Abstract
A number of meiosis-specific transcripts are selectively eliminated during the mitotic cell cycle in fission yeast. Mmi1, an RNA-binding protein, plays a crucial role in this selective elimination. Mmi1 recognizes a specific region, namely, the determinant of selective removal (DSR) on meiotic transcripts and induces nuclear exosome-mediated elimination. During meiosis, Mmi1 is sequestered by a chromosome-associated dot structure, Mei2 dot, allowing meiosis-specific transcripts to be stably expressed. Red1, a zinc-finger protein, is also known to participate in the Mmi1/DSR elimination system, although its molecular function has remained elusive. To uncover the detailed molecular mechanisms underlying the Mmi1/DSR elimination system, we sought to identify factors that interact genetically with Mmi1. Here, we show that one of the identified factors, Iss10, is involved in the Mmi1/DSR system by regulating the interaction between Mmi1 and Red1. In cells lacking Iss10, association of Red1 with Mmi1 is severely impaired, and target transcripts of Mmi1 are ectopically expressed in the mitotic cycle. During meiosis, Iss10 is downregulated, resulting in dissociation of Red1 from Mmi1 and subsequent suppression of Mmi1 activity.
INTRODUCTION
Cells drastically change their gene expression profiles to adapt to changes in their environment. Previous experiments in yeast have demonstrated that hundreds of transcripts are upregulated when cells enter the meiotic program from mitotic growth (1,2). This global change of gene expression is carried out through posttranscriptional regulation in addition to transcriptional regulation. In the fission yeast Schizosaccharomyces pombe, an RNA-binding protein, Mmi1, plays a crucial role in selectively eliminating meiosis-specific transcripts in mitotic cells (3,4). Mmi1 belongs to the YTH family (5) and recognizes a region termed the determinant of selective removal (DSR), which is enriched with repeats of hexanucleotide motifs (6). Meiotic transcripts targeted by Mmi1 are removed from cells via nuclear exosome- and polyadenylation-dependent degradation (3,7). The canonical poly(A)-polymerase, Pla1, and the fission yeast homolog of poly(A)-binding protein nuclear 1 (PABPN1), Pab2, play a pivotal role in Mmi1-mediated elimination of meiotic transcripts (7,8). The zinc finger protein, Red1, is also known to be involved in the Mmi1/DSR system, although its molecular function remains unknown (9). Pla1, Pab2, exosome subunits such as Rrp6, and Red1 colocalize with Mmi1 foci present in the nuclei of mitotically growing cells (7,9).
Mmi1 inhibits the progression of meiosis because Mmi1 regulates meiotic transcripts that are essential for meiosis (3). Moreover, Mmi1 overexpression impairs meiosis (3). To overcome Mmi1-mediated suppression of meiosis, another RNA-binding protein, Mei2, and its binding partner, meiRNA, suppress Mmi1 (3). In meiotic prophase, Mei2 and meiRNA form a dot structure at the sme2 gene locus on chromosome II, which encodes meiRNA (10–12). meiRNA carries multiple copies of the DSR motif and is degraded via the Mmi1-mediated degradation machinery (6). On the basis of these observations, we proposed that meiRNA serves as a decoy substrate for Mmi1.
Mmi1 also induces heterochromatin formation at a subset of its target genes (13–15). Red1 is required for Mmi1-mediated heterochromatin formation. Red1 associates with the loci of Mmi1 targets that are methylated on lysine 9 of histone H3, the hallmark of heterochromatin (13). Furthermore, interaction between Red1 and Clr4, the histone methyltransferase homologous to SUV39h (16), has been previously observed (13), suggesting that Red1 is a determinant to select a subset of genes from Mmi1 target genes for facultative heterochromatin formation.
The importance of the Mmi1/DSR system is unambiguous, as loss of Mmi1 activity causes ectopic expression of meiotic transcripts and is toxic to cell growth (3). To further elucidate the molecular mechanisms underlying Mmi1-mediated RNA degradation, we screened for factors that are involved in the Mmi1/DSR system (7). In this study, we characterized the factors identified in the screen, and we herein describe a novel factor that regulates Red1.
MATERIALS AND METHODS
Fission yeast strains, genetic analysis and media
The S. pombe strains used in this study are listed in Supplementary Table S1. General genetic procedures used for the analyses of the S. pombe strain have been previously described (17). A standard protocol was used for deletion and gene tagging (18). Growth media used in the study included complete medium Yeast Extract (YE), Minimal Medium (MM) (19), synthetic sporulation medium Synthetic Sporulation Agar (SSA) (20) and sporulation medium SPorulation Agar (SPA) (17).
Genetic screen
Suppressors of JZ464 were isolated using random insertion of a G418-resistant cassette as previously described (21). The G418-resistant cassette was amplified by PCR with primers N18-CGGATCCCCGGGTTAATTAA and N18-GAATTCGAGCTCGTTTAAAC (N18: 18 nt random DNA sequence). The PCR products were introduced into JZ464 cells. G418-resistant transformants were replica plated on SSA to induce spore formation. Colonies stained by iodine vapors, a stain specific for spores, were selected, and the site of the cassette insertion was determined by sequencing of inverse PCR products.
Plasmid construction
The iss10 open reading frame (ORF) was PCR-amplified with a pair of primers, one carrying a SalI site at the initiation codon and a NotI site at the stop codon. PCR products were digested with SalI and NotI and cloned into pREP41 (22), carrying the GFP ORF or three copies of the HA epitope so that GFP or the HA epitope was fused to the C terminus of the iss10 ORF. The red1 ORF was similarly cloned with a pair of primers, one carrying an NdeI site and a BamHI site.
Fluorescence microscopy
For observations of mitotically growing cells, cells were grown in MM medium at 30°C. To induce meiosis, cells were grown in MM medium at 30°C, washed, spotted on SPA medium and incubated for 4–6 h at 30°C. The DeltaVision/SoftwoRx system (Applied Precision) was used for fluorescence microscopy. Images were taken along the z-axis at 0.2 µm intervals, deconvolved and merged into a single projection.
Northern and western blot analyses
Northern blot analysis was performed as previously described (23) by using DNA probes for transcripts of mei4, ssm4 and iss10. Immunoprecipitation and western blot analysis were performed as previously described (24) by using anti-Mmi1 antibodies (our laboratory preparation), anti-myc antibody (9E10; Santa Cruz Biotechnology), anti-GFP antibody (clones 7.1 and 13.1; Roche Applied Science), and anti-α-tubulin antibody (TAT-1; a gift from Dr. Keith Gull).
Quantitative RT-PCR analysis
cDNA was synthesized using total RNA treated with DNase I (Turbo DNA-free kit; Ambion) according to the manufacturer’s instructions (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems). Quantitative PCR was performed using the 7300 Real Time PCR System and Power SYBR Green PCR Master Mix (Applied Biosystems). The act1 gene encoding actin was used for normalization. Primers used in this study are listed in Supplementary Table S2.
Two-hybrid assay
The iss10 ORF and the red1 ORF were cloned in pGAD424 and pGBKT7 (Clontech), respectively. The Saccharomyces cerevisiae strain AH109 was transformed with both plasmids. pGAD-T-antigen, pGBK-p53 and pGBK-lamin were used as controls.
RESULTS
Identification of factors involved in Mmi1-driven selective elimination
The sme2Δ strain of fission yeast cannot proceed to meiosis due to retention of Mmi1 activity during meiosis (Supplementary Figure S1) (3,6,25). To identify factors that might be related to the DSR/Mmi1-dependent elimination system, we screened for mutations that could recover meiotic arrest in the sme2Δ strain by randomly inserting a G418-resistant kanR cassette into the genome and determining the sites of the insertion (7). In addition to the previously reported factors, pab2 and pla1 (7), we isolated six mutations designated as iss (insertional suppressor of sme2Δ) (Figure 1A and B). The iss1 gene (SPAC22G7.10) encodes a poly(A) polymerase-binding protein, which is homologous to budding yeast Fip1 (26). The iss3 gene (SPAC1006.93c) is identical to red1, which encodes a conserved zinc-finger protein and has been shown to be involved in Mmi1-mediated selective elimination (9,14). The iss4 gene (SPBC337.03) encodes an RNA polymerase II transcription termination factor, which is homologous to budding yeast RTT103 (27). Recently, the same gene was reported as rhn1 and was found to suppress the expression of meiotic mRNAs (28). The iss6 gene (SPAC19G12.17) is identical to erh1/new10, which encodes an enhancer of rudimentary homolog (ERH)-like protein (29,30). The iss9 gene (SPBC2A9.11c) encodes a protein homologous to budding yeast Thp3, which is involved in transcription elongation (31), whereas the iss10 gene (SPAC7D4.14c) encodes a protein with no homology to any known protein. The insertion sites of the kanR cassette of each mutant are shown in Supplementary Figure S2. In the iss4 and iss6 mutants, the cassette was inserted into the upstream region of the genes, whereas the sites of insertion in the other mutants were within the ORFs (Supplementary Figure S2).
Figure 1.

Suppressor mutants of meiotic arrest in sme2Δ. (A) Suppressor genes of sme2Δ and their gene products. (B) Recovery of sporulation in the sme2Δ mutant by iss mutations. JY450 (wt), JZ464 (sme2Δ) and sme2Δ iss double mutants (JT549 [pab2], JT551 [pla1], JT552 [iss1], JT550 [iss3], JT954 [iss4], JT955 [iss6], JT956 [iss9] and JT957 [iss10]) were incubated on SSA medium at 30°C for 4 days, and sporulation frequency was measured (n > 500).
Involvement of the iss genes in the Mmi1/DSR elimination system
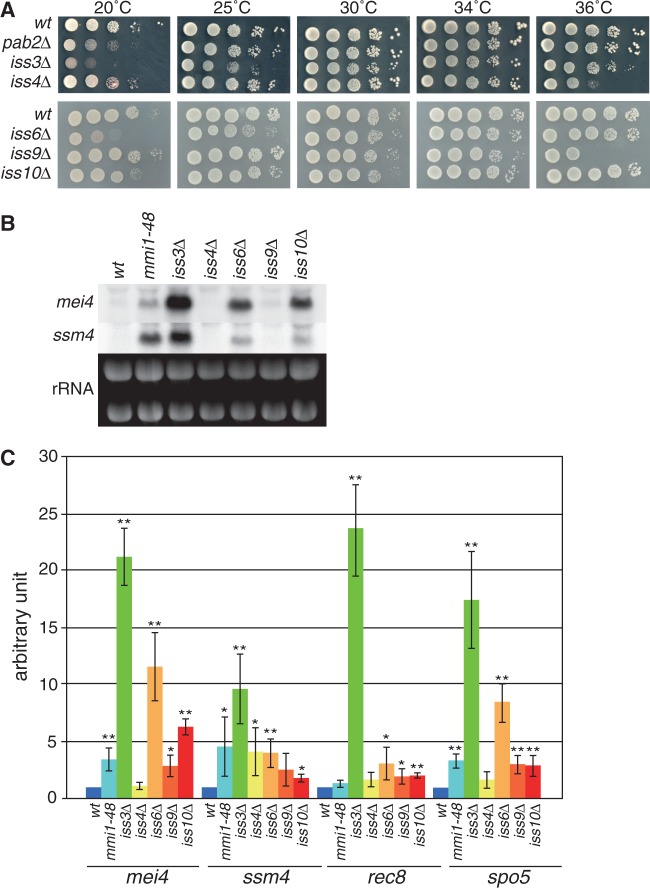
We examined whether the iss genes identified in our screen were involved in the Mmi1-dependent elimination system. We made deletion mutants of each gene and observed the resulting phenotypes. pla1 and iss1, whose product is predicted to interact with Pla1 based on its homology, were determined to be essential genes. Deletion of other genes had no impact on cell growth at normal temperature (Figure 2A). However, the iss4Δ strain exhibited weak cold-sensitive and temperature-sensitive phenotypes, the iss3Δ strain showed cold sensitivity and weak temperature sensitivity, the iss6Δ strain showed growth defects at lower temperatures. Deletion of iss9 conferred temperature sensitivity. The iss10Δ strain showed a subtle cold-sensitive phenotype.
Figure 2.
iss genes contribute to Mmi1-mediated mRNA elimination. (A) Growth profiles of deletion mutants of each iss gene. The 10-fold serial dilutions of the wild-type (JY450), pab2Δ (JV833), iss3Δ (JV832), iss4Δ (JV835), iss6Δ (JT958), iss9Δ (JV967) and iss10Δ (JV969) strains were spotted on YE media and incubated at the indicated temperatures. (B) Expression of mei4 and ssm4 mRNAs in deletion mutants of iss genes under vegetative growth conditions. Transcripts of mei4 and ssm4 were analyzed by northern blot analysis in the wild-type (JY450), mmi1-48 (JT221), iss3Δ (JT959), iss4Δ (JV835), iss6Δ (JT958), iss9Δ (JV967) and iss10Δ (JV969) strains. rRNAs stained with ethidium bromide are shown in the bottom of the panel as loading controls. (C) Expression of Mmi1-target genes was analyzed by quantitative RT-PCR in the same conditions as (B). Transcripts of mei4, ssm4, rec8 and spo5 were quantified and normalized to actin. Results represent the mean ± standard deviation from three independent samples. *P < 0.05; **P < 0.01 compared with the wild-type strain (Student’s t-test).
We next investigated the expression of Mmi1-target genes in mitotic cells by using northern blot analysis (Figure 2B) and quantitative RT-PCR (Figure 2C). In each of the iss deletion mutants, DSR-carrying mRNAs were accumulated in variable amounts, as in the previously reported hypomorphic mmi1 mutant (mmi1-48) (3). Accumulation was most prominent in the iss3Δ/red1Δ strain, suggesting the importance of Iss3/Red1 in the pathway. Expression of mei4, which encodes a transcription factor essential for the progression of meiosis (32), was not detected in the iss4Δ strain. We tested meiotic expression of mei4 in the sme2Δ background by using the iss insertional mutants isolated in our screen. Expression of mei4 is restricted to meiotic cells (3,32), and this meiosis-specific induction was abolished in the sme2Δ strain due to failure of Mmi1 activity inhibition (Supplementary Figure S3). Expression of mei4 in the iss3, iss4, iss6, iss9 and iss10 original mutants during mitotic growth was similar to that in the respective deletion mutants, irrespective of the presence of the sme2 gene (Supplementary Figure S3, +N lanes). Meiotic expression of mei4 in the sme2Δ mutant was recovered by the iss mutations, including iss4, although less completely than the other mutations (Supplementary Figure S3, -N lanes). Transcriptional activation of the mei4 promoter during meiosis (3) may facilitate the accumulation of mei4 transcripts in the iss mutants, which exhibited almost no mei4 expression during mitotic growth.
These observations suggest that the newly identified Iss proteins participate in the Mmi1/DSR elimination system in a similar fashion as Iss3/Red1. Subsequent studies were focused on the characterization of Iss10, as it was found to function in close proximity to Iss3/Red1.
Iss10 localizes to the same cellular compartment as Mmi1
We have previously reported that Mmi1 and its related factors, including Rrp6, form several dot structures in the nucleus of mitotically growing cells (3,7). Red1 was also shown to localize to the Mmi1 foci (Figures 3 and 4) (9). We examined the subcellular distribution of Iss10 in a strain expressing GFP-tagged Iss10. Iss10 was observed at several nuclear foci, similarly to Mmi1 (Figure 3A). We compared the localization of Iss10-GFP with CFP-tagged Mmi1 and mCherry-tagged Red1 and found that Iss10 colocalized with Mmi1 and Red1 in the nucleus of mitotically growing cells (Figure 3B).
Figure 3.
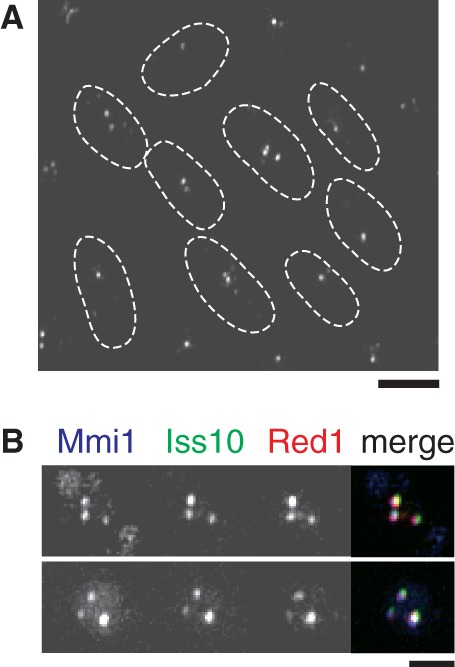
Interaction between Iss10 and Mmi1. (A) Live observation of Iss10-GFP in cells in vegetative growth (JT971). The dotted lines indicate the shape of cells. Bar, 5 µm. (B) Colocalization of Iss10 with Mmi1 and Red1. JT960 cells expressing CFP-Mmi1, Iss10-GFP and Red1-mCherry from the respective endogenous promoters were examined by fluorescence microscopy. Images of the nuclear region are shown. Merged images are shown in the right panels: blue, CFP-Mmi1; green, Iss10-GFP; red, Red1-mCherry. Bar, 2 µm.
Figure 4.
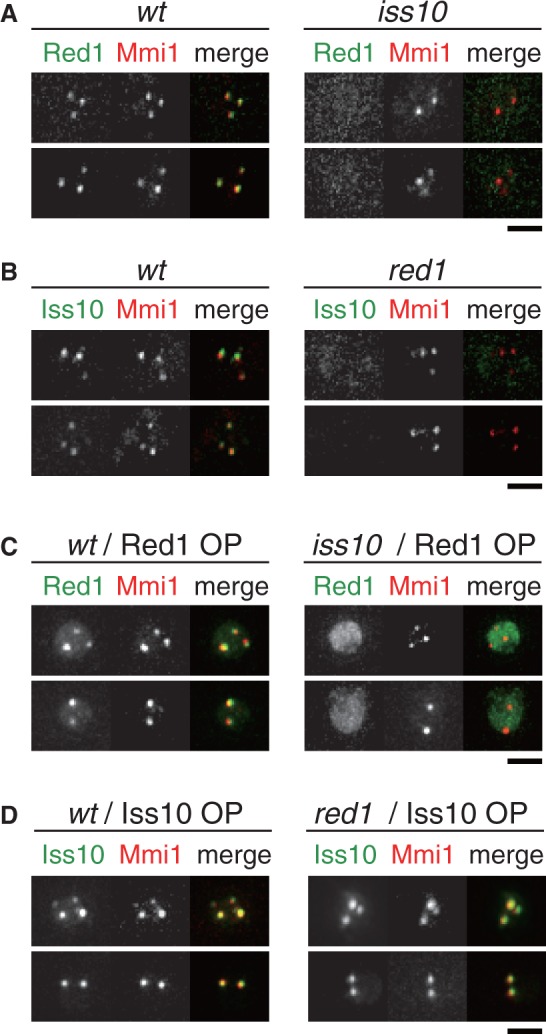
Iss10 is required for the proper localization of Red1 to Mmi1 foci. (A) Localization of Red1 in iss10Δ cells. Wild-type (JT961) and iss10Δ (JT962) cells expressing Red1-GFP and CFP-Mmi1 from the respective endogenous promoters were examined by fluorescence microscopy. Images of the nuclear region are shown. Merged images are shown in the right panels: green, Red1-GFP; red, CFP-Mmi1. (B) Localization of Iss10 in red1Δ cells. Wild-type (JT963) and red1Δ (JT964) cells expressing Iss10-GFP and CFP-Mmi1 were examined. Merged images: green, Iss10-GFP; red, CFP-Mmi1. (C) Localization of overexpressed Red1 in iss10Δ cells. Wild-type (JT965) and iss10Δ (JT966) cells expressing Red1-YFP from an expression plasmid and CFP-Mmi1 from an endogenous promoter were examined. Merged images: green, Red1-YFP; red, CFP-Mmi1. (D) Localization of overexpressed Iss10 in red1Δ cells. Wild-type (JT965) and red1Δ (JT967) cells expressing Iss10-GFP from an expression plasmid and CFP-Mmi1 from an endogenous promoter were examined. Merged images: green, Iss10-GFP; red, CFP-Mmi1. Bars, 2 µm.
Iss10 and Red1 are mutually dependent on each other for localization to Mmi1 foci
To examine whether Iss10 is directly associated with Red1, we observed localization of Red1 in the iss10 deletion mutant and vice versa. In iss10Δ cells, Red1 did not localize to Mmi1 dots (Figure 4A). Localization of Iss10 to Mmi1 foci was also dependent on Red1 (Figure 4B). When overexpressed in each deletion mutant, the two factors behaved differently. Overexpressed Red1 could not localize properly but accumulated in the nucleoplasm in the iss10Δ cells (Figure 4C). In contrast, overexpressed Iss10 was able to properly colocalize with Mmi1 in red1Δ cells (Figure 4D).
We further tested interactions between Mmi1 and Red1 or Iss10 in each deletion mutant by immunoprecipitation. In wild-type cells, Red1 weakly co-precipitated with Mmi1 (Figure 5A) (9). In iss10Δ cells, the interaction between Red1 and Mmi1 was severely compromised (Figure 5A, lane 3 versus 4). In contrast, the amount of Iss10 was drastically reduced in red1Δ cells (Figure 5B). These observations suggest that Iss10 regulates the association of Red1 to Mmi1 foci, and that Red1 has an important role in the stabilization of Iss10. This finding is consistent with the aforementioned experiments, in which localization of Iss10 in red1Δ cells could be restored by overexpressing Iss10, but not vice versa (Figure 4C and D). We also confirmed direct interaction of Iss10 with Red1 by two-hybrid assays (Figure 5C).
Figure 5.
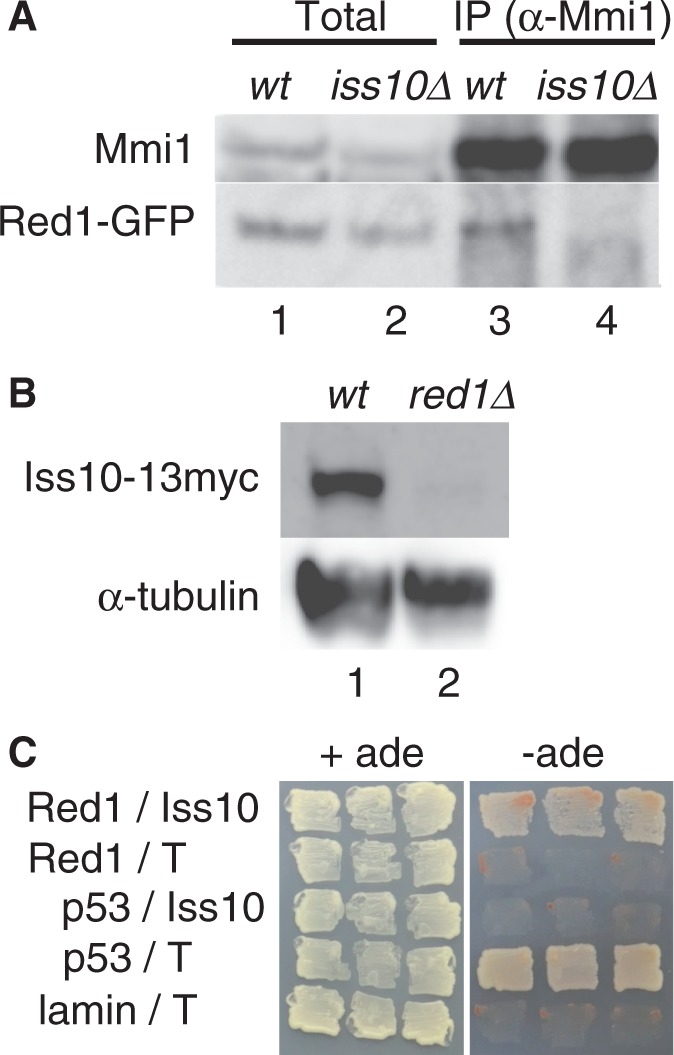
Iss10 facilitates the interaction between Red1 and Mmi1. (A) Co-immunoprecipitation of Red1 and Mmi1 in iss10Δ cells. Native cell extracts prepared from exponentially growing wild-type (JT961) and iss10Δ (JT968) cells expressing Red1-GFP from an endogenous promoter were subjected to immunoprecipitation with anti-Mmi1 antibodies. Precipitates (lanes 3 and 4) and 10% equivalent cell extracts (lanes 1 and 2) were immunoblotted with anti-Mmi1 antibodies and an anti-GFP antibody. (B) Iss10 expression levels in red1Δ cells. Native cell extracts prepared from exponentially growing wild-type (JT969) and red1Δ (JT970) cells expressing Iss10-13myc from an endogenous promoter were subjected to western blot analysis by using an anti-myc antibody. α-Tubulin was used as a loading control. (C) Two-hybrid interaction of Iss10 with Red1. Growth was monitored on an adenine-depleted medium. The set of p53 and T-antigen (T) was used a positive control, whereas the other sets, including p53, T-antigen and lamin, were used as negative controls.
Destabilization of Iss10 leads to disappearance of Red1 during meiosis
We next observed localization of Iss10 during meiosis. In meiotic prophase cells, Mmi1 converges into a single dot structure composed of Mei2 and meiRNA and is inactivated (Figure 6A) (3). We found that Iss10 foci disappeared at this stage (Figure 6A). Iss10 protein levels were greatly reduced in nitrogen-depleted diploid cells that underwent meiosis (Figure 6B). Northern blot analysis and quantitative RT-PCR showed that iss10 transcript levels were comparable in mitotic and meiotic cells (Figure 6C, Supplementary Figure S4), indicating that iss10 expression is regulated at the protein level. The localization shift of Iss10 is reminiscent of Red1, which also disappeared during meiotic prophase (Supplementary Figure S5A). In contrast to Iss10, Red1 was still expressed during meiosis (Supplementary Figure S5B) (9). These observations led us to believe that the loss of Red1 from Mmi1 during meiosis might be triggered by the destabilization of Iss10. To test this hypothesis, we overexpressed Iss10 and observed the localization of Red1 in meiotic cells. When overexpressed from the expression plasmid, Iss10 could colocalize with Mmi1 even in meiotic prophase cells (Figure 6D). Moreover, overexpressed Iss10 could cause ectopic localization of Red1 at the Mmi1 dot, where Mmi1 is sequestered by Mei2 and meiRNA (Figure 6D).
Figure 6.

Reduction of Iss10 levels leads to disappearance of Red1 during meiosis. (A) Localization of Iss10 during meiosis. Wild-type (JT971) cells expressing CFP-Mmi1, Iss10-GFP and Mei2-mCherry from the respective endogenous promoters were examined by fluorescence microscopy under mitotically growing and meiotic conditions. Merged images: blue, CFP-Mmi1; green, Iss10-GFP; red, Mei2-mCherry. The dotted lines indicate the shape of cells. Bar, 5 µm. (B) Iss10 expression levels during meiosis. Native cell extracts prepared from exponentially growing (+N) and meiotic (−N) wild-type (JT973) cells expressing Iss10-13myc from the endogenous promoter were subjected to western blot analysis by using an anti-myc antibody. α-Tubulin was used as a loading control. (C) Expression of iss10 mRNAs during meiosis. Transcripts of iss10 were analyzed by northern blot analysis in exponentially growing (+N) and meiotic (−N) wild-type (JY362) cells. rRNAs stained with ethidium bromide are shown in the bottom panel as loading controls. (D) Localization of Red1 in cells overexpressing Iss10. Meiotic wild-type (JT972) cells expressing Iss10-GFP from the expression plasmid and CFP-Mmi1 and Red1-mCherry from endogenous promoters were examined. Merged images: blue, CFP-Mmi1; green, Iss10-GFP; red, Red1-mCherry. The dotted lines indicate the shape of cells. Bar, 5 µm. (E) Inhibition of sporulation by Iss10 overexpression in temperature-sensitive mei2 mutants. JY450 (wt) and JV393 (mei2-ts) were transformed with a multicopy plasmid expressing Iss10 (iss10) or an empty plasmid (-). Transformants were incubated on SSA medium at the indicated temperatures for 4 days, and sporulation frequency was measured. Error bars indicate standard deviations from three measurements (total n > 500). **P < 0.01 (Student’s t-test).
Interestingly, Iss10 overexpression had no effect on meiosis and sporulation in wild-type cells (Figure 6E, Supplementary Figure 6). We hypothesized that Mei2 and meiRNA could suppress Mmi1 activity even when Iss10 and Red1 colocalized with Mmi1 during meiosis. To confirm this, we overexpressed Iss10 in cells with reduced Mei2 activity. In temperature-sensitive mei2 mutant cells (25), Iss10 overexpression impaired sporulation at semirestrictive temperatures (Figure 6E, 28 and 30°C), but it did not cause deficiencies in sporulation at the permissive temperature as in the wild-type cells (Figure 6E, 25°C). These observations indicate that the disappearance of Iss10 during meiosis contributes to the suppression of Mmi1 activity, although the inhibitory function of the Mei2 and meiRNA complex is predominant.
DISCUSSION
In our present study, we identified novel components of the Mmi1-meditated degradation system and characterized Iss10 as a regulator of Red1. In addition to Pab2 and Pla1, which have already been reported as Mmi1-related factors (7), we identified factors with homology to components of the transcription cycle of RNA polymerase II: poly(A) polymerase-binding protein Iss1, transcription termination factor Iss4/Rhn1 and transcription elongation factor Iss9. Iss6/Erh1 is a fission yeast ERH protein. ERH is a highly conserved small protein and is known to be related to various processes such as regulation of pyrimidine metabolism, cell cycle progression and transcription (33–36). It has recently been suggested that erh1 is required for cellular responses to various stresses, including nitrogen depletion (30). The molecular function of Erh1/Iss6 in the Mmi1 system remains to be clarified. Detailed analysis of the iss genes not addressed in this study will be fully described in future experiments.
We demonstrated that Iss10 is important for the proper localization of Red1 to the Mmi1 foci, and that Red1 is required for the stable expression of Iss10. During meiosis, neither Iss10 nor Red1 show any specific localization. In contrast to Red1, the levels of which do not change between mitotic and meiotic cells, the levels of Iss10 drop drastically when cells enter meiosis. This reduction is likely due to posttranslational modification, as the levels of transcript are constant. Several signaling pathways such as the TOR kinase pathway and the stress-responsive MAP kinase pathway are known to have important functions in the initiation of meiosis (37,38). Whether these pathways participate in the destabilization of Iss10 remains an intriguing question.
Mmi1, which causes the degradation of meiotic transcripts, is also deleterious for meiosis (3). To overcome this, Mmi1 is sequestered by the Mei2 dot and is inactivated (3,6). On the basis of the findings of our current study, we propose a novel mechanism used to suppress Mmi1 function during meiosis, as depicted in Supplementary Figure S7. In mitotic cells, Iss10 participates in the Mmi1/DSR system by supporting the interaction between Mmi1 and Red1. During meiosis, Iss10 levels are decreased. This may prompt detachment of Red1 from Mmi1 and promote inactivation of the Mmi1/DSR system, in parallel with inhibition by Mei2 and meiRNA. In agreement with this model, we found that Iss10 overexpression during meiosis was able to restore the localization of Red1 to the Mmi1 dot and decreased sporulation frequencies when Mei2 activity was reduced.
The functional importance of Red1 in the Mmi1/DSR system is evident because deletion of the red1 gene causes high-level expression of DSR-containing meiotic transcripts. We have described a novel mechanism for Red1 regulation. However, the molecular function of Red1 remains enigmatic. Deciphering the role of Red1 will be necessary for the comprehensive understanding of the Mmi1/DSR system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research (C) [23570223 to A.Y.] and (S) [21227007 to M.Y.]; Japan Society for the Promotion of Science. Funding for open access charge: Grants-in-Aid for Scientific Research (S) [21227007].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Taro Nakamura and Piotr Kozlowski for communication of results before publication. They thank Dr Yuki Naito for helpful discussions.
REFERENCES
- 1.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 2.Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 3.Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 4.Harigaya Y, Yamamoto M. Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res. 2007;15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- 5.Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem. Sci. 2002;27:495–497. doi: 10.1016/s0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita A, Shichino Y, Tanaka H, Hiriart E, Touat-Todeschini L, Vavasseur A, Ding DQ, Hiraoka Y, Verdel A, Yamamoto M. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol. 2012;2:120014. doi: 10.1098/rsob.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J. Biol. Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiyama T, Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30:1027–1039. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe Y, Yabana S, Chikashige Y, Hiraoka Y, Yamamoto M. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature. 1997;386:187–190. doi: 10.1038/386187a0. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita A, Watanabe Y, Nukina N, Yamamoto M. RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell. 1998;95:115–123. doi: 10.1016/s0092-8674(00)81787-0. [DOI] [PubMed] [Google Scholar]
- 12.Shimada T, Yamashita A, Yamamoto M. The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell. 2003;14:2461–2469. doi: 10.1091/mbc.E02-11-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiriart E, Vavasseur A, Touat-Todeschini L, Yamashita A, Gilquin B, Lambert E, Perot J, Shichino Y, Nazaret N, Boyault C, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tashiro S, Asano T, Kanoh J, Ishikawa F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells. 2013;18:327–339. doi: 10.1111/gtc.12038. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 17.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: Handbook of Genetics. New York: Plenum Publishing Corporation; 1974. [Google Scholar]
- 18.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 20.Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp. Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- 21.Sukegawa Y, Yamashita A, Yamamoto M. The fission yeast stress-responsive MAPK pathway promotes meiosis via the phosphorylation of Pol II CTD in response to environmental and feedback cues. PLoS Genet. 2011;7:e1002387. doi: 10.1371/journal.pgen.1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita A, Watanabe Y, Yamamoto M. Microtubule-associated coiled-coil protein Ssm4 is involved in the meiotic development in fission yeast. Genes Cells. 1997;2:155–166. doi: 10.1046/j.1365-2443.1997.1100307.x. [DOI] [PubMed] [Google Scholar]
- 24.Niccoli T, Yamashita A, Nurse P, Yamamoto M. The p150-Glued Ssm4p regulates microtubular dynamics and nuclear movement in fission yeast. J. Cell Sci. 2004;117:5543–5556. doi: 10.1242/jcs.01475. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe Y, Yamamoto M. S. pombe mei2 encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- 26.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 27.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama T, Sugioka-Sugiyama R, Hada K, Niwa R. Rhn1, a nuclear protein, is required for suppression of meiotic mRNAs in mitotically dividing fission yeast. PLoS One. 2012;7:e42962. doi: 10.1371/journal.pone.0042962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bitton DA, Wood V, Scutt PJ, Grallert A, Yates T, Smith DL, Hagan IM, Miller CJ. Augmented annotation of the Schizosaccharomyces pombe genome reveals additional genes required for growth and viability. Genetics. 2011;187:1207–1217. doi: 10.1534/genetics.110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzyzanowski MK, Kozlowska E, Kozlowski P. Identification and functional analysis of the erh1(+) gene encoding enhancer of rudimentary homolog from the fission yeast Schizosaccharomyces pombe. PLoS One. 2012;7:e49059. doi: 10.1371/journal.pone.0049059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimeno S, Tous C, Garcia-Rubio ML, Ranes M, Gonzalez-Aguilera C, Marin A, Aguilera A. New suppressors of THO mutations identify Thp3 (Ypr045c)-Csn12 as a protein complex involved in transcription elongation. Mol. Cell. Biol. 2011;31:674–685. doi: 10.1128/MCB.01188-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI. Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics. 1994;138:1163–1170. doi: 10.1093/genetics/138.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelsthorpe M, Pulumati M, McCallum C, Dang-Vu K, Tsubota SI. The putative cell cycle gene, enhancer of rudimentary, encodes a highly conserved protein found in plants and animals. Gene. 1997;186:189–195. doi: 10.1016/s0378-1119(96)00701-9. [DOI] [PubMed] [Google Scholar]
- 35.Lukasik A, Uniewicz KA, Kulis M, Kozlowski P. Ciz1, a p21 cip1/Waf1-interacting zinc finger protein and DNA replication factor, is a novel molecular partner for human enhancer of rudimentary homolog. FEBS J. 2008;275:332–340. doi: 10.1111/j.1742-4658.2007.06203.x. [DOI] [PubMed] [Google Scholar]
- 36.Amente S, Napolitano G, Licciardo P, Monti M, Pucci P, Lania L, Majello B. Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett. 2005;579:683–689. doi: 10.1016/j.febslet.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto M. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:788–797. doi: 10.2183/pjab.86.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otsubo Y, Yamamoto M. Signaling pathways for fission yeast sexual differentiation at a glance. J. Cell Sci. 2012;125:2789–2793. doi: 10.1242/jcs.094771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.