Abstract
Transcription activator-like effector (TALE) proteins can be designed to bind virtually any DNA sequence. General guidelines for design of TALE DNA-binding domains suggest that the 5′-most base of the DNA sequence bound by the TALE (the N0 base) should be a thymine. We quantified the N0 requirement by analysis of the activities of TALE transcription factors (TALE-TF), TALE recombinases (TALE-R) and TALE nucleases (TALENs) with each DNA base at this position. In the absence of a 5′ T, we observed decreases in TALE activity up to >1000-fold in TALE-TF activity, up to 100-fold in TALE-R activity and up to 10-fold reduction in TALEN activity compared with target sequences containing a 5′ T. To develop TALE architectures that recognize all possible N0 bases, we used structure-guided library design coupled with TALE-R activity selections to evolve novel TALE N-terminal domains to accommodate any N0 base. A G-selective domain and broadly reactive domains were isolated and characterized. The engineered TALE domains selected in the TALE-R format demonstrated modularity and were active in TALE-TF and TALEN architectures. Evolved N-terminal domains provide effective and unconstrained TALE-based targeting of any DNA sequence as TALE binding proteins and designer enzymes.
INTRODUCTION
Transcription activator-like effector (TALE) proteins can be designed to bind virtually any DNA sequence of interest (1). The DNA binding sites for natural TALE transcription factors (TALE-TFs) that target plant avirulence genes have a 5′ thymidine.(1–3) Synthetic TALE-TFs also have this requirement. Recent structural data indicate that there is an interaction between the N-terminal domain (NTD) and a 5′ T of the target sequence.(4) A survey of the recent TALE nuclease (TALEN) literature yielded conflicting data regarding the importance of the first base of the target sequence, the N0 residue.(5–8) Additionally, there have been no studies regarding the impact of the N0 base on the activities of TALE recombinases (TALE-Rs). Here, we quantified the impact of the N0 base in the binding regions of TALE-Rs, TALE-TFs, TALE DNA-binding domains expressed as fusions with maltose binding protein (MBP-TALEs) and TALENs. Each of these TALE platforms have distinct N- and C-terminal architectures, but all demonstrated highest activity when the N0 residue was a thymidine. To simplify the rules for constructing effective TALEs in these platforms, and allow precision genome engineering applications at any arbitrary DNA sequence, we devised a structure-guided activity selection using our recently developed TALE-R system. Novel NTD sequences were identified that provided highly active and selective TALE-R activity on TALE binding sites with 5′ G, and additional domain sequences were selected that permitted general targeting of any 5′ N0 residue. These domains were imported into TALE-TF, MBP-TALE and TALEN architectures and consistently exhibited greater activity than did the wild-type NTD on target sequences with non-T 5′ residues. Our novel NTDs are compatible with the golden gate TALEN assembly protocol and now make possible the efficient construction of TALE transcription factors, recombinases, nucleases and DNA-binding proteins that recognize any DNA sequence allowing for precise and unconstrained positioning of TALE-based proteins on DNA without regard to the 5′ T rule that limits most natural TALE proteins.
MATERIALS AND METHODS
Oligonucleotides
Primers and other oligonucleotides (Supplementary Information) were ordered from Integrated DNA Technologies (San Diego, CA).
Generation of TALE-R NTD evolution plasmids
The TALE-R system previously reported by Mercer et al. (9) was adapted for this study. Briefly, pBCS (containing chloramphenicol and carbenicillin resistance genes) was digested with HindIII/Spe1. The stuffer (Avr X, where X is the N0 base), containing twin recombinase sites, was digested with HindIII/Xba1 and ligated into the vector to create a split beta-lactamase gene. pBCS AvrX was then digested with BamH1/Sac1, and Gin127-N-stuffer-Avr15 was digested with BamH1/Sac1 and ligated into the vector to create Gin127-N-stuffer-Avr15-X. The stuffer was digested with Not1/Stu1 for evolutions at the N-1 TALE hairpin and Not1/Sph1 for evolutions at the N0 TALE hairpin.
Generation of TALE NTD evolution libraries
Primer ptal127 Not1 fwd and reverse primers KXXG lib rev or KXXXX lib rev were used to generate N-terminal variants at the N-1 TALE hairpin and were subsequently digested with Not1/Stu1 then ligated into digested Gin127-AvrX. Forward primer ptal127 Not1 fwd and reverse primer KRGG Lib Rev were used to PCR amplify a library with mutations in the N0 TALE hairpin. This was subsequently digested with Not1/Sph1 and ligated into Not1/Sph1-digested Gin127-AvrX.
TALE-R NTD evolution assay
Round 1 ligations were ethanol precipitated and transformed into electrocompetent Top10 F’ cells then recovered in SOC for 1 h. The cells were grown overnight in 100 ml Super Broth (SB) media containing 100 µg/ml chloramphenicol. DNA was isolated via standard procedures. The resulting plasmid DNA (Rd 1 input) was transformed into electrocompetent Top10F’ cells; cells were grown overnight in 100 ml of SB containing 100 µg/ml carbenicillin and 100 µg/µl chloramphenicol. Plasmid DNA was isolated via standard procedures. Round 1 output was digested with Not1/Xba1 and ligated into the Gin127-AvrX vector with complementary sticky ends. This protocol was repeated three to four times when a consensus sequence was observed and clones were characterized.
Measurement of N-terminal TALEN activity
Four TALEN pairs containing each possible 5′ base were generated using the golden gate protocol (3,10). Fusion A and B plasmids were directly ligated via second golden gate reaction into the Goldy TALEN (N Δ152/C +63) framework. The NTD was modified by digesting the pCAG vector with BglII/Nsi1 and ligating with PCR-amplified NTD digested with BglII/Nsi1. TALEN pairs (50–75 ng each TALEN/well) were transfected into HeLa cells in wells of 96-well plates at a density of 1.5 × 104 cells/well. After transfection, cells were placed in a 37°C incubator for 24 h, then were moved to 30°C for 2 days and then moved to 37°C for 24 h. Genomic DNA was isolated according to a published protocol, and DNA mutation rates were quantified with the CelI Surveyor assay and by sequencing (11). For CelI assays, genomic DNA was amplified by nested PCR, first with primers CCR5 outer fwd/CCR5 outer rev and then with CCR5 inner fwd/CCR5 inner rev. For sequencing of indels, the second PCR was performed with CCR5 indel fwd/CCR5 indel rev. Fragments were then digested with BamH1/EcoR1 and ligated into pUC19 with complementary digestion.
TALE-TFs and luciferase assay
Variant NTDs from the recombinase selection were PCR amplified with primers ptal127 SFI fwd and N-Term Sph1. The PCR product was amplified and digested with Not1/Stu1 and ligated into pTAL127-SFI Avr15, which contains twin SFI-1 digestion sites facilitating transfer of the N-terminal-modified TALE from pTAL127-SFI Avr15 into pcDNA 3.0 VP64. Corresponding TALE binding sites were cloned into the pGL3 Basic vector (Promega) upstream of the luciferase gene. For each assay, 100 ng of pcDNA was co-transfected with 5 ng of pGL3 vector and 1 ng of pRL Renilla luciferase control vector into HEK293t cells in a well of a 96-well plate using Lipofectimine 2000 (Life Technology) according to manufacturer’s specifications. After 48 h, cells were washed, lysed and luciferase activity assessed with the Dual-Luciferase reporter system (Promega) on a Veritas Microplate luminometer (Turner Biosystems). Transfections were done in triplicate and results averaged.
MBP-TALE assay
Affinity assays of MBP-TALE binding to biotinylated oligonucleotides were performed using the protocol described by Segal et al. (12). Briefly, AvrXa7 TALE domains were expressed from pMAL MBP-AvrXa7 plasmid in XL1-Blue cells and purified on amylose resin. Biotinylated oligonucleotides containing the target AvrXa7 target site with modified 5′ residues were used to determine TALE-binding activity in sandwich enzyme-linked immunosorbent assay format. Antibodies targeting the MBP substituent were used for assay development.
RESULTS
Preliminary analysis of the 5′ T rule
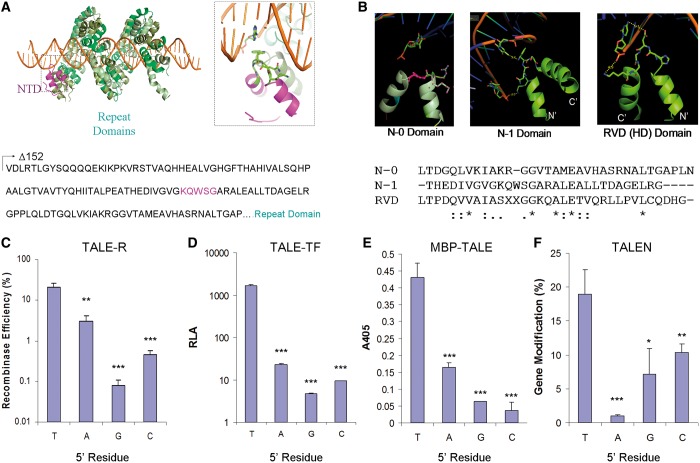
A recent crystal structure of a TALE protein bound to PthXo7 DNA sequence revealed a unique interaction between W232 in the N-1 hairpin with a thymidine at the 5′ end of the contacted region of the DNA substrate (the N0 base) (4). This study provided a structural basis for the previously established 5′ T rule reported when the TALE code was first deciphered (Figure 1a and b) (2). There are conflicting data regarding the importance of the first base of the target sequence of TALENs (5–8). We initially assessed the requirement for a 5′ T in the target DNA in the context of TALE-Rs using four split beta lactamase TALE recombinase selection vectors containing four AvrXa7 binding sites with all possible 5′ residues flanking a Gin32G core (Figure 1c). We then evaluated recognition of the N0 residue by TALE-TFs using four luciferase reporter vectors containing a pentamer AvrXa7 promoter region with recognition sites containing each possible 5′ residue (Figure 1d) (9,13). With bases other than a 5′ T, we observed decreases in activity up to >100-fold in TALE-Rs and 1000-fold in TALE-TFs relative to the sequence with a 5′ T (Figure 1c and d). These reductions were observed despite variations in the C-terminal architectures of these chimeras that reportedly remove the 5′ T bias, especially in the presence of a greatly shortened C-terminal domain (CTD) (7,14). Enzyme-linked immunosorbent assay also indicated decreased affinity of MBP-TALE DNA-binding proteins toward target oligonucleotides with non-T 5′ residues (Figure 1e). Finally, examination of the activity of designed TALENs with wild-type NTDs on targets with non-T 5′ nucleotides showed up to 10-fold decrease in activity versus those with a 5′ T (Figure 1f). Our results indicate that a 5′ T is an important design parameter for maximally effective TALE domains in the context of recombinases, transcription factors, nucleases and simple DNA-binding proteins.
Figure 1.
The specificity of the TALE NTD. (A) Illustration of a TALE bound to its target DNA (green, RVD domain; magenta, N-1 hairpin domain). (B) Structural analysis suggests contact of the 5′ T by W232 of the N-1 hairpin. This hairpin shares significant sequence homology with RVD hairpins. (C–F) Analyses of NT-T (wt) NTD in the context of (C) AvrXa7 TALE-R, (D) AvrXa7 TALE-TF, (E) AvrXa7 MBP-TALE and (F) a CCR5 targeting TALEN. (* = P < 0.05, ** = P < 0.01, *** = P < 0.001 compared with 5′ T).
Evolution of the TALE NTD to accommodate non-T 5′ residues
To create a more flexible system for DNA recognition, we hypothesized that we could use our recently developed TALE-R selection system to evolve the NTD of the TALE to remove the 5′ T constraint (Supplementary Scheme S1) (9). Libraries were generated with residues K230 through G234 randomized, and TALE-Rs with activity against each possible 5′ base were isolated after several rounds of selection (Figure 2a–c). The most active selected clones exhibited strong conservation of K230 and G234; the former may contact the DNA phosphate backbone, and the latter may influence hairpin loop formation (Supplementary Figure S2) (4). In the case of library K230-W232, K230S was frequently observed but had much lower activity than K230R or K230 variants in nearly all variants assayed individually. One clone (NT-G) of several observed with a W232 to R232 mutation demonstrated a significant shift of selectivity from 5′ T to 5′ G; the sequence resembles that of the NTD of a recently described Ralstonia TALE protein in this region. The Ralstonia NTD, in the context of plant transcription factor reporter gene regulation, has been reported to prefer a 5′ G in its substrate (see Supplementary Figure S3 for a protein alignment) (15). Residue R232 may contact the G base specifically, as indicated by the stringency of NT-G for 5′ G. The preference of NT-G for a 5′ G was comparable with the specificity of the wild-type domain for 5′ T. We were unable to derive NTD variants specific for 5′ A or 5′ C, but a permissive NTD, NT-αN, was obtained that resembles the K265-G268 N0 hairpin that accepts substrates with any 5′ residue and maintains high activity. We hypothesize that this variant makes enhanced non-specific contacts with the DNA phosphate backbone compared with the wild-type NTD, enhancing the overall binding of the TALE-DNA complex without contacting a specific 5′ residue.
Figure 2.
(A–C) Activities of recombinase selection variants against substrates with (A) 5′ G, (B) 5′ A and (C) 5′ C. (D) Alignment of optimized TALE NTDs, illustrating sequence differences in the N-1 hairpin. (E) Comprehensive comparison of optimized NTD activities in the context of MBP-TALE AvrXa7. (* = P < 0.05, ** = P < 0.01, *** = P < 0.001, compared with wild-type and 5′ A/G/C).
We hypothesized that a shortened hairpin structure would allow selection of variants with specificity for 5′ A or 5′ C residues. A library with randomization at Q231-W232 and with residue 233 deleted was designed to shorten the putative DNA-binding loop. Recombinase selection revealed a highly conserved Q231Y mutation that had high activity in a number of clones (Figure 2d). In particular, NT-βN demonstrated improved activity on substrates with 5′ A, C and G but diminished activity on 5′ T substrates compared with TALEs with the wild-type NTD (Figure 2e).
Applications of evolved TALE NTDs
To assess the portability of the evolved NTDs in designer TALE fusion protein applications, optimized NTDs were incorporated into TALE-TFs, MBP-TALEs and TALENs. TALE-TFs with NT-G, NT-αN and NT-βN domains demonstrated 400–1500-fold increases in transcriptional activation of a luciferase target gene bearing operator sites without a 5′ T residue when compared with the TALE-TF with the NT-T domain. The NT-G-based TF retained the 5′ G selectivity as observed in the TALE-R selection system. The activities of NT-αN- and NT-βN-based TFs against all 5′ nucleotides tracked the relative activity observed in the recombinase format (Figure 3). MBP-TALEs also exhibited greater relative binding affinity for target oligonucleotides with sites that did not have a 5′ T than did the wild-type MBP-TALE (Supplementary Figure S4), providing further evidence that the selected domains enhanced recognition of or tolerance for non-thymine 5′ bases.
Figure 3.

Analysis of selected NTDs in the context of TALE-TFs. (A) Illustration of 5xAvr promoter region on the luciferase reporter plasmid used for transcription activation experiments. (B) Relative luciferase activation of substrates with indicated 5′ residues by TALE-TFs with NT-T, NT-G, NT-αN and NT-βN domains. (* = P < 0.05, ** = P < 0.01, *** = P < 0.001, compared with NT-T and respective 5′ A/G/C/T).
Four of the optimized NTDs were then imported into the Goldy TALEN framework (10). For these experiments, four substrates were constructed within the context of the Δ32 locus of the CCR5 gene (Figure 4a). Each substrate contained a different 5′ residue. Experiments included TALENs with wild-type (NT-T) and dHax3 NTDs (dHax3 is commonly used NTD variant isolated from Xanthomonas campestris) with specificity for 5′ T (5,14,16), to benchmark gene editing activity. The substrate TALEN pairs were designed to retain as much RVD homology (50–90%) as possible to determine the activity enhancing contributions of the variant NTDs (Figure 4a).
Figure 4.
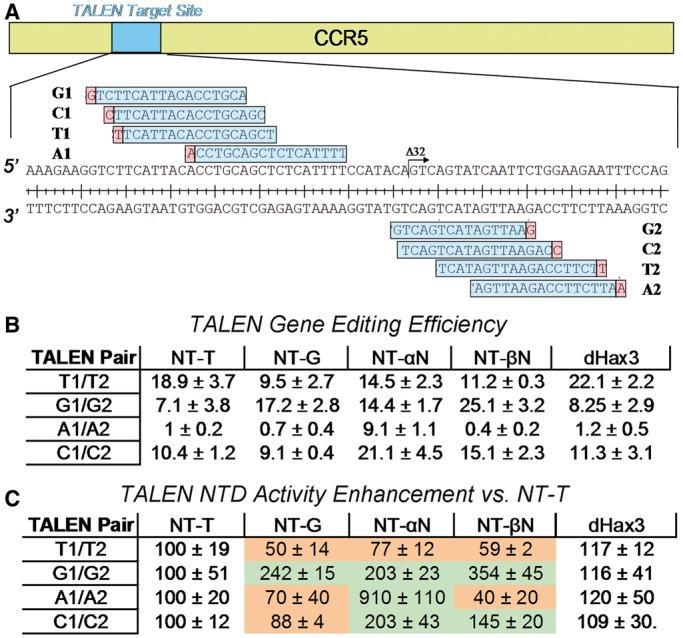
Design and activity of TALEN pairs with wild-type and evolved NTD’s with varying 5′ bases. (A) The CCR5 gene expanded to highlight the target site for induction of the Δ32 mutation. (B) Gene editing efficiency of the wild-type (NT-T) TALEN, TALENs with domains optimized for non-T 5′ residues, and dHax3 NTD. (C) Fold enhancement of the TALEN pairs with optimized NTD versus TALENs with 5′ T specificity. The activity of each NTD is shown on each TALEN pair substrate.
Activities of the TALENs were analyzed both by sequencing and by using the Cel1 assay (11). The selected domains exhibited increases in gene editing activity between 2- and 9-fold for the non-T 5′ residues when compared with activities of the TALEN containing the wild-type domain (Figure 4 and Supplementary Figure S5). Activity was highest on TALEN pair T1/T2 with wild-type or dHax3 NTD. The TALEN pair substrate G1/G2 was processed most effectively by TALENs with NT- αN, NT-βN and NT-G, with 2.0–3.5-fold enhancement versus NT-T. NT-αN had activity 9- and 2-fold higher than the wild-type NT-T on TALEN pairs A1/A2 and C1/C2, respectively. Although the impact of a mismatch at the 5′ residue is more modest in TALENs than in TALE-TF and TALE-R frameworks, the optimized NTDs greatly improved TALEN activity when used in gene editing experiments.
DISCUSSION
Most, but not all, previous studies have suggested that a thymidine is required as the 5′-most residue in design of optimal TALE DNA-binding domains (3,5–7,10,13,14,17). The analyses described here indicate that a thymidine is optimal, and in some cases critical, for building functional TALE fusion proteins. This requirement therefore imposes limitations on the sequences that can be effectively targeted with TALE transcription factor, nuclease and recombinase chimeras. Although this requirement theoretically imposes minor limitations on the use of TALENs for inducing gene knockout, given their broad spacer region tolerance, NTD’s that can accommodate any 5′ residue would further simplify the rules for effective TALE construction and greatly enhance applications requiring precise TALE placement for genome engineering and interrogation (e.g. precise cleavage of DNA at a defined base pair using TALENs, seamless gene insertion and exchange via TALE-Recombinases, displacement of natural DNA-binding proteins from specific endogenous DNA sequences to interrogate their functional role, the development of orthogonal transcription factors for pathway engineering, the synergistic activation of natural and synthetic genes wherein transcription factor placement is key (18,19) and many other applications). Other uses in DNA-based nanotechnology include decorating DNA nanostructures/origami with specific DNA-binding proteins (20,21). Here, targeting to specific sites is constrained based on DNA folding/structure and thus being able to bind any site is critical. Elaboration of these structures and devices with DNA-binding proteins could be a fascinating approach to expanding function. Indeed, it is not difficult to imagine many applications for DNA binding proteins and their fusions when all targeting constraints are removed. Encouraged by these potential applications, we aimed to develop NTDs that enable targeting of sites initiated at any base.
We used our recently developed TALE-R system to evolve the NTD of the TALE to remove the 5′ T constraint. In three rounds of selection, we obtained an NTD with specificity for a 5′ G. Numerous selections were performed in attempts to obtain variants that recognized either 5′ A or 5′ C. We inverted the G230-K234 hairpin, extended the K230-G234/ins232 hairpin, attempted modification of the K265-G268 N0 hairpin, and evaluated random mutagenesis libraries. None of these strategies yielded NTDs with affinity for target sequences with 5′ A or 5′ C, although we did identify an NTD, NT-βN, with a deletion that recognized substrates with both 5′ A and 5′ C residues with acceptable affinity.
The strong selection preference exhibited by the NTDs NT-T and NT-G and the importance of W232 in NT-T and R232 in NT-G are likely due to specific interactions of these amino acids with the 5′ terminal residue of the DNA recognition sequence. It was recently reported that the Ralstonia solanacearum TALE stringently requires a 5′ G, and a sequence alignment with NT-G shows what appears to be a comparable N-1 hairpin containing an arginine at the position analogous to 232 in NT-G (Supplementary Figure S3). Owing to the high structural homology between the NTDs Brg11 and NT-T, it may be possible to modify the preference of the Ralstonia TALE NTD to thymine by a simple arginine to tryptophan mutation or to eliminate specificity by grafting NT-αN or NT-βN domains into this related protein. It is also interesting to note that arginine–guanine interactions are common in evolved zinc finger domains (22).
The variant NTDs selected were successfully imported into TALE-TFs, MBP-TALEs and TALENs and generally conferred the activity and specificity expected based on data from the recombinase evolution system. TALE-TFs with optimized NTDs enhanced TALE activation between 400- and 1500-fold relative to the activity of NT-T against AvrXa7 promoter sites with non-T 5′ residues. When incorporated into TALENs, our NTD with non-T selectivity enhanced activity 2–9-fold relative to that of the NT-T domain on substrates with 5′ A, C or G. The increases in TALEN gene editing generally correlated with increases in activity observed in TALE-R and TALE-TF constructs. The specificity and high activity of NT-G was maintained, as evidenced by the lower activity in assays with TALEN pairs A1/A2, C1/C2, and T1/T2, and the generally high activity of NT-αN and NT-βN was also imparted into the TALEN Δ152/+63 architecture.
It was recently reported that alternatively truncated TALEs with synthetic TALE RVD domains do not require a 5′ T in the DNA substrate (7). We constructed the reported Δ143, +47 truncation as a Goldy TALE-TF and observed substantially lower activity on the AvrXa7 substrate than we observed with the Δ127, +95 truncation, which has been most commonly used by others and which is the truncation set used in our study (Supplementary Figure S6) (7,14). Thus, the difference in reported outcomes could be due to the truncated architectures used.
In summary, we confirmed the importance of a 5′ thymidine in the DNA substrate for binding and activity of designed TALEs in the context of TALE-R, TALE-TF, MBP-TALEs and TALEN chimeras. Targeted mutagenesis and TALE-R selection were applied to engineer TALE NTDs that recognize bases other than thymine as the 5′ most base of the substrate DNA. The engineered TALE domains developed here demonstrated modularity and were highly active in TALE-TF and TALEN architectures. These novel NTDs expand by ∼15-fold the number of sites that can be targeted by current TALE-Rs, which have strict geometric requirements on their binding sites and which are highly sensitive to the identity of the N0 base (9). Furthermore, they now allow for the precise placement of TALE DBDs and TALE-TFs at any DNA sequence to facilitate gene regulation, displacement of endogenous DNA-binding proteins and synthetic biology applications where precise binding might be key. Although TALENs based on the native NTD show varying degrees of tolerance of N0 base substitutions, our data indicate that the novel NTDs reported here also facilitate higher efficiency gene editing with any N0 base as compared with natural NTD-based TALENs. With the removal of all sequence targeting restraints from TALE-based proteins, we envision the ever-expansive use of this technology in genome engineering, synthetic biology, medicine and nanotechnology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes for Health (NIH) grant [DP1CA174426].
Conflict of interest statement. Authors are inventors on a patent application covering this work.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Thomas Gaj and Jia Liu for discussion. B.M.L., A.C.M. and C.F.B. designed research; B.M.L. and A.C.M. performed experiments; B.M.L., A.C.M. and C.F.B. analyzed data; and B.M.L. and C.F.B. wrote the manuscript.
REFERENCES
- 1.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 2.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 3.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012;40:e117. doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao H, Wu X, Chai J, Han Z. Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res. 2012;22:1716–1720. doi: 10.1038/cr.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsaesser J, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Rese. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol. Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 9.Mercer AC, Gaj T, Fuller RP, Barbas CF., III Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 2012;40:11163–11172. doi: 10.1093/nar/gks875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, II, Tan W, Penheiter SG, Ma AC, Leung AY, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF., III Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods. 2012;9:805–807. doi: 10.1038/nmeth.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. the USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 15.de Lange O, Schreiber T, Schandry N, Radeck J, Braun KH, Koszinowski J, Heuer H, Strauß A, Lahaye T. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytologist. 2013;199:773–786. doi: 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- 16.Kay S, Boch J, Bonas U. Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant Microbe Interact. 2005;18:838–848. doi: 10.1094/MPMI-18-0838. [DOI] [PubMed] [Google Scholar]
- 17.Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beerli RR, Dreier B, Barbas CF., III Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl Acad. Sci. USA. 2000;97:1495–500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacca B, Niemeyer CM. DNA origami: the art of folding DNA. Angew Chem. Int. Ed. Engl. 2012;51:58–66. doi: 10.1002/anie.201105846. [DOI] [PubMed] [Google Scholar]
- 21.Simmel FC. DNA-based assembly lines and nanofactories. Curr. Opin. Biotechnol. 2012;23:516–521. doi: 10.1016/j.copbio.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 22.Segal DJ, Crotty JW, Bhakta MS, Barbas CF, III, Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J. Mol. Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.