD (n = 3).
HeLa cells were transfected with expression constructs encoding Flag-ADAR1p150 and Flag-ADAR1p110 (Figure 3a), and after 24 h, they were cultured in the absence or presence of arsenite. Immunoblotting using a Flag antibody confirmed that expression of Flag-ADAR1p150 and Flag-ADAR1p110 was similar (Figure 3b). Cells were subsequently fixed and stained with antibodies specific for TIAR (red) or Flag (green) and visualized using fluorescent microscopy. In the absence of arsenite, ADAR1p150 was predominantly cytoplasmic while ADAR1p110 was found exclusively in the nucleus, where it was enriched in the nucleoli (Figure 3c and d; ii and iii). Nuclear localization of ADAR1p110 was due to the absence of the NES located within the Zα domain of ADAR1p150 (Figure 3a), as reported previously (5,6). When HeLa cells were treated with arsenite, ADAR1p150 co-localized with TIAR in cytoplasmic stress granules (Figure 3c; iv–vi). In contrast, ADAR1p110 remained in the nucleus and did not localize to stress granules following arsenite treatment (Figure 3d; iv–vi). These data are therefore in keeping with the idea that the Zα domain is required for localization to stress granules. On the other hand, an alternative explanation was that ADAR1p110 was not recruited to stress granules due to its nuclear localization. However, this seemed unlikely as other nuclear proteins re-localize to stress granules during stress (e.g. HuR, TIAR) (25). Nevertheless, to address this possibility, a new construct was prepared in which a deletion was made within Flag-ADAR1p150 that essentially removed the majority of the N-terminal region that is not present in ADAR1p110 (aa 6–237), while retaining the short sequence that contains the NES (aa 121–136), giving rise to Flag-ΔN-NES-ADAR1 (Figure 3a). HeLa cells were subsequently transfected with Flag-ΔN-NES-ADAR1, and after 24 h, they were cultured in the absence or presence of arsenite. Expression of Flag-ΔN-NES-ADAR1 (ΔN-NES) was verified using immunoblotting (Figure 3b). Cells were then fixed and stained with TIAR (red) or Flag (green) antibodies and visualized using fluorescent microscopy. In the absence of arsenite, Flag-ΔN-NES-ADAR1 was found predominantly in the cytoplasm (Figure 3e; ii and iii). Retention of the N-terminal NES was therefore effective in directing nuclear export, in contrast to what was seen with Flag-ADAR1p110 (compare Figure 3e and d; ii). When HeLa cells were treated with arsenite, TIAR localized to stress granules as expected (Figure 3e; iv). In contrast, Flag-ΔN-NES-ADAR1 did not co-localize with TIAR but remained distributed throughout the cytoplasm (Figure 3e; iv–vi). These data therefore demonstrated that despite its cytoplasmic localization, Flag-ΔN-NES-ADAR1, which was largely equivalent to Flag-ADAR1p110, did not localize to stress granules. Importantly, these data supported the idea that the Zα domain is required for localization to stress granules.
The Zα domain is sufficient for stress granule localization
The data described in Figures 2 and 3 suggested that the Zα domain is required for localization of ADAR1 to stress granules. We next directly tested whether the minimal Zα domain from ADAR1p150 (ZαADAR1) was sufficient for localization of proteins to stress granules.
An expression construct was prepared in which the short sequence encoding the ZαADAR1 domain was placed downstream of the coding region for GFP (Figure 4a; GFP-ZαADAR1). The ZαADAR1 domain boundaries chosen were based on those used in previous studies (13,15,36). HeLa cells were subsequently transfected with GFP-ZαADAR1, and after 24 h, they were cultured in the absence or presence of arsenite. Immunoblotting confirmed that GFP-ZαADAR1 was expressed at a level comparable with GFP-ADAR1p150 (data not shown). Cells were subsequently fixed and processed for visualization of TIAR (red) and GFP-ZαADAR1 (green) using fluorescence microscopy. In the absence of arsenite, TIAR was predominantly nuclear, whereas GFP-ZαADAR1 was largely cytoplasmic (Figure 4b; i–iii). In the presence of arsenite, both TIAR and GFP-ZαADAR1 co-localized to stress granules (Figure 4b; iv–vi). These data therefore confirmed that the ZαADAR1 domain was sufficient to direct localization of GFP to stress granules. Moreover, equivalent observations were made when the ZαADAR1 domain was fused to mCherry (data not shown).
Figure 4.
The Zα domain of ADAR1p150 is sufficient for localization to stress granules. (a) A schematic diagram of GFP-ZαADAR1. The position of the NES is shown as a white circle. The Flag epitope tag (F) and Zα domain are indicated. (b) HeLa cells were transiently transfected with the expression vector GFP-ZαADAR1. After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) or GFP-ZαADAR1 (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (c) A schematic diagram of Flag-PTB4, Flag-PTB4-ZαADAR1 and Flag-ZαADAR1-PTB4. The position of the NES is shown as a white circle. The Flag epitope tag (F) and Zα domains are indicated. (d) HeLa cells were transiently transfected with Flag-PTB4 (PTB4), Flag-PTB4-ZαADAR1 (PTB4-Zα) and Flag-ZαADAR1-PTB4 (Zα-PTB4), and lysates were prepared after 24 h. Immunoblotting with a Flag antibody was used to analyze expression. Actin was a loading control. (e–g) HeLa cells were transiently transfected with expression vectors for Flag-PTB4. (e) Flag-PTB4-ZαADAR1 (f) and Flag-ZαADAR1-PTB4 (g). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i, iv) or Flag-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (h) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for Flag-PTB4 (PTB4), Flag-PTB4-ZαADAR1 (PTB4-Zα) or Flag-ZαADAR1-PTB4 (Zα-PTB4) was then determined. Error bars are mean ± SD (n = 3).
To corroborate our findings, we went on to create additional Flag-tagged constructs where the ZαADAR1 domain was placed either downstream or upstream of the coding region for the polypyrimidine tract binding protein 4 (PTB4) (32) (Figure 4c; Flag-PTB4-ZαADAR1 and Flag-ZαADAR1-PTB4, respectively). A construct encoding Flag-PTB4 was used as a control (Figure 4c). HeLa cells were transfected with the various PTB4 expression constructs, and after 24 h, they were cultured in the absence or presence of arsenite. Immunoblotting was used to confirm equal expression (Figure 4d). Cells were then fixed and stained with antibodies specific for TIAR (red) or Flag (green). In untreated cells, Flag-PTB4 exclusively localized to the nucleus (Figure 4e; ii and iii). In contrast, addition of the ZαADAR1 domain either downstream or upstream of PTB4 (Flag-PTB4-ZαADAR1 or Flag-ZαADAR1-PTB4, respectively) resulted in both nuclear and cytoplasmic localization (Figure 4f and g; ii and iii). This altered localization of PTB4 was likely to result from inclusion of the NES within the ZαADAR1 domain. In arsenite-treated cells, Flag-PTB4 remained in the nucleus while TIAR localized to stress granules (Figure 4e; iv–vi). In contrast, Flag-PTB4-ZαADAR1 and Flag-ZαADAR1-PTB4 efficiently co-localized with TIAR in stress granules following arsenite treatment (Figure 4f, g; iv–vi, and h). The ZαADAR1 domain is thus sufficient to direct localization of PTB4 to stress granules. The observation that both Flag-PTB4-ZαADAR1 and Flag-ZαADAR1-PTB4 localized to stress granules additionally revealed that this function of the ZαADAR1 domain is independent of its relative position. Moreover, similar observations were made when the Zα domain was fused to the coding region for the RNA-binding protein Raver1 (31) (data not shown). These data together therefore demonstrated that the Zα domain from ADAR1p150 was sufficient to direct localization of various proteins to cytoplasmic stress granules.
The Zα domain is sufficient for localization to stress granules induced by poly(IC)
The Zα domain from ADAR1p150 was sufficient for localization of various proteins to arsenite-induced stress granules in HeLa cells (Figures 2–4). We next asked whether the ZαADAR1 domain was sufficient for localization to stress granules induced by transfection of long dsRNA [poly(IC)], which triggers an interferon response. In this case, long dsRNA typically induces stress granules via activation of PKR (protein kinase R), which phosphorylates eIF-2α and thereby inhibits translation (37). We have previously shown that ADAR1p150 localizes to stress granules that assemble in response to long dsRNA (24).
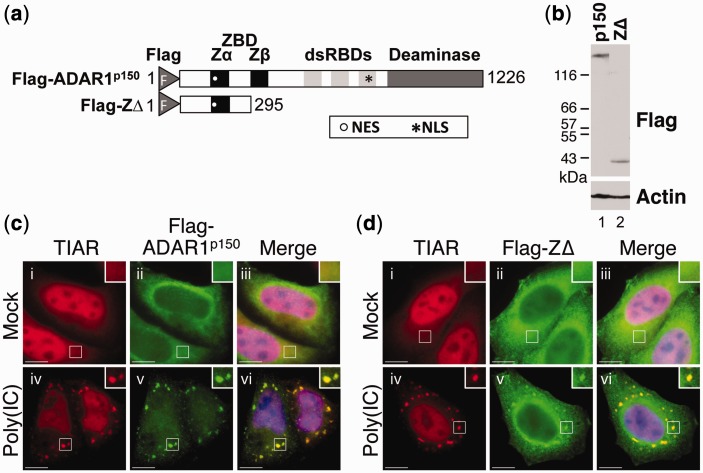
Expression constructs encoding either Flag-ADAR1p150 or Flag-ZΔ, which comprised the N-terminal region of ADAR1p150 and thus contained the ZαADAR1 domain (Figure 5a), were used to transfect HeLa cells. Immunoblotting using a Flag antibody confirmed that expression of Flag-ADAR1p150 and Flag-ZΔ was comparable (Figure 5b). After 24 h, the HeLa cells were either mock-transfected or transfected with poly(IC) to induce an interferon response, as described previously (24). After a further 7 h, the cells were fixed and stained for visualization of TIAR (red) or Flag-tagged proteins (green) using fluorescence microscopy. In mock-transfected cells, both Flag-ADAR1p150 and Flag-ZΔ were largely cytoplasmic (Figure 5c and d; ii and iii). Following transfection of cells with poly(IC), both Flag-ADAR1p150 and Flag-ZΔ co-localized with TIAR in stress granules (Figure 5c and d; iv–vi). These data therefore demonstrated that the Zα domain from ADAR1p150 was sufficient for localization to stress granules induced by poly(IC).
Figure 5.
The Zα domain of ADAR1p150 localizes to stress granules induced by poly(IC). (a) A schematic diagram of Flag-ADAR1p150 and Flag-ZΔ. The position of the NES and NLS in each construct is shown as a white circle and a black asterisk, respectively. The Flag epitope tag (F), the Z-DNA-binding domains (ZBD; Zα and Zβ), dsRBDs and deaminase domain are indicated. (b) HeLa cells were transfected with the expression constructs Flag-ADAR1p150 (p150) and Flag-ZΔ (ZΔ), and lysates were prepared after 24 h. Immunoblotting with a Flag antibody was used to analyze expression. Actin was a loading control. (c and d) HeLa cells were transiently transfected with expression vectors for Flag-ADAR1p150 (c) and Flag-ZΔ (d). After 24 h, the cells were either mock transfected (i–iii) or transfected with poly(IC) (iv–vi) before processing for visualization of TIAR (red; i and iv) and Flag-tagged proteins (green; ii and v) using fluorescence microscopy DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm.
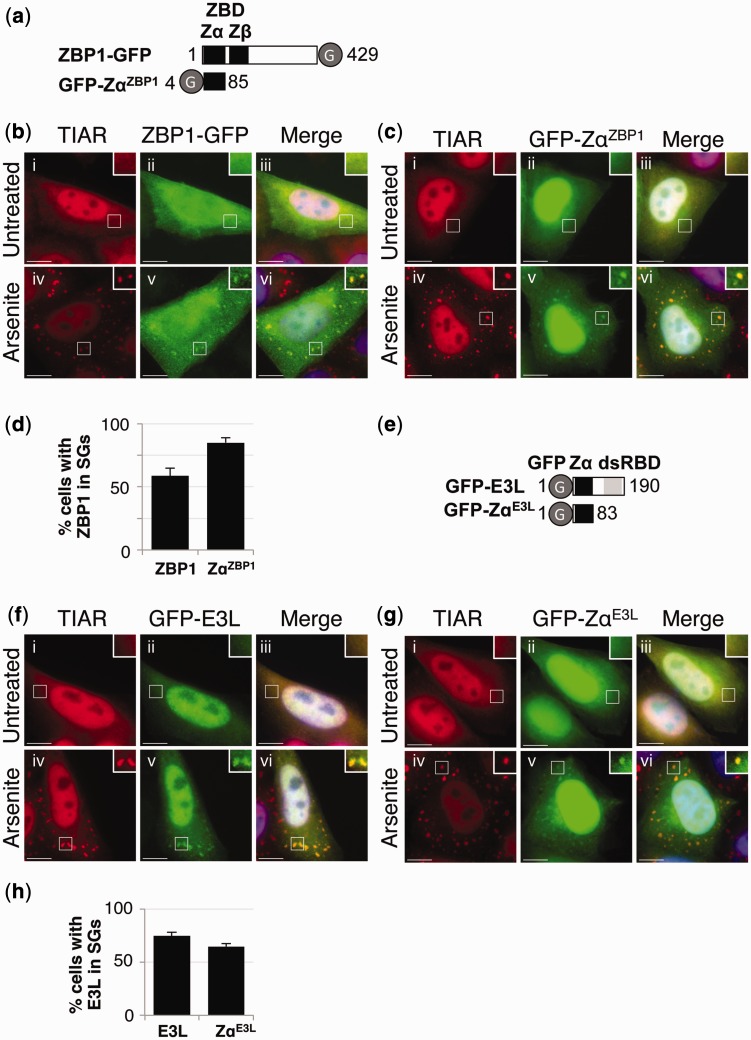
Zα domains from ZBP1 and E3L proteins also result in stress granule localization
The Zα domain from ADAR1p150 was sufficient to target various proteins to stress granules (Figures 4 and 5). We went on to investigate whether the Zα domain found in other Z-DNA binding proteins (ZBP1 and E3L) also gives rise to stress granule localization. Despite the fact that the Zα domains from ADAR1p150, ZBP1 and E3L share only ∼25% sequence identity, they are all functional Z-DNA-binding domains (14,22,23).
Expression constructs were initially prepared where GFP was fused to either full-length ZBP1 or the Zα domain from ZBP1 (ZαZBP1) to give ZBP1-GFP and GFP-ZαZBP1, respectively (Figure 6a). The boundaries of the ZαZBP1 domain were as described previously (23). HeLa cells were transfected with ZBP1-GFP or GFP-ZαZBP1, and after 24 h, cells were treated with or without arsenite. Cells were subsequently fixed and processed for visualization of TIAR (red) or GFP-tagged proteins (green) using fluorescent microscopy. In the absence of arsenite treatment, ZBP1-GFP was distributed throughout the cell, whereas GFP-ZαZBP1 was enriched in the nucleus (Figure 6b and c; ii and iii). In the presence of arsenite, both ZBP1-GFP and GFP-ZαZBP1 co-localized with TIAR in stress granules (Figure 6b, c; iv–vi, and d). The relatively weak appearance of the stress granules observed with GFP-ZαZBP1 was due to the strong nuclear signal. Although localization of GFP-ZαZBP1 to stress granules appeared to be slightly better than the full-length protein (ZBP1-GFP; Figure 6d), this may simply reflect a difference in expression. Localization of full-length ZBP1-GFP in stress granules was consistent with previous data (23). However, localization of the ZαZBP1 domain alone to stress granules had not previously been shown. This observation was nevertheless consistent with previous findings where deletion of the ZαZBP1 domain abolished stress granule localization (23). Our data together now confirmed that the ZαZBP1 domain was sufficient to localize GFP to stress granules following oxidative stress.
Figure 6.
The Zα domain from ZBP1 and E3L localizes to stress granules. (a) A schematic diagram of ZBP1-GFP and GFP-ZαZBP1. GFP (G) and the Z-DNA-binding domains (ZBD; Zα and Zβ) are indicated. (b and c) HeLa cells were transiently transfected with expression vectors for ZBP1-GFP (b) and GFP-ZαZBP1 (c). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) and GFP-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (d) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for ZBP1-GFP (ZBP1) or GFP-ZαZBP1 (ZαZBP1) was then determined. Error bars are mean ± SD (n = 3). (e) A schematic diagram of GFP-E3L and GFP-ZαE3L. GFP (G) and the Zα domain are indicated. (f and g) HeLa cells were transiently transfected with an expression vector for GFP-E3L (f) or GFP-ZαE3L (g), and after 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) and GFP-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (h) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for GFP-E3L (E3L) or GFP-ZαE3L (ZαE3L) was then determined. Error bars are mean ± SD (n ≥ 3).
We next investigated whether the Zα domain from the vaccinia virus E3L protein (ZαE3L) also resulted in stress granule localization following arsenite treatment. Expression vectors were prepared where the coding region for GFP was inserted upstream of the sequence for either full-length E3L or ZαE3L (GFP-E3L and GFP-ZαE3L, respectively; Figure 6e). GFP-E3L and GFP-ZαE3L were subsequently used to transfect HeLa cells, and after 24 h, the cells were cultured in the absence or presence of arsenite, then fixed and processed for visualization of TIAR (red) and GFP-tagged proteins (green) using fluorescence microscopy. In the absence of arsenite, GFP-E3L and GFP-ZαE3L were found in both the nucleus and cytoplasm, although they were enriched in the nucleus (Figure 6f and g; ii and iii). In the presence of arsenite, both GFP-E3L and GFP-ZαE3L co-localized with TIAR in stress granules (Figure 6f, g; iv–vi, and h). These data therefore confirmed that the ZαE3L domain was also sufficient for localization to stress granules.
These data together enabled us to conclude that all of the Zα-containing proteins tested (ADAR1p150, ZBP1 and E3L) localize to stress granules, and that the Zα domain was sufficient for their localization.
Amino acids required for Z-RNA binding are required for stress granule localization
We have convincingly demonstrated that the presence of a Zα domain is sufficient for localization of proteins to cytoplasmic stress granules (Figures 2–5). We now wanted to determine whether this function of the Zα domain depended on its ability to bind either Z-RNA or Z-DNA.
Structural analyses previously identified a number of highly conserved amino acid residues in the ZαADAR1 domain that are critical for interacting with both Z-DNA and Z-RNA (13,38). Moreover, co-crystal structures of ZαZBP1 and ZαE3L have shown that similar residues are likewise important for Z-DNA binding in other Zα domains (14,17). Mutagenesis further highlighted the importance of these amino acid residues for Z-DNA binding, as exemplified by various studies (22,23,39). We next tested whether mutation of key residues in the ZαADAR1 domain that are essential for Z-DNA/Z-RNA binding would interfere with localization to stress granules.
Expression constructs were therefore prepared in which single point mutations were introduced into full-length Flag-ADAR1p150 to give Flag-ADAR1p150 E171A, Flag-ADAR1p150 K169A and Flag-ADAR1p150 Y177A (Figure 7a). Although mutation of E171 was predicted to be a neutral mutation that would not affect nucleic acid binding, mutation of either Lys169 or Tyr177 was expected to disrupt binding of Flag-ADAR1p150 to Z-DNA or Z-RNA (13,39). HeLa cells were transfected with the various ADAR1p150 expression constructs, and after 24 h, they were cultured in the absence or presence of arsenite. Immunoblotting with a Flag antibody was used to show that expression was comparable (Figure 7b). Cells were subsequently fixed and stained with antibodies to TIAR (red) and Flag (green) for visualization by fluorescence microscopy. In the absence of arsenite, Flag-ADAR1p150, Flag-ADAR1p150 E171A, Flag-ADAR1p150 K169A and Flag-ADAR1p150 Y177A were all predominantly cytoplasmic (Figure 7c–f; ii and iii). Following arsenite treatment, both Flag-ADAR1p150 and Flag-ADAR1p150 E171A efficiently co-localized with TIAR in stress granules (Figure 7c, d; iv–vi and g). In contrast, the proportion of cells in which either Flag-ADAR1p150 K169A or Flag-ADAR1p150 Y177A co-localized with TIAR in stress granules was significantly less (Figure 7e, f; iv–vi and g). Moreover, when co-localization in stress granules was observed, the stress granules generally appeared much weaker than those seen with wild-type ADAR1p150 (data not shown). These data therefore confirmed that mutation of the amino acid residues necessary for binding to Z-RNA or Z-DNA substantially impaired localization of ADAR1p150 to stress granules. Moreover, similar observations were made when the same mutations were introduced into Flag-ZR, Flag-Z and Flag-ZΔ expression constructs (Supplementary Figures S1–S3). In addition, when equivalent mutations were made in the vaccinia virus E3L protein, a reduction in stress granule localization was also observed (Supplementary Figure S4).
Figure 7.
Z-RNA-binding residues are required for localization to stress granules. (a) A schematic diagram of Flag-ADAR1p150, Flag-ADAR1p150 K169A, Flag-ADAR1p150 Y177A and Flag-ADAR1p150 E171A. White stars indicate the positions of the point mutations. The Flag epitope tag (F), Z-DNA-binding domains (ZBD; Zα and Zβ), dsRBDs and deaminase domain are indicated. (b) HeLa cells were transfected with expression vectors for wild-type Flag-ADAR1p150 (WT), Flag-ADAR1p150 K169A (K169A) Flag-ADAR1p150 E171A (E171A) and Flag-ADAR1p150 Y177A (Y177A), and lysates were prepared after 24 h. Immunoblotting with a Flag antibody was used to analyze expression. Actin was a loading control. (c–f) HeLa cells were transiently transfected with expression vectors for Flag-ADAR1p150 (c), Flag-ADAR1p150 E171A (d), Flag-ADAR1p150 K169A (e) and Flag-ADAR1p150 Y177A (f). After 24 h, the cells were cultured in the absence (i–iii) or presence of arsenite (iv–vi) before processing for visualization of TIAR (red; i and iv) or Flag-tagged proteins (green; ii and v) using fluorescence microscopy. DAPI staining is in blue (Merge; iii and vi). Bar = 10 μm. (g) TIAR was used as a marker to identify stress granule-containing cells, and the proportion of cells with stress granules positive for wild-type Flag-ADAR1p150 (WT) or Flag-ADAR1p150 mutants (K169A, Y177A, and E171A) was then determined. P ≤ 1 × 10−3 (double asterisk). Error bars are mean ± SD (n = 3).
These data together therefore allowed us to conclude that binding of ADAR1p150 to Z-DNA or Z-RNA is necessary for localization to stress granules. As stress granules are cytoplasmic, it is plausible that Z-RNA is the most likely candidate to fulfill this role. Although many proteins have been shown to localize to stress granules as a result of mRNA binding (25), localization of specific proteins to stress granules via Z-RNA binding represents a novel method of recruitment.
DISCUSSION
We have conclusively demonstrated that the Zα domain from ADAR1p150 is necessary and sufficient for localization of ADAR1p150 to cytoplasmic stress granules in HeLa cells following either oxidative stress or transfection of long dsRNA. Furthermore, we have shown that the Zα domain from both ZBP1 and E3L is also able to direct localization to stress granules. Finally, our data suggest that localization of the ZαADAR1 domain to stress granules is dependent on its ability to interact with either Z-RNA or Z-DNA. We have thus identified a novel role for the Z-DNA-binding domains that are found within a limited number of proteins in eukaryotic cells.
We previously speculated that ADAR1p150 may localize to cytoplasmic stress granules following editing of cytoplasmic RNA targets, whereby the IU-dsRNA generated could trigger stress granule formation following specific interaction with a stress-granule-like complex (24,33). In this scenario, ADAR1p150 would localize to stress granules by virtue of association with the edited dsRNA. However, our data now rule out this possible explanation for how ADAR1p150 localizes to stress granules, as localization does not require a functional deaminase domain (Figure 1). Rather, our data suggest that recruitment of ADAR1p150 to stress granules depends on its interaction with Z-RNA within stress granules. Although we are unable to rule out the possibility that ADAR1p150 also interacts with Z-DNA, this seems an unlikely scenario due to its cytoplasmic location. The idea that Z-RNA exists in stress granules is in keeping with previous studies that showed that Z-RNA is abundant in both the cytoplasm and the nucleolus (40). The idea that ADAR1p150 localizes to stress granules by interacting with Z-RNA is intriguing. This scenario represents not only a novel method for localization of proteins to stress granules but also one that will result in rigorous selection of the few proteins that contain a Z-DNA-binding domain (9,41). It is therefore essential to consider the identity of the Z-RNAs that may be present in stress granules.
Various genome-wide studies have been undertaken to identify potential Z-DNA-forming regions (ZDRs) within eukaryotic genomes (19,42,43). Although in silico predictions have dominated these studies, one recent study additionally used a Z-DNA-specific probe (ZαADAR1) to obtain direct evidence for the existence of ZDRs in Z-conformation within human cells (19). In silico predictions alone suggested that ZDRs occur preferentially within sequences in close proximity of transcription start sites, where the Z-DNA would be stabilized by negative supercoiling that occurs in the wake of the translocating RNA polymerase (15,42,43). In contrast, when a Z-DNA-specific probe was used in conjunction with computational analyses, of the 186 ZDR hotspots identified, only two mapped to transcription start sites (19). Instead, many of the hotspots identified were found within tandem repeat sequences, including ALR/alpha satellite sequences and Alu subfamilies. The remaining ZDRs were predicted within introns and exons, consistent with previous data (43). Although these data focused on putative Z-DNA-forming sequences, it is likely that Z-RNA occurs within equivalent sequences. We therefore speculate that Z-RNA exists within the population of cellular RNAs sequestered within stress granules, which may include Alu sequences (25,26,44). It is likely that similar sequences exist within other mammalian genomes that also have the potential to form Z-DNA, including those found within introns and exons, as well as alternative tandem repeat sequences. It is therefore likely that Z-RNA would similarly exist in stress granules found in other mammalian cells. As discussed earlier in the text, we anticipate that any Z-RNA found within stress granules will interact specifically with proteins that contain Z-DNA-binding domains, such as ADAR1p150, and that this interaction will be essential for their recruitment (Figure 7). Interaction with other stress granule proteins may subsequently contribute to the stable association of ADAR1p150 in stress granules (24).
It is interesting to speculate that Z-RNA may form within Alu sequences that localize to stress granules in human cells (44). Previous data have shown that the majority of editing occurs within non-coding regions of RNA, particularly within Alu repeat sequences (45–48). These observations together lead us to speculate that editing of Alu sequences may occur following recruitment of ADAR1p150 to stress granules by Z-RNA binding. This idea is in keeping with a previous study that showed that editing of an RNA target was markedly enhanced when it contained a sequence that favored Z-RNA formation (11). Editing of the Alu sequence would thus give rise to IU-dsRNA, which would in turn suppress the induction of interferon and apoptosis triggered by stress (49). Cell survival would thus be enhanced following recruitment of ADAR1p150 to stress granules.
We have shown that full-length ADAR1p150, ZBP1 and E3L localize to stress granules (Figures 1–3 and 5–7). Localization of ZBP1 to stress granules is consistent with previous findings (23). However, we have gone further to show that the minimal Zα domain from ADAR1p150 (ZαADAR1), ZBP1 (ZαZBP1) or E3L (ZαE3L) fused to other proteins is sufficient to direct their localization to stress granules (Figures 4 and 6). The observation that the ZαZBP1 domain is sufficient for stress granule localization is in keeping with previous findings where deletion of ZαZBP1 from ZBP1 abolished stress granule localization (23). It is intriguing that each of the Z-DNA-binding domain-containing proteins plays a role in the immune response in mammalian cells. As mentioned earlier in the text, ADAR1p150 and ZBP1 are upregulated by interferon and have been implicated in host response mechanisms (16,21). Moreover, ADAR1p150 appears to play both antiviral and pro-viral roles in mammalian cells depending on the virus-host combination (50). The E3L protein from vaccinia virus antagonizes the actions of interferon and is thus essential for viral pathogenicity (22). Moreover, this observation depends on the presence of a functional Zα domain within E3L. Previous studies have postulated that the Zα domain of E3L may compete with host proteins such as ADAR1p150 and ZBP1 for binding nucleic acids in the Z-conformation (23). In light of our current data, we can now extend this hypothesis to propose that these proteins will compete for binding Z-RNA present in stress granules, which may in turn interfere with the role they play during the immune response in mammalian cells. In addition, co-localization of E3L and ADAR1 is likely to antagonize the enzymatic function of ADAR1p150 (41). We now intend to identify the Z-RNAs that bind ADAR1p150, ZBP1 and E3L in stress granules to understand the potential interplay between these proteins and how this impacts on the immune response.
The data we have described herein have identified a novel role for the Zα domain found in ADAR1p150 and other proteins that contain Z-DNA-binding domains. We have demonstrated that the Zα domain is necessary and sufficient for localization to cytoplasmic stress granules. This unexpected observation raises some interesting questions as to why Z-RNA should be used to localize proteins containing Z-DNA-binding domains to stress granules. Further studies will be therefore be key to fully understand how these proteins contribute to the immune response in mammalian cells. Nevertheless, our findings describe a hitherto unknown function for Z-DNA-binding domains.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council [BB/F018347/1 to A.D.J.S.]. Funding for open access charge: University of Cambridge.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Chris Smith and Stefan Rothenburg for plasmids, and Barbara Blacklaws for providing vaccinia virus genomic DNA.
REFERENCES
- 1.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl Acad. Sci. USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desterro JMP, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 7.Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol. Biol. Cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Athanasiadis A, Placido D, Maas S, Brown Ii BA, Lowenhaupt K, Rich A. The crystal structure of the Zβ domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005;351:496–507. doi: 10.1016/j.jmb.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Koeris M, Funke L, Shrestha J, Rich A, Maas S. Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res. 2005;33:5362–5370. doi: 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rich A. Z-DNA: the long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Mol. Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 15.Brown BA, Lowenhaupt K, Wilbert CM, Hanlon EB, Rich A. The Zalpha domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl Acad. Sci. USA. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishhi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 17.Ha SC, Lokanath NK, Van Quyen D, Wu CA, Lowenhaupt K, Rich A, Kim Y-G, Kim KK. A poxvirus protein forms a complex with left-handed Z-DNA: crystal structure of a Yatapoxvirus Zα bound to DNA. Proc. Natl Acad. Sci. USA. 2004;101:14367–14372. doi: 10.1073/pnas.0405586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: implications for development of a therapy for poxvirus infection. Proc. Natl Acad. Sci. USA. 2004;101:1514–1518. doi: 10.1073/pnas.0308260100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Xiao J, Li J, Lu L, Feng S, Dröge P. Human genomic Z-DNA segments probed by the Zα domain of ADAR1. Nucleic Acids Res. 2009;37:2737–2746. doi: 10.1093/nar/gkp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomé AR, Kus K, Correia S, Paulo LM, Zacarias S, de Rosa M, Figueiredo D, Parkhouse RME, Athanasiadis A. Crystal structure of a poxvirus-Like zalpha domain from cyprinid Herpesvirus 3. J. Virol. 2013;87:3998–4004. doi: 10.1128/JVI.03116-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barraud P, Allain F. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y-G, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl Acad. Sci. USA. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deigendesch N, Koch-Nolte F, Rothenburg S. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 2006;34:5007–5020. doi: 10.1093/nar/gkl575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissbach R, Scadden ADJ. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA. 2012;18:462–471. doi: 10.1261/rna.027656.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 27.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Revised edn. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 31.Gromak N, Rideau A, Southby J, Scadden ADJ, Gooding C, Huttelmaier S, Singer RH, Smith CWJ. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson F, Smith CWJ. A splicing repressor domain in polypyrimidine tract-binding protein. J. Biol. Chem. 2006;281:800–806. doi: 10.1074/jbc.M510578200. [DOI] [PubMed] [Google Scholar]
- 33.Scadden ADJ. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol. Cell. 2007;28:491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 35.Maas S, Kawahara Y, Tamburro KM, Nishikura K. A-to-I RNA editing and human disease. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schade M, Turner CJ, Kühne R, Schmieder P, Lowenhaupt K, Herbert A, Rich A, Oschkinat H. The solution structure of the Zα domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc. Natl Acad. Sci. USA. 1999;96:12465–12470. doi: 10.1073/pnas.96.22.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Placido D, Brown Ii BA, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schade M, Turner CJ, Lowenhaupt K, Rich A, Herbert A. Structure-function analysis of the Z-DNA-binding domain Zα of dsRNA adenosine deaminase type I reveals similarity to the (α + β) family of helix-turn-helix proteins. EMBO J. 1999;18:470–479. doi: 10.1093/emboj/18.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zarling DA, Calhoun CJ, Feuerstein BG, Sena EP. Cytoplasmic microinjection of immunoglobulin Gs recognizing RNA helices inhibits human cell growth. J. Mol. Biol. 1990;211:147–160. doi: 10.1016/0022-2836(90)90017-G. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Wolff KC, Jacobs BL, Samuel CE. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology. 2001;289:378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- 42.Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc. Natl Acad. Sci. USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroth GP, Chou PJ, Ho PS. Mapping Z-DNA in the human genome. Computer-aided mapping reveals a nonrandom distribution of potential Z-DNA-forming sequences in human genes. J. Biol. Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 44.Fitzpatrick T, Huang S. 3′-UTR-located inverted Alu repeats facilitate mRNA translational repression and stress granule accumulation. Nucleus. 2012;3:359–369. doi: 10.4161/nucl.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morse DP, Aruscavage PJ, Bass BL. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl Acad. Sci. USA. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barak M, Levanon EY, Eisenberg E, Paz N, Rechavi G, Church GM, Mehr R. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res. 2009;37:6905–6915. doi: 10.1093/nar/gkp729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 49.Vitali P, Scadden ADJ. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology. 2011;411:180–193. doi: 10.1016/j.virol.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.