Abstract
The absence of a quality control (QC) system is a major weakness for the comparative analysis of genome-wide profiles generated by next-generation sequencing (NGS). This concerns particularly genome binding/occupancy profiling assays like chromatin immunoprecipitation (ChIP-seq) but also related enrichment-based studies like methylated DNA immunoprecipitation/methylated DNA binding domain sequencing, global run on sequencing or RNA-seq. Importantly, QC assessment may significantly improve multidimensional comparisons that have great promise for extracting information from combinatorial analyses of the global profiles established for chromatin modifications, the bindings of epigenetic and chromatin-modifying enzymes/machineries, RNA polymerases and transcription factors and total, nascent or ribosome-bound RNAs. Here we present an approach that associates global and local QC indicators to ChIP-seq data sets as well as to a variety of enrichment-based studies by NGS. This QC system was used to certify >5600 publicly available data sets, hosted in a database for data mining and comparative QC analyses.
INTRODUCTION
The recent development of high-throughput sequencing technologies has led to a rapid expansion of studies analyzing the genome-wide patterns of gene regulatory events and features, such as epigenetic DNA and histone modification, and the binding patterns of transcription factors and their co-regulatory complexes, (posttranslationally) modified chromatin-associated factors and chromatin- or transcription-modulatory multi-subunit machineries (1–9). Moreover, the mapping of transcriptomes by RNA-seq (10–13), global nascent RNA sequencing or global run on sequencing (GRO-seq) (14) or ribosome-associated (‘ribosome footprinting’) RNAs (15), and technologies revealing chromatin conformation are also based on massive parallel sequencing (16–18). A particular challenge is the comparison of multidimensional profiles for several factors, their posttranslational modifications and/or chromatin marks. Indeed, such studies are not easily comparable, as they are performed in different settings by different individuals using different cells and antibodies. Moreover, profiles are established at different platforms with highly variable sequencing depths. As a result, studies performed even with the same cells in different laboratories can differ extensively (3). This presents serious limitations for the interpretation of such global comparative studies and reveals the need for a quantifiable system for assessing the quality and comparability of next-generation sequencing (NGS)-derived profiles and moreover the robustness of local features, such as peaks at particular loci, which are derived from the mapping of read-count intensities (RCIs).
A large number of factors can influence the quality of NGS-based profilings. Particularly in the case of immunoprecipitation-based approaches [e.g. chromatin immunoprecipitation (ChIP-seq), methylated DNA immunoprecipitation (19,20), GRO-seq (21)], experimental parameters like cross-linking efficiencies in different cell types or tissues, shearing or digestion of chromatin or the selectivity and affinity of an antibody (batch) can vary substantially between experiments and different experimenters and will ultimately impact on the overall quality of the final readout. Currently, quality assessment is performed by visual profile inspection of defined chromatin regions and complemented by peak caller predictions. In addition, a number of analytical methods have been described [for a recent summary of the methodologies used by the ENCODE consortium see (22)]. However, none of them has been shown to be applicable to the large variety of ChIP-seq and enrichment-related NGS profiling assays. For instance, methods like fraction of mapped reads retrieved into peak regions (FRiP) (23) or irreproducibility discovery rate (IDR) (24) require prior use of peak calling algorithms for evaluation and are therefore dependent on peak-calling performance of a given tool with the user-defined parameters. Consequently, they cannot be easily used for multi-profile comparisons when different peak callers are required (e.g. transcription factors (TFs) and histone modifications with ‘broad’ profiles).
In addition to the performance of the immunoprecipitation/enrichment assays, the rapid technological progress provided NGS platforms with largely different sequencing capacities ranging from tens of millions (e.g. Illumina Genome analyzer v1, hereafter referred to as ‘GA1’) to >3 billion (HiSeq2000) reads per flow cell. As a consequence, the public databases hosting NGS-generated data sets are populated with ChIP-seq profiles presenting a large variety in sequencing depth. Importantly, previous studies have demonstrated that by increasing the sequencing depth, the number of discovered binding sites increases accordingly. Intuitively, it is expected that the number of sequenced reads required to discover all binding events is directly related to their total number and to their binding pattern (i.e. ‘broad’ regions covering large parts of a genome will require more reads to be properly identified than ‘sharp’ patterns with few target sites). When evaluating the quality of NGS-based profiling, it is therefore important to assess if a given ChIP-seq profile is performed under optimal sequencing conditions, including the minimal sequencing depth required to discover most of the relevant binding events of a given factor.
For all the above reasons, we have developed a bioinformatics-based quality control (QC) system that uses raw NGS data sets to (i) infer a set of global QC indicators (QCis), which reveal the comparability of different enriched-NGS data sets, (ii) provide local QCis to judge the robustness of cumulative read counts (‘peaks or islands’) in a particular region, (iii) provide guidelines for the choice of the optimal sequencing depth for a given target and, finally, (iv) to have quantitative means of comparing different antibodies and antibody batches for ChIP-seq and related antibody-driven studies. In addition, we have established a QC indicator database that will be expanded to cover virtually all publicly available enrichment-related NGS profiling assays. Thus, users can compare the quality indicators computed by the NGS-QCi Generator for a given ChIP-seq experiment with the quality indicators for published data sets present in the QC indicator database. This information will guide users toward optimization of the ChIP-seq process, if the QC is lower than that achieved previously by others and/or with other antibodies. Moreover, this QC system will be useful for antibody development and certification. We discuss the simplicity and versatility of the present QC method and database in view of currently existing QC assessment procedures and guidelines. The NGS-QC Database of QC indicators for publicly available profiles and the NGS-QC Generator tool are freely accessible through a customized Galaxy instance at http://igbmc.fr/Gronemeyer_NGS_QC.
MATERIALS AND METHODS
Data sets
Publicly available data sets were downloaded from GEO (25). When available, aligned files (either in BED or BAM format) were used; otherwise sequence data sets, available through the short read archive database, were first aligned to the corresponding reference genome using Bowtie2 under standard alignment options (26).
Assessment of the inherent robustness of ChIP-seq profiles
Based on the rationale that beyond a sequencing depth threshold a ChIP-seq profile changes only in amplitude but not in pattern, we evaluated this property by monitoring the changes of its RCIs after read-subsampling. For this, aligned reads were randomly sampled at three distinct densities (90, 70 and 50%; referred to as s90, s70 and s50 subsets, respectively). To avoid bias, random sampling was performed without replacement; each separately sampled density subset was generated from the original read data set. RCI profiles were constructed by counting the overlaps within a defined window (‘bin’). With the aim of having no more than one binding event per bin, it is currently fixed to 500 bp. An empirical evaluation of the influence of this parameter on the assessment of the quality indicators confirmed our initial choice (Supplementary Figure S1d).
Reconstructed profiles from randomly sampled subsets are then compared with that constructed from the initial total mapped reads (TMRs) by computing the recovered RCI (recRCI) per bin after sampling as follows:
Where samRCI is the RCI/bin retrieved after sampling and oRCI is that found in the original profile. Under the working hypothesis that, as a consequence of random sampling, recRCI is directly proportional to the sampling density, the divergence from the expected RCI behavior is measured as follows:
where samd corresponds to the random sampling density; i.e. 90, 70 and 50% for s90, s70 and s50, respectively. Importantly, the RCI dispersion or δRCI is inversely proportional to the original RCI (Supplementary Figure S1C) and it has been empirically observed to present a direct correlation with the quality of ChIP-seq profiles (Supplementary Figure S2). Thus, for providing a quantitative assessment of the changes of RCI dispersion in a given data set, we have evaluated the fraction of bins displaying a δRCI within in a given interval, which has been defined as the global density QC indicator ‘denQCi’. This global indicator—evaluated in conditions where only a half of the initial sequenced reads are available (s50)—is systematically used in this study to measure the degree of robustness of the evaluated profile to the read-subsampling treatment (i.e. high denQCi corresponds to low RCI dispersion). In addition, the changes in robustness on subsequent read subsampling has been evaluated by comparing the denQCi for the sampling closest to the original profile (s90) with that sampling only half of the sequenced reads (s50). This is defined as the similarity QC (simQCi) indicator, computed as ratio between denQCis for the s90 and s50 sampling subsets. The current version of NGS-QCi Generator provides both global quality indicators (denQCi and simQCi) for dispersion intervals of 2.5, 5 and 10%. Further details concerning the assessment of these indicators are provided in the QC report (see Supplementary File S1 and Supplementary Figure S4).
Local QCis
Given that the above analyses were computed for 500-bp bins, the δRCI/bin data can be used as local QCis. The NGS-QCi Generator provides such information in either wiggle or BED formats; the default condition identifies bins with δRCI ≤ 10%. Local QCis in wiggle file format can be uploaded in the Integrated Genome Browser (IGB) and displayed as a heat-map together with standard RCI wiggle files (as illustrated in Figure 3B). In a similar manner, the corresponding BED file can be uploaded in the UCSC Genome Browser. This display option is useful to visualize predicted δRCIs associated to a given chromatin region of interest. Furthermore, 500-bp chromatin regions with δRCIs thresholds of 2.5, 5 or 10% can be downloaded as a table in BED format. The data sets facilitate comparative analyses of multiple profiles in the context of defined δRCI thresholds.
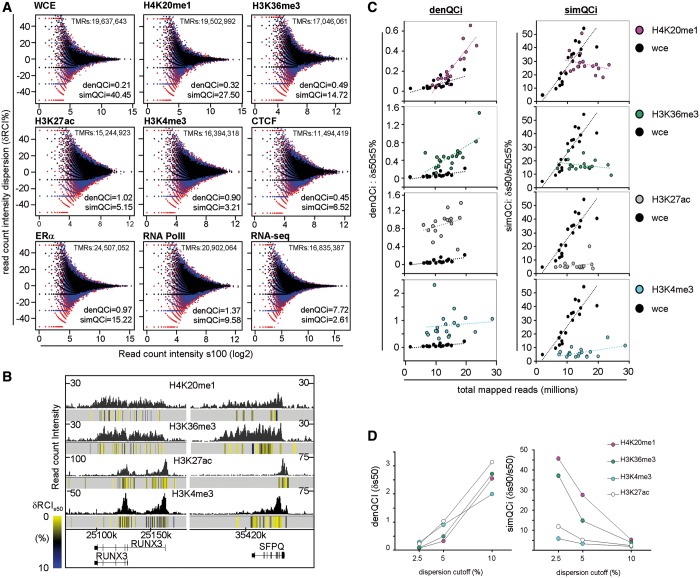
Figure 3.
QCis for several types of ChIP-seq and enrichment-related NGS profiles. (A) Scatterplots illustrating the RCI dispersion (δRCI%) after sampling for different types of NGS profiles (overlays of s90, black; s70, blue; s50, red). TMR, density (denQCi, δs50 ≤ 5%) and similarity (simQCi, δs90/s50 ≤ 5%) QCis are indicated. Note that the input profile has the lowest denQCi and highest simQCi (WCE; top left), whereas the highest denQCi and lowest simQCi were measured for an RNA-seq profile (bottom right). (B) RCI dispersion per 500-bp bins is illustrated as color-coded heat-map (indicated at left) below the corresponding ChIP-seq profiles. (C) Density and similarity QCis for different profiles of the indicated histone modifications are compared with input WCE profiles. Note the different characteristics of the target profiles on increasing TMRs, which reveals that for H4K20me1 and H3K36me3 profiles presenting TMRs <15 million present QCis similar to the input. (D) Density and similarity QCis are displayed at stringent (δs50 ≤ 2.5%), intermediate (δs50 ≤ 5%) and relaxed (δs50 ≤ 10%) dispersion intervals.
QC-STAMP and NGS-QCi database
The contribution of the two QCis to the single descriptor QC-STAMP was defined by following equation:
To evaluate the divergence of this global descriptor over all enrichment-related NGS profiles currently compiled in the NGS-QC database, the QC-STAMP distributions assessed for three different RCI dispersion intervals were subdivided in four quantiles to which the following grades have been attributed: ‘D’, lower quartile (<25%); ‘C’, interquartile 25–50%; ‘B’, interquartile 50–75% and ‘A’ upper quartile (>75%). The NGS-QCi Generator database associates these grades for 2.5, 5 and 10% δRCI to each profile as a three-letter symbol, such that, for example AAA (‘triple A’) reveals an A grade for all three δRCIs. All available profiles are displayed as a dynamic QC-STAMP versus TMR scatterplot, which allows judging of their QCi similarities in the context of the sequencing depth. Note that the global QC-STAMP descriptor will be dynamically reevaluated when novel entries are provided to the database.
Peak detection approach
In addition to the well-described peak caller MACS (27), peak calling has been performed with MeDiChI, a model-based deconvolution approach originally developed for ChIP-chip assays (28), which we have adapted to ChIP-seq analyses. MeDiChI computes a model from a randomly selected subset of the multiple binding events present in a genome-wide profile. This model is then used as a deconvolution kernel for genome-wide prediction of likely binding events, which are further validated by nonparametric bootstrapping. As we compared ChIP-seq profiles generated at different sequencing depths, we have included a P-value/peak intensity product ranking-based approach for defining a common false discovery rate (FDR) during comparison. For this, a ranking coefficient (RC) for the ith peak identified by MeDiChI was calculated by the following equation:
This RC was sorted from the highest to the lowest value, and the FDR was assessed as follows:
Where i* is the ranking position based on the RC, and N is the total number of peaks. Thus, all ERα ChIP-seq profiles have been compared at a FDR threshold ≥45 or FDR adjusted P-value threshold 10−4.5.
RESULTS
Previous studies described the concept of a ‘saturation point’ as the sequencing depth after which no new binding sites are identified by a given peak caller with additional sequenced reads (5,29). This concept has been initially evaluated in a retrospective manner by assessing the number of significant binding sites retrieved when only a subset of the original sequenced reads was used for profile reconstruction (random subsampling approach). Intuitively the ‘saturation point’ concept predicts that beyond such threshold no further binding sites would be discovered and by consequence, the increased sequencing depth should only influence the overall read-count intensity of the corresponding profile.
Following the same concept, the QC system presented here evaluates the stability of the pattern of a given profile beyond the saturation point by measuring the reproducibility of ChIP-seq and enrichment-related NGS profiles under conditions where only a subset of the TMRs are used for reconstruction. In the ideal ‘saturation’ condition, such a reconstruction will generate a profile with the same read distribution pattern across the genome but with a decrease of the RCIs according to the percentage of TMRs used (Figure 1A). The extent to which this reproducibility is attained is defined as ‘robustness’ of the original profile and is assessed by the resampling of a given data set at the level of half of the original TMRs (referred to as ‘s50’). Whereas none of the currently available profiles displays ideal robustness at s50, the evaluation of the deviation from such ideal behavior reflects the degree of robustness and represents a quantitative method for assigning a set of quality descriptors to any NGS-generated profile.
Figure 1.
Assessing quality descriptors for ChIP-seq profiles. (A) Based on the rationale that a robust profile displays a proportional decrease of its RCIs along the genome when a randomly sampled population of its TMRs is used for profile reconstruction, the present quality assessment method quantifies the deviation from the expected RCI decrease within defined thresholds. (B) TMRs are randomly sampled into three distinct populations (90, 70 and 50%), which are used for profile reconstruction by computing the RCIs in 500-bp bins. The RCI divergence from expectation (δRCI) is measured relative to the original profile (s100). This information generates local QCis and is displayed together with the original RCI profile to identify robust chromatin regions (δRCI heat-map below the bottom profile). In addition, two global QCis are calculated, comprising the density QCi [denQCi, defined as the fraction of bins displaying <5% δRCI after 50% TMRs sampling (‘δs50/5’)] and the similarity QCi (simQCi), defined as ratio of denQCi after 90% sampling over that after 50% sampling (‘δs90/s50/5’). (C) Genome-browser screenshots of three different H3K4me3 ChIP-seq profiles. In addition, the RCI dispersion per 500-bp bins (local QCi) is illustrated as color-coded heat-map below the corresponding ChIP-seq profiles. Note that while all three profiles present ∼19 million TMRs, they differ significantly in their global RCI amplitudes. Furthermore, their corresponding global QCis assessed from 5 random sampling assays are displayed (average ± standard deviation).
ChIP-seq profile’s robustness dispersion provides quality descriptors
This QC system evaluates the robustness of RCI dispersion for any given ChIP-seq and enrichment-related NGS profiles by comparing distinct randomly sampled populations derived from the primary data set (Figure 1B). Specifically, TMRs are first resampled at 90, 70 and 50% (referred to as s90, s70 and s50, respectively) of the original data set. The genome-wide read-count distribution within 500 bp bins is then evaluated for the sampled subsets and compared with that observed for the original profile (s100) (for the effect of bin size on measuring profile robustness see Supplementary Figure S1). Under the assumption of a proportional RCI decrease on read subsampling (saturation concept), the bin RCI divergence from expectation is calculated (δRCI or local divergence; defined as the difference between the theoretically expected RCI and that observed after resampling). Furthermore, a global quantitative assessment of the changes in bin RCI dispersion is given by the evaluation of the total bins presenting a defined RCI dispersion. This global indicator, defined as density Quality indicator (denQCi), evaluated in conditions where only a half of the initial sequenced reads are available (s50), is systematically used in this study to illustrate the degree of robustness of the evaluated profile to the reads-subsampling treatment (i.e. δs50 ≤ 5% makes reference to the fraction of bins with δRCI ≤ 5% when half of the TMRs are used for profile reconstruction).
Furthermore, the changes in robustness on successive read subsampling has been evaluated by comparing the denQCi obtained for the subset closest to the original profile (s90) relative to that assessed from half of all sequenced reads (s50). This second global indicator has been defined as the ‘similarity QC indicator’ (simQCi) because it reveals the similarity between the robustnesses assessed at s90 and s50. Overall, the higher the denQCi and the lower the simQCi, the more ‘robust’ is the evaluated profile.
ChIP-seq profiles established from similar TMRs can lead to variable quality patterns as revealed by visual inspection of three ChIP-seq profiles of the tri-methylation of lysine 4 of histone 3 (H3K4me3) generated with antibodies obtained from the same supplier and with similar (∼19 millions) TMR levels (Figure 1C). Yet, they present major differences of global RCIs and background levels (note the different scales). Indeed, the computing of the QCis provides quantitative descriptors (denQCi, δs50 ≤ 5% and simQCi, δs90/s50 ≤ 5%) for the relative quality of the three profiles, which fully comply with the visual quality assessment, thus illustrating the usefulness of this approach in providing quantitative QC values for comparing different ChIP-seq data sets. Note that multiple random TMR samplings performed for each of the illustrated profiles revealed a coefficient of variation of <2% for the computed QCis. This demonstrates a high stability of the measurement of global QCis even when derived from a single random drawing (Figure 1C and Supplementary Figure S2).
Sequencing-depth influences the quality of ChIP-seq profiles
ChIP-seq and related assays are in most cases based on reads obtained from a single flow cell channel. Importantly, read densities of flow cells have largely increased over the past few years, ranging from <40 million for the first Genome Analyzer from Illumina (GA1) to >3 billion reads (300 Gb) for the Hiseq2000 platform. Consequently, the TMRs used for profile reconstruction can vary dramatically, inducing questions concerning the comparability of profiles that were constructed with different amounts of TMRs.
To evaluate the direct influence of sequencing depth on NGS-profiling robustness, we performed an analysis of biological replicates for ERα binding in H3396 breast cancer cells (3), which was performed by using one channel of the GA1, GA2X or HiSeq2000 platforms. We also included a comparison with half of a HiSeq channel by using multiplex technology. As expected, the sequencing depth provided by the different sequencing platforms, correlates well with the overall RCIs (Figure 2A). Importantly, TMR sampling analysis revealed a 16.2-fold increase of denQCi and, thus, global profile ‘robustness’, with increased sequencing depth (‘δs50 ≤ 5%’ in Figure 2A).
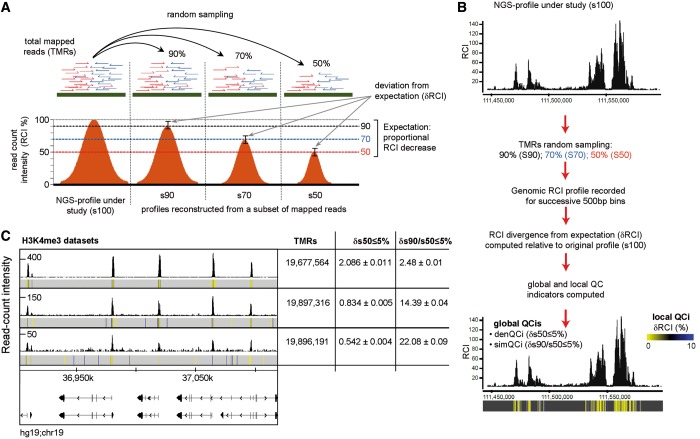
Figure 2.
ERα binding sites detection assessed for different sequencing depths. (A) ERα RCI profiles obtained from different sequencing platforms [i.e. Genome Analyser 1 (GA1); GA2X and Hiseq2000] are illustrated. Each of the displayed ChIP-seq profiles was obtained by sequencing a single channel of the corresponding platform except for Hiseq2000, where half a channel or a full one was used. The corresponding mapped reads and their associated denQCi (δs50 ≤ 5%) are displayed. (B) Total ERα binding sites identified in ChIP-seq profiles generated at different sequencing depths compared with those that complied with the δs50 ≤ 5% criterion. ERα binding sites were predicted with MeDiChI (FDR adjusted P-values threshold 10−4.5; see methods for details). (C) Venn diagrams illustrating overlap and outlier populations for ERα binding sites retrieved from sequencing a full HiSeq2000 channel compared with those identified at lower sequencing depths. This analysis was performed for total ERα sites (top panel) and those complying with δs50 ≤ 5% (bottom panels). (D) Boxplots displaying peak intensity and FDR adjusted P-value associated to overlap and outlier populations displayed in (C). Note that the ERα sites in the overlaps show systematically higher intensities and confidence than the outliers and that this difference is decreased for the δs50 ≤ 5% populations. (E) Considering the sites identified with the full HiSeq2000 channel as ‘true’ sites, the fraction of true sites recovered in the compared profiles (top panel), as well as the false calls, estimated from the outlier population (bottom panel) are illustrated. Note the increase of true sites and a concomitant decrease of false calls in the population that complies with δs50 ≤ 5%.
As expected, the number of TMRs used for ERα profile construction strongly influenced the total number of predicted statistically significant binding sites. In fact, with >50 million reads for the Hiseq2000 profile, 22 150 ERα sites were predicted (FDR adjusted P-value threshold 10−4.5; for peak detection algorithm, see ‘Materials and Methods’ section). In contrast, only 2038 sites were predicted from ∼5 million reads obtained with one GA1 channel (Figure 2B). Albeit the total number of predicted peaks increased strongly with increasing sequencing depths, the number of sites that complied with δs50 ≤ 5% shows a much slower increase and entered a plateau phase above 24 million TMRs. This indicates that the ‘robust’ ERα binding sites approach saturation as defined in previous studies on sequencing depth and de novo discovery of transcription factor binding sites (5,29,30).
As we have profiled ERα binding under identical treatment conditions, it was reasonable to assume that the sites identified at low sequencing depth constitute a subpopulation of those identified in the high TMR profiles. In fact, when comparing the ERα binding sites predicted at highest sequencing depth with those derived from the other profiles, not only the number but also the robustness of peaks in the overlapping population increased with increasing sequencing depth. From 1321 ERα sites in the overlap between GA1 and the full channel HiSeq2000 profile, >80% of them (1096 sites) comply with δs50 ≤ 5% (Figure 2C). Similarly, the number of ERα binding sites overlapping with the GA2X or half channel HiSeq2000 data sets increased strongly over that obtained with GA1, as did the number of robust peaks.
The above comparison revealed also a significant number of nonoverlapping sites (Figure 2C). While it is reasonable to assume that the outliers of the HiSeq2000 profile (red) result mainly from the incomplete binding site recovery from the other profiles, those outliers that are seen in the low TMR profiles but not in the HiSeq2000 are more likely ‘false positives’. Indeed, the number of such sites is variable and does not follow a common trend as the increase of the overlap population with increasing sequencing depth; in this respect, the GA2X data set is suboptimal with 4- to 5-times more outliers (green) than the GA1 (gray) and 1/2Hiseq (blue) ones. Importantly, when considering only the robust peak population, the GA2X outliers were significantly reduced to about the level seen with GA1 and 1/2Hiseq ones. In addition, the nonoverlapping sites, including those of the full channel HiSeq2000, showed consistently lower peak intensities and weaker confidence P-values relative to overlapping population (Figure 2D).
Considering the full channel HiSeq data set as ‘gold standard’, the number of recovered ‘true’ ERα binding sites increased from <5% for the GA1 data set to ∼60% for the half channel HiSeq2000 profile (Figure 2E). Importantly, 80% ‘true positive’ binding sites were recovered when only robust ERα sites are considered, indicating that the denQCi criterion identifies the highly reliable sites when comparing ChIP-seqs with largely differing sequencing depths.
The QCis are universally applicable to all ChIP-seq and enrichment-related NGS profiling assays
While in previous studies profile saturation has been defined after peak calling (5,29,30), the present QC evaluation system evaluates robustness directly from the raw pattern of genome-aligned reads. Therefore, QCis can be established for any type of enrichment-related NGS profiles, including ChIP-seq, RNA-seq, GRO-seq and others, making this methodology a universal tool for multidimensional quality profile comparison. Indeed, we have computed QCis for several types of publicly available NGS-generated profiles and observed a high variability between the corresponding QCis even when data sets with similar TMRs were compared (Figure 3A and Supplementary Figure S3). RNA-seq, which does not involve manipulations like cross-linking and immunoselection, generated the most robust profiles, while a nonenriched input profile (whole-cell extract, WCE) constructed from ∼19 million TMRs displayed the worst quality indicators. For nearly identical TMRs, the ChIP-seq profile of H4K20me1 revealed significantly improved QCis, as expected for the immunoselection of specific chromatin regions. Importantly, other histone modification profiles constructed from similar or even lower TMRs displayed better QCis than either H4K20me1 or WCE, thereby revealing that the robustness of a profile depends not only on the sample preparation and sequencing depth but also on the nature of the immunoprecipitated target. Note that H4K20me1 and H3K36me3 generate rather broad enrichment profiles revealing a spread of the mark over a large chromatin region, while those established for H3K27ac or H3K4me3 exhibit more discrete patterns of locally confined marks (Figure 3B). Our observation that the 500-bp RCI dispersion is generally higher in the H4K20me1 or H3K36me3 profiles compared with those of H3K27ac or H3K4me3 (see heat-map δRCI dispersion in Figure 3B) is likely to originate from the combination of several effects, including (i) the spread, local density and accessibility of the marks and (ii) the quality (i.e. affinity and selectivity) of the antibodies.
In addition to revealing quality differences between data sets for different targets at similar TMRs, the QCi computation also provides important quality information about data sets for the same target at different sequencing depths. Indeed, comparing the QCis for several H4K20me1 data sets generated from largely different TMRs reveals that below 15 million TMRs the QCis become indistinguishable from the WCE profiles, strongly arguing that significantly higher sequencing depths are essential to establish accurate profiles for such targets (Figure 3C). In contrast, H3K4me3 or H3K27ac ChIP-seq profiles have good QCis even for TMRs below 15 million reads.
That we observe major QCi differences between the various data sets reported for similar TMRs indicates that—in addition to the inherent pattern of the evaluated target—other factors, involving most likely all the experimental steps that generate the ultimate DNA library for sequencing, influence the quality of the profile (Figure 3C and Supplementary Figure S3).
Whereas most of the above described QCis have been established for a dispersion interval of 5% (δs50 ≤ 5%), different dispersion thresholds (e.g. δs50 ≤ 2.5% or δs50 ≤ 10%) may reveal additional characteristics of the studied profiles. Indeed Figure 3D illustrates that the QCis determined for different dispersion intervals do not necessarily show a linear relationship. This information has been used as an additional source for quality evaluation (see below QC-STAMP) and represents a potential method for defining common QCi conditions in the case of multi-profile comparisons by allowing variable robustness dispersion cutoffs (Supplementary File S1).
NGS-QCi Generator: a stand-alone in silico platform for computing QCis
The above methodology infers local and global quality indicators for any available NSG-generated profile following a stand-alone approach, as it does not require additional wet-lab efforts. It has been implemented in the NGS-QCi Generator, a computational tool that is accessible at a customized cloud of the web-based platform Galaxy (31–33) (Supplementary File S1). The NGS-QCi Generator provides a comprehensive report summarizing the global QCis (Supplementary Figure S4) and provides access to the computed RCI dispersion per 500-bp bins (wiggle or BED format) defined as local QCis, which can be used to identify the robustness of specific regions of interest (Figure 3B and Supplementary Figure S5). Using the NGS-QCi Generator we have created a QCis database, which comprises at present the QC analysis of >5600 NGS data sets, including ChIP-seq profiles of histone modifications and variants, transcription factors, as well as GRO-seq and RNA-seq profiles (Figure 4A). This QCi database will be expanded to cover virtually all of the publicly available NGS profiles.
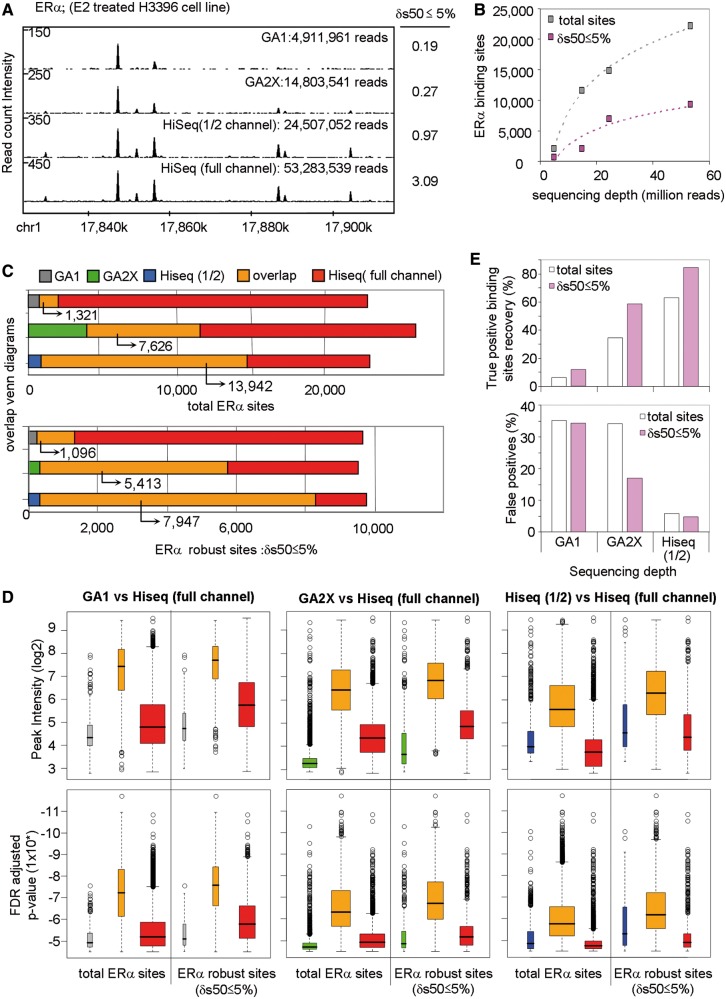
Figure 4.
A universal NGS-QCi database for comparative analysis. (A) Cloud of NGS-QCis for multiple profiles present in the NGS-QCi database (http://igbmc.fr/Gronemeyer_NGS_QC). Density (left) and similarity (right) QCis are displayed relative to the TMRs; color codes are indicated at the right. QCis of input (WCE) profiles are displayed as black circles; the dashed line is the corresponding fitted curve. Arrows indicate the location of the data sets specified in (C). (B) QCis of the evaluated NGS profiles displayed in (A) are expressed in a single term, QC-STAMP, and represented as boxplots for different RCI dispersion intervals (2.5, 5 and 10%). Discrete quality grades ‘A’ to ‘D’ were associated with different quantiles (QC-STAMP dist > 75%; >75% QC-STAMP dist > 50%; >50% QC-STAMP dist > 25%; QC-STAMP dist < 25% associated to A, B, C and D qualitative indicators, respectively). (C) Examples of NGS profiles associated to different QC-STAMPs.
To facilitate and simplify the recognition of QCi divergence between profiles we have defined QC-STAMP, a global descriptor that combines the information provided by denQCi and simQCi. The QC-STAMP corresponds to a three-letter code composed of A, B, C and D that is derived from the position of a given profile QCi within the distribution of compiled QCis in the database. The first letter reveals this position for a δRCI dispersion threshold of 2.5%, the second and third letter for 5% and 10% δRCI, respectively. A to D grading was done to specify the following intervals: D, lower quartile (<25%); C, interquartile (25–50%); B, interquartile (50–75%); A, upper quartile (>75%) (Figure 4B). As an example, the H3K4me3 profile derived from 10 007 440 TMRs [arrow (3) in Figure 4A] classified as ‘triple A’ profile, while nonenriched WCE profiles were, as expected, of the lowest possible quality, ‘triple D’ (Figure 4C). Similarly expected was the high QC performance of RNA-seq, which does not involve the complex experimentation and immunoprecipitation procedures as ChIP-seq, and consequently received ‘triple A’ rating [arrow (1) in Figure 4A]. Note that these ratings are meant to provide a simplified view of the evaluated profile’s robustness but not to replace the QCis, which provide more specific information.
As the quality of a ChIP-seq profile is the direct consequence of a rather large number of factors (e.g. cross-linking efficiency, chromatin shearing, antibody affinity and selectivity, variability between experiments, experimenters and platforms), the QCis cannot per se identify the source for the bad quality of a given profile. However, it does allow identifying data sets of divergent quality, which cannot be compared with each other, even though they might have been generated under similar conditions. Importantly, in contrast to current practice, the sequencing depth applied for generating NGS profiles is a tunable parameter to generate profiles of similar quality. As illustrated in Figures 3 and 5 for similar TMR levels, H4K20me1 or H3K36me3 profiles display in general poorer quality than those of H3K27ac or H3K4me3. However, increasing the sequencing depth will improve their quality descriptors to attain comparable levels, such that, for example, only ‘triple A’ data sets can be compared (Figure 5). In this respect, we believe that the QCi database will become an important reference to perform a priori predictions of the minimal sequencing depth required for a given target to reach a predefined quality.
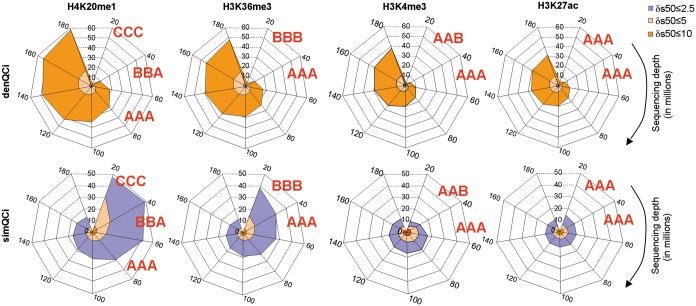
Figure 5.
Meta-analysis illustrating the influence of the sequencing depth on the density and similarity QCis. Meta-analysis performed by compiling several profiles and subsequently sampled at defined TMRs ranging from 20 to 180 million. For each resampled subset the corresponding QCis were computed and displayed in spider-web charts, in which denQCi and simQCi are displayed for different δRCI thresholds (color-coded as indicated at the top left). QC-STAMPs have been associated to the evaluated profiles as illustrated. Note that for H4K20me1 sequencing depths of up to 60 million reads are required to obtain a ‘triple A’ grade, while H3K27ac and H3K4me3 receive this grade with 20 million TMRs.
The NGS-QCis in the context of previously described working standards and guidelines for ChIP-seq assays
Multidimensional comparative analyses of ChIP-seq profiles require prior quality assessment. Currently, this is done by visual inspection of profiles in a genome browser (for instance by evaluating the pattern in regions previously described as containing a chromatin enrichment) and complemented peak caller predictions based on (some) user-defined parameters.
In addition to visual inspection, analytical methods have been developed with the aim of providing quantitative quality assessments of NGS-generated profiles [for a recent summary of the methodologies used by the ENCODE consortium see (22)]. Methods like FRiP (23) or IDR (24) require prior use of peak calling algorithms for evaluation and are therefore dependent on peak-calling performance of a given tool with the user-defined parameters. Consequently, they cannot be easily used for multi-profile comparisons when different peak callers are required. This is for example the case when transcription factor profiles are compared with epigenetic profiles that display broad RCI patterns. Note that the IDR approach can only be used when replicate profiles are available, which is strongly suggested but not a routine procedure (see GEO entries). Furthermore, the criteria used for reproducibility by the IDR analysis can be misleading in cases where compared profiles present broad enrichment patterns (Supplementary Figure S6; see also below).
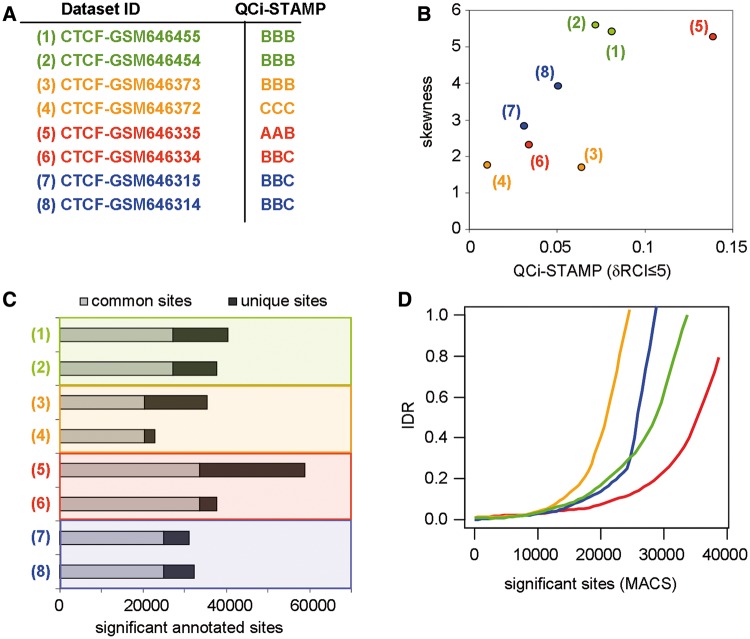
Two other methods; signal distribution skewness (34) and strand cross-correlation analysis (SCC) (22) operate in a peak caller-independent manner. Signal distribution skewness evaluates the asymmetry of genome-wide tag-count distribution, while SCC measures the quality of evaluated ChIP-seq profiles from the sequence tag density on forward and reverse strand reads at target sites. SCC is thus applicable mainly, if not exclusively, to ‘sharp’ patterns like those observed for transcription factor ChIP-seq data sets. It is rather evident that SCC cannot be used for quality assessment of broad patterns, as significantly enriched reads of such profiles cover large areas. Thus, from the conceptual point of view in addition to the present QCi system, signal distribution skewness appears to constitute the only other universal quality measurement method. To compare signal distribution skewness and our NGS-QC we have evaluated the degree of skewness in four publicly available CTCF ChIP-seq data sets (each of them represented by two biological replicates) and compared it with QCi-STAMP (Figure 6A and B). Both methods provide similar quality predictions, with the important exception that the difference in quality of one pair of the evaluated replicates (GSM646372 and GSM646373 data sets) was predicted by the QCi-STAMP but not by the skewness analysis (Figure 6B). To understand the origin of this discrepancy, we assessed the number of common and unique sites for each pair of replicate data sets [peak caller MACS (27); default P-value threshold conditions: 1 × 10−5), followed by IDR analysis for the predicted binding sites (Figure 6C and D, respectively). Interestingly, this complementary analysis revealed a lower number of significant common sites for replicate GSM646372 (‘triple C’) and GSM646373 (‘triple B’) than for the other replicate data sets. This IDR-defined differential quality of the two pairs of replicates was equally well detected by the QCi-STAMP (but not the skewness) approach. Overall, these comparisons show that QCi-STAMP provides a more versatile and reliable quality discrimination of NGS-generated profile than the skewness approach. Moreover, in contrast to IDR, QCi-STAMP reveals which of the replicates should be repeated to increase the overall quality without the necessity of using peak caller approaches.
Figure 6.
Comparison of QCi-STAMP performance with other analytical methodologies. (A) A set of four biological duplicates was selected from publicly available CTCF ChIP-seq profiles (pairs are enhanced by color code) and their corresponding QCi-STAMP descriptors were inferred (‘A’ for highest and ‘D’ for lowest quality). (B) The skewness of the read-count signal distribution of the biological replicates compared with the predicted QCi-STAMP (dRCI ≤ 5%). Note that the QCi-STAMP descriptors discriminate between data set (3) and (4), while their skewness evaluation does not. (C) Significant binding sites were predicted by MACS (default P-value threshold: 1 × 10−5) and classified based on their overlap between CTCF replicates (common and unique sites). Common sites were assessed by accepting up to 40-nt distance between MACS-predicted summits. (D) ‘IDR’ among CTCF replicates assessed by sorting significant binding sites according to the corresponding P-value. Note that in agreement with the QCi-STAMP descriptors, but differing with the skewness analysis (see panel C), data sets (3) and (4) present the worst IDR, while data sets (5) and (6) present the best IDR pattern.
An additional limitation of the IDR analysis, namely the dependence on peak caller performance, becomes apparent from analysing CTCF (Figure 6; sharp peaks) and H3K4me3 data sets (Supplementary Figure S6; broad peaks). While IDR analysis of CTCF can be done with 40 nt summit distance overlaps (i.e. the maximal distance between predicted summits to consider two binding events as reproduced), such conditions are noninformative for the H3K4me3 data set. To overcome this limitation, larger summit distance thresholds (e.g. 500 nt) have to be used to get informative results (Supplementary Figure S6). It is thus unlikely that comparisons between ChIP-seq profiles presenting different enrichment patterns can be done with IDR. In contrast, the QCi-STAMP reliably predicts the different qualities for the ‘triple A’ and ‘triple B’ pair of replicates and the common quality for the two ‘triple B’ replicates in the case of the evaluated H3K4me3 data sets (Supplementary Figure S6A), as illustrated for the CTCF profiles (Figure 6A).
DISCUSSION
The assessment of the quality of ChIP-seq data sets has been mostly performed by visual inspection in a genome browser and/or by the capacity of peak/island/pattern caller algorithms to predict locally enriched sequence counts. In both cases, it is a rather subjective analysis relying on user-defined criteria, such as the choice of ‘representative’ regions or thresholds for peak detection, and the statistical models and/or parameters used for assessment of enriched patterns. Only recently, methods are being developed that aim at providing a quantitative measure for the quality of ChIP-seq assays but so far there is no tool that provides a universal quality assessment for past and present NGS-generated profiles.
The present NGS-QC approach provides quantitative QCis generated from the evaluation of a feature common to all NGS-generated profiles, namely the profile construction from sequenced read overlaps. Conceptually, the QC Generator interrogates the robustness of such a profile when fewer sequenced reads are available, irrespective of the underlying experimental approach; simplistically this can be described as a numerical analysis similar to the visual inspection of Figure 2A, which displays RCIs at different TMRs but for the entire genome-aligned profile and not only for a selected region.
This concept has an inherent universal dimension, which is essential for comparative purposes and considering that the public GEO repository represents a powerful source for performing in silico data set comparisons, we have established a database of QCis for >5600 profiles. Our ultimate goal is to cover all publicly available ChIP-seq and enrichment-related NGS data sets to provide a comprehensive QCi library to the scientific community. Moreover, we invite all our colleagues to use the QC Generator for evaluation of their own profiles and suggest that all newly reported IP-based NGS profiles (which show the largest variability) are provided with the corresponding global QCis. We also invite the community to import all newly defined QCis into the global QCi database. Collectively, this database will be a highly valuable source of information about the quality that can be achieved, for example, for ChIP-seq of a certain target with a given (batch of) antibodies.
We believe that the universality, together with its simplicity and broad accessibility, makes the present system an attractive tool for QC analysis of profiles before engaging peak detection algorithms. Once a profile has been QCed, the QC descriptors provide objective numerical criteria to any NGS-generated profile that is provided to the community. Thus, existing profiles can be compared with others in multidimensional studies and meta-analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ligue National Contre le Cancer (laboratoire labelisé); the Association pour la Recherche sur le Cancer; the Institut National du Cancer (INCa); the European Community contracts [LSHC-CT-2005-518417 ‘EPITRON’ and HEALTH-F4-2009-221952 ‘ATLAS’]; the Alliance Nationale pour les Sciences de la Vie et de la Santé (AVIESAN)/Institut multi-organismes cancer (ITMO Cancer); D.G.C. was fellow of the Fondation pour la Recherche Médicale (FRM); FRM (aide aux projets innovants) (to W.v.G.). Funding for open access charge: Alliance Nationale pour les Sciences de la Vie et de la Santé (AVIESAN)/Institut Thématique Multi-Organismes Cancer (ITMO Cancer).
Conflict of interest statement. A patent application (EP123406478.4) describing the use of the NGS-QC system has been filed and the software has been deposited at the Agence Pour le Protection des Programmes (Paris).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of our laboratory and the computational IT support services at the IGBMC, especially Jean-Luc Toussaint, for support in generating the Web site interface. Furthermore, we would like to thank Malgorzata Nowicka for her support on statistical issues related to this study and Pierre-Etienne Cholley for the current maintenance and content curation of the NGS-QC database, and for the implementation of an executable version of the NGS-QC Generator. M.A.M.P developed the concept of Quality Control indicators and generated all data set comparisons; W.v.G implemented the NGS-QC-Generator, the NGS-QC database and its web interface together with M.A.M.P; D.C provided statistical support during data sets processing. M.A.M.S worked with W.v.G in the implementation of the customized Galaxy instance and is currently responsible for its maintenance. M.A.M.P and H.G. wrote the manuscript and the user tutorial.
REFERENCES
- 1.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 3.Ceschin DG, Walia M, Wenk SS, Duboe C, Gaudon C, Xiao Y, Fauquier L, Sankar M, Vandel L, Gronemeyer H. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25:1132–1146. doi: 10.1101/gad.619211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims RJ, III, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, III, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mamanova L, Andrews RM, James KD, Sheridan EM, Ellis PD, Langford CF, Ost TW, Collins JE, Turner DJ. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat. Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 19.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 21.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011;5:1752. [Google Scholar]
- 25.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiss DJ, Facciotti MT, Baliga NS. Model-based deconvolution of genome-wide DNA binding. Bioinformatics. 2008;24:396–403. doi: 10.1093/bioinformatics/btm592. [DOI] [PubMed] [Google Scholar]
- 29.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19 10 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho JW, Bishop E, Karchenko PV, Negre N, White KP, Park PJ. ChIP-chip versus ChIP-seq: lessons for experimental design and data analysis. BMC Genomics. 2011;12:134. doi: 10.1186/1471-2164-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.