Abstract
Lysine is catabolized via the saccharopine pathway in plants and mammals. In this pathway, lysine is converted to α-aminoadipic-δ-semialdehyde (AASA) by lysine-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH); thereafter, AASA is converted to aminoadipic acid (AAA) by α-aminoadipic-δ-semialdehyde dehydrogenase (AASADH). Here, we investigate the occurrence, genomic organization and functional role of lysine catabolic pathways among prokaryotes. Surprisingly, only 27 species of the 1478 analyzed contain the lkr and sdh genes, whereas 323 species contain aasadh orthologs. A sdh-related gene, identified in 159 organisms, was frequently found contiguously to an aasadh gene. This gene, annotated as lysine dehydrogenase (lysdh), encodes LYSDH an enzyme that directly converts lysine to AASA. Pipecolate oxidase (PIPOX) and lysine-6-aminotransferase (LAT), that converts lysine to AASA, were also found associated with aasadh. Interestingly, many lysdh–aasadh–containing organisms live under hyperosmotic stress. To test the role of the lysine-to-AASA pathways in the bacterial stress response, we subjected Silicibacter pomeroyi to salt stress. All but lkr, sdh, lysdh and aasadh were upregulated under salt stress conditions. In addition, lysine-supplemented culture medium increased the growth rate of S. pomeroyi under high-salt conditions and induced high-level expression of the lysdh–aasadh operon. Finally, transformation of Escherichia coli with the S. pomeroyi lysdh–aasadh operon resulted in increased salt tolerance. The transformed E. coli accumulated high levels of the compatible solute pipecolate, which may account for the salt resistance. These findings suggest that the lysine-to-AASA pathways identified in this work may have a broad evolutionary importance in osmotic stress resistance.
Keywords: lysine catabolism, osmoprotection, prokaryote, stress resistance
Introduction
The saccharopine pathway is the main irreversible pathway for lysine catabolism in higher eukaryotes. It has an essential role in homeostasis and the regulation of cellular lysine levels (Deleu et al., 1999; Papes et al., 1999). The two initial steps of this pathway are catalyzed by lysine-ketoglutarate reductase (LKR), which condenses lysine and α-ketoglutarate into saccharopine and saccharopine dehydrogenase (SDH), which hydrolyzes saccharopine into α-aminoadipic-δ-semialdehyde (AASA) and glutamic acid (Figure 1) (Arruda et al., 2000). AASA is then oxidized to aminoadipic acid (AAA) by α-aminoadipic-δ-semialdehyde dehydrogenase (AASADH) (Arruda et al., 2000). In plants and animals, the genes encoding LKR/SDH and AASADH are induced in response to osmotic and oxidative stresses (Guerrero et al., 1990; Stroeher et al., 1995; Deleu et al., 1999; Buchanan et al., 2005; Kirch et al., 2005; Rodrigues et al., 2006; Brocker et al., 2010) but the mechanism by which the enzymes protect against these stress remains to be elucidated.
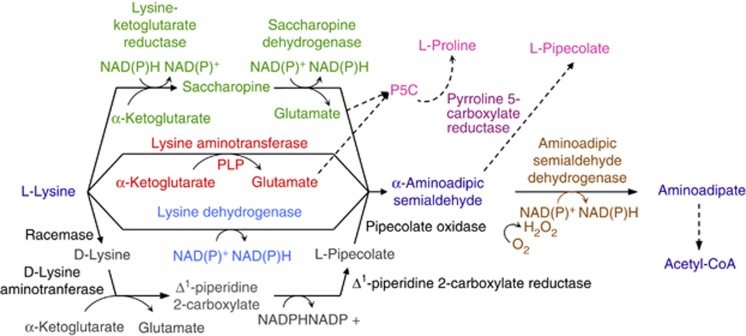
Figure 1.
The four pathways used by prokaryotes to catabolize lysine to AASA. In green, lysine is converted to AASA via the saccharopine pathway. The two-step reaction involves lysine-ketoglutarate reductase, which condenses lysine and α-ketoglutarate into saccharopine, and SDH, which hydrolyzes saccharopine into AASA and glutamate. In red, lysine is directly converted into AASA by LAT, which catalyzes the transamination of α-ketoglutarate, yielding AASA and glutamate. In blue, lysine is also directly converted into AASA by LysDH, which catalyzes the oxidative deamination of lysine. In gray, a multi-step reaction starts with the conversion of L-lysine to D-lysine by a racemase. D-lysine is then deaminated by an aminotransferase to form Δ1-piperidine-2-carboxylate, which is then converted to pipecolate by Δ1-piperidine-2-carboxylate reductase. Pipecolate is finally oxidized to AASA by pipecolate oxidase. All four pathways use AASADH (brown) to convert AASA to aminoadipate. P6C, glutamate and AAA can be used as substrates for several cellular pathways, leading to the production, for example, of pipecolate and proline (pink) by P5CR (dark pink).
Recently, we identified LKR and SDH in the marine bacterium Silicibacter pomeroyi (Serrano et al., 2012). An interesting aspect of the LKR- and SDH-encoding genes in S. pomeroyi is their colocalization in an operon together with the gene encoding glutathione-S-transferase (GST) (Serrano et al., 2012). GST is an important enzyme associated with cellular detoxification, xenobiotics clearance and protection against chemical and heavy-metal toxicity, and oxidative stresses (Veal et al., 2002; Gratão et al., 2005; Allocati et al., 2009). This genomic organization suggests a possible connection between stress responses and the saccharopine pathway.
In this work, we investigate the occurrence and organization of the genes encoding LKR, SDH and AASADH in the sequenced genomes of prokaryotic organisms. We examined the breadth of distribution of the saccharopine pathway and the organization of the genes encoding its enzymes within the prokaryotic genomes. Surprisingly, we discovered that prokaryotic organisms use other lysine catabolic enzymes rather than the saccharopine pathway to convert lysine to AASA. The genes encoding these other enzymes are frequently organized in operons. One of these operons, lysdh–aasadh, contains the gene encoding lysine dehydrogenase (LYSDH), an enzyme that directly transforms lysine into AASA (Misono and nagasaki, 1982). The role of this operon in the salt stress response was investigated in S. pomeroyi. Furthermore, we expressed this operon in Escherichia coli to assay the involvement of LYSDH along with AASADH in protection against high-salt stress. We discuss the role for the lysine-to-AASA pathways in the stress response of prokaryotes.
Materials and methods
Bioinformatics
The prokaryote genome sequences were retrieved from the NCBI Microbial Genomes database (ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria) as of October 2011. We retrieved 2712 sequences comprising completely sequenced bacterial genomes and plasmids from 1362 bacteria and 116 archaea. The nucleotide sequences encoding LKR, SDH and AASADH from S. pomeroyi, yeast, human, mouse and plants were used as true positive queries. Nucleotide sequences encoding other enzymes that share conserved domains with the enzymes of the saccharopine pathway were used as true negative queries.
The query nucleotide sequences were tBLASTn (Altschul et al., 1997) against prokaryotic genomes using a 10−10 e-value cut-off. The NCBI taxonomy database was used to cross-information through the ‘GI' and ‘TaxID' associated with each organism group. The ‘best hit' obtained in the tBLASTn search was mapped to the corresponding full-length protein sequence using BLASTx. To avoid misinterpretation of the BLAST results, the bidirectional best hit function was used to select true positives. All the data were parsed into MySQL tables and, using SQL queries, crossed with data containing the organism information. The information contained in the ‘.gff' file was used to determine the distance between genes, orientation and genomic organization.
The phylogenetic analysis was performed using ClustalW for the multiple sequence alignments and MEGA 5 (Tamura et al., 2011) for the creation of maximum likelihood phylogenetic trees using JTT model (Jones et al., 1992) with 1000 bootstrap replicates.
Bacterial strain and culture conditions
S. pomeroyi DSS-3 were obtained from the American Type Culture Collection (www.atcc.org). The bacteria were plated on solid Difco Marine Broth 2216 (MB) containing 0.8% agar and grown for 2 days at 30 °C. Individual colonies were inoculated in half-strength MB containing 5 g l−1 sucrose and grown in a rotatory shaker at 200 rpm at 30 °C for 36 h. Liquid cultures were obtained by inoculating 2 ml of 12.5% MB containing various concentrations of NaCl with 100 μl of the primary cultures.
Transcript quantification
Total RNA was isolated from bacterial cultures using the Illustra RNAspin Mini Isolation Kit (GE Healthcare, Buckinghamshire, UK). The RNA quality and integrity was verified by gel electrophoresis. First-strand-complementary DNA was synthesized from 1 μg total RNA using the RevertAid H Minus First Strand complementary DNA Synthesis Kit (Fermentas, Vilnius, LT, USA). Quantitative real-time PCR was conducted using defined primers (Supplementary Table S1) and the SYBR GREEN PCR master mix (Applied Biosystems, Warrington, UK). Relative mRNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA levels using the ΔΔCT method. The PCR amplification was performed in triplicate. The Student's t-test was used to evaluate the significance of the differential mRNA levels.
Construction of vectors for LYSDH, AASADH and LYSDH/AASADH and expression in E. coli BL21 DE3
The lysdh and aasadh gene sequences (separately or together as an operon unit) were amplified from the S. pomeroyi genomic DNA using the following forward primers: 5′-ggtcagccatgggcatgcgctggaacatttgtgt-3′ (lysdh) 5′-ggtcagccatgggcatgaaagatatcgtcatgac-3′ (aasadh), which contain NcoI restriction site, and reverse primers: 5′-ggagttgctcagctcaagcggccttgttttggg-3′ (lysdh) 5′-ggctacgctcagctcagatgtcgaacttcaccc-3′ (aasadh), which contain BlpI restriction site. The amplified fragments were cloned into pET28a (Novagen, Darmstadt, DE) that had been double-digested with NcoI and BlpI. The restriction endonucleases and the T4 DNA ligase were obtained from New England Biolabs (Ipswich, MA, USA). The pET28a–lysdh, pet28a–aasadh and pET28a–lysdh–aasadh constructs and the empty vector were transformed into the BL21 DE3 pRIL E. coli strain (Stratagene, San Diego, CA, USA). The positive colonies were verified by PCR, digestion and sequencing.
To induce recombinant protein expression the four clones were grown in LB medium containing 50 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) (inducer) or 5 mM D-glucose (repressor). RNA and protein were extracted using the NucleoSpin RNA/Protein kit (Macherey–Nagel, Duren, DE). complementary DNA was synthesized and RT-PCR was performed to confirm the presence of lysdh and aasadh transcripts. Protein extraction and quantification was performed as described (Karlsson et al., 1994). Expression of the LYSDH and AASADH proteins was verified by SDS-PAGE.
Stress treatment of E. coli transformed with the expression vectors
E. coli lines harboring the four constructs were grown overnight in 2.5 ml LB containing 50 μM kanamycin in a rotatory shaker at 250 r.p.m. and 37 °C. Fifty microliters of each pre-inoculum was used to inoculate 1.2 ml LB with or without 400 mM NaCl and either 50 μM IPTG or 5 mM D-glucose. The effect of lysine was analyzed under the same conditions with or without 5 mM L-lysine.
Solutes extraction
E. coli transformed with empty pET28a or the recombinant plasmids were grown in triplicate for 8 h in LB medium containing 50 μM kanamycin and 50 μM IPTG. Bacteria were pelleted by centrifugation and washed three times with glucose-tris-EDTA buffer. The pellet was frozen in liquid nitrogen, and the cells were disrupted by freezing and thawing three times. The solutes were extracted with 80% ethanol (Gouesbet et al., 1992). For S. pomeroyi, bacterial cultures were pelleted after 24 h growth in 12.5% MB containing 400 mM NaCl in the presence of 0, 5, 10 and 20 mM lysine or pipecolate. The ethanol extract was evaporated to dryness, and the dry residue was dissolved in deionized water.
Pipecolate determination
The samples were analyzed in a Waters Acquity UPLC coupled to a TQD triple quadrupole mass spectrometer (Micromass–Waters, Manchester, UK). Five microliters of each sample was injected into a Waters Acquity C18-BEH (2.1 × 50 mm2, 1.7 μm) column at 30 °C. The column was eluted using a 0–10% acetonitrile gradient in 50 mM ammonium acetate pH 6.5 at a flow rate of 0.15 ml min−1. Electrospray ionization in the positive ion mode was performed under the following parameters: ion source temperature 150 °C, desolvation temperature 300 °C, capillary 3.0 kV and cone 50 V. The MS/MS spectra of the detected peaks were obtained using collision-induced dissociation with a collision energy of 15 V. Pure L-pipecolic acid (Sigma, St Louis, MO, USA) was used as standard. The retention time and fragmentation pattern were used to identify L-pipecolic acid in the bacterial samples.
Results
Identification of lysine catabolic enzymes linked to AASA synthesis in prokaryotes
tBLASTn searches using the LKR, SDH and AASADH nucleotide sequences against 2712 nucleotide sequence files comprising chromosomal and plasmidial genome sequences corresponding to 1478 microbe organisms identified orthologs for both the lkr and sdh genes in a restricted set of 27 organisms (Table 1).
Table 1. Distribution of enzymes of the Lysine-to-AASA catabolic pathways among prokaryotes.
| Kingdom | Phylum | Number of species containing a distinct enzyme of the four lysine-to-AASA pathways | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
AASADH |
LKR |
SDH |
LysDH |
LAT |
PIPOX |
LKR and SDH |
LysDH and AASADH |
LAT and AASADH |
Pip2C, PIPOX and AASADH |
| Archea | Euryarchaeotes | 19 | — | — | 12 | 1 | — | — | 2 | — | — |
| Crenarchaeotes | — | — | — | 12 | — | — | — | — | — | — | |
| Unclassified | 1 | — | — | 1 | — | — | — | — | — | — | |
| Eubacteria | Actinobacteria | 45 | 4 | 4 | 2 | 39 | — | 4 | 2 | 38 | — |
| Bacteroidetes | 31 | 20 | 19 | 7 | 9 | — | 19 | 7 | 8 | — | |
| Cyanobacteria | 6 | 1 | — | 3 | — | — | — | 1 | — | — | |
| Deinococcus/Thermus | 9 | — | — | — | — | — | — | — | — | — | |
| Firmicutes | 19 | — | — | 23 | — | — | — | 12 | — | — | |
| Proteobacteria | 177 | 14 | 5 | 96 | 4 | 61 | 4 | 88 | 3 | 25 | |
| Other | 16 | — | — | 3 | 3 | — | — | 3 | 2 | — | |
| Total number of taxa | 323 | 39 | 28 | 159 | 56 | 61 | 27 | 115 | 51 | 25 | |
Enzymes: AASADH, α-aminoadipic-δ-semialdehyde dehydrogenase; LAT, lysine-6-aminotransferase; LKR, lysine-ketoglutarate reductase; LysDH, lysine dehydrogenase; SDH, saccharopine dehydrogenase; PIPOX, pipecolate oxidase; Pip2C, Δ1-piperidine-2-carboxylate reductase.
We found a large number of SDH-related sequences, harboring around 40% amino-acid similarity to the first two-thirds of the SDH amino-acid sequence that were contiguously located to an aasadh gene. BLASTp searches against the protein database identified a partial 3-dimensional structure for the N-terminal portion of the enzyme encoded by the SDH-related sequence from S. pomeroyi (PDB code: 3IC5). We used PyMol (Delano, 2002) to structurally align this N-terminal structure with the fungal SDH structure (PDB 2Q99), but this resulted in an alignment with RMSD >3.5 Å (Supplementary Figure S1A). A valid and low-RMSD alignment was obtained with an Archaea LYSDH enzyme (PDB 3ABI) (Yoneda et al., 2010) (Supplementary Figure S1B). We also found a LYSDH in Agrobacterium tumefaciens (Misono and Nagasaki, 1982) whose amino-acid sequence is 70% similar to the amino-acid sequences of the SDH-related sequences found in S. pomeroyi. Thus, we annotated these sdh-related genes as lysdh (Table 1). LYSDH directly converts lysine into AASA in a single-step reaction using NAD(P)+ as a cofactor (Misono and Nagasaki, 1982; Yoneda et al., 2010) (Figure 1). We found lysdh homologs in 159 bacterial species, and the gene was colocalized in an operon with the aasadh gene in 36 of them (Supplementary Table S2). Interestingly, 29 of these 36 organisms live or are able to live in highly saline, drought, heavy-metal and oxidative stress environments (Supplementary Table S2).
We then searched for other genes that frequently colocalize with aasadh as this gene was found in a large number of organisms. We identified the genes encoding lysine-6-aminotransferase (LAT) and pipecolate oxidase (PIPOX) colocalized with aasadh. Similar to LYSDH, LAT catalyzes the direct conversion of lysine to AASA using α-ketoglutarate as the lysine ɛ-amino group aceptor for the production of glutamate and AASA (Fujii et al., 2000) (Figure 1). This enzyme was identified in 56 prokaryotic organisms, in 35 of which it was located in an operon with the aasadh gene (Supplementary Table S3). Twenty-nine of these 35 organisms live or are able to live in highly saline, drought, heavy metal and oxidative stress environments. PIPOX catalyzes the final reaction step of a pathway, described in the Pseudomonas genus, which starts with the isomerization of L-lysine to D-lysine by a racemase. Then, D-lysine is deaminated by D-lysine aminotransferase to form Δ1-piperidine-2-carboxylate, which is used by Δ1-piperidine-2-carboxylate reductase to form L-pipecolate. The pipecolate is then converted to AASA by PIPOX (Revelles et al., 2005) (Figure 1). Δ1-piperidine-2-carboxylate reductase, PIPOX and AASADH were found in 25 organisms, and genes similar to Pseudomonas pipox were found in 61 organisms, 48 of which showed colocalization of pipox and aasadh in operons (Supplementary Table S4).
Thus, in a large number of prokaryotes, LKR/SDH, LYSDH, LAT and PIPOX are the alternative pathways for the production of AASA from lysine. All the four pathways have AASADH as a common enzyme to convert AASA to AAA (Figure 1). The most abundant and widely distributed lysine catabolic enzymes found in the prokaryotes, analyzed in this work, are LYSDH and LAT (Table 1).
Lysine catabolism genes are linked to stress response genes in operons
Lkr, sdh, lysdh, lat and aasadh colocalize in operons in the genomes of diverse prokaryotes (Supplementary Figure S2). Interestingly, phylogenetically distant taxa conserve similar operon configurations. Additionally, genes encoding transcription factors involved in the regulation of amino-acid metabolism, such as asnC, are frequently located adjacent to lysdh or lat. It is likely that these operons allow the co-expression of functionally related genes in response to a range of stress conditions.
The lkr–sdh configuration was found in the genomes of nine organisms (four actinobacteria, four α-proteobacteria and one γ-proteobacteria) (Supplementary Table S5). Five of these bacteria are considered moderately halophilic (from the Silicibacter, Roseobacter, Roseovarius and Salinispora genera), one is a psychrophilic and piezophilic deep-sea bacterium (Shewanella genus) and the remaining three are actinobacteria found in dry soil (Supplementary Table S5). The lysdh–aasadh configuration was found in 36 organisms, 22 of which are proteobacteria (two α-, seven β- and 12 γ-proteobacteria), 11 firmicutes, 2 bacteroidetes and 1 putative nitrospira bacterium (Supplementary Table S2). Twenty-nine of these 36 species live or are able to live in hyperosmotic or oxidative stress environments (Supplementary Table S2). The lat–aasadh configuration was found in 35 organisms, 32 being actinobacteria, 2 bacteroidetes and s1 δ-proteobacteria (Supplementary Table S3). Interestingly, there are also some cases of single gene duplication (that is aasadh gene in Rhodococcus jostii RHA1 and Rhodococcus opacus B4) or operon duplication, in Mycobacterium bovis. In several Mycobacterium, the lat gene is localized in an operon with asnc and the sigma factor sigf (Supplementary Figure S2B), which is known to be essential for survival in conditions of heat shock, pH and oxidative stress (Gebhard et al., 2008). The gene encoding AsnC is upregulated by starvation and has a significant role during the persistent/latent phase of mycobacterial infection (Betts et al., 2002). The sigf–asnc–lat configuration was found in 18 of the 22 Mycobacteria that have completely sequenced genomes (Supplementary Table S3). Other actinomycetes that contain the LAT pathway produce pipecolate to serve as a substrate for antibiotic synthesis. In Streptomyces, for example, the LAT enzyme is considered to be the first step in the β-lactamic biosynthetic pathway (such as in cephamycin production) (Alexander et al., 2007). There is no evidence that the LAT pathway is involved in abiotic stress responses in these organisms.
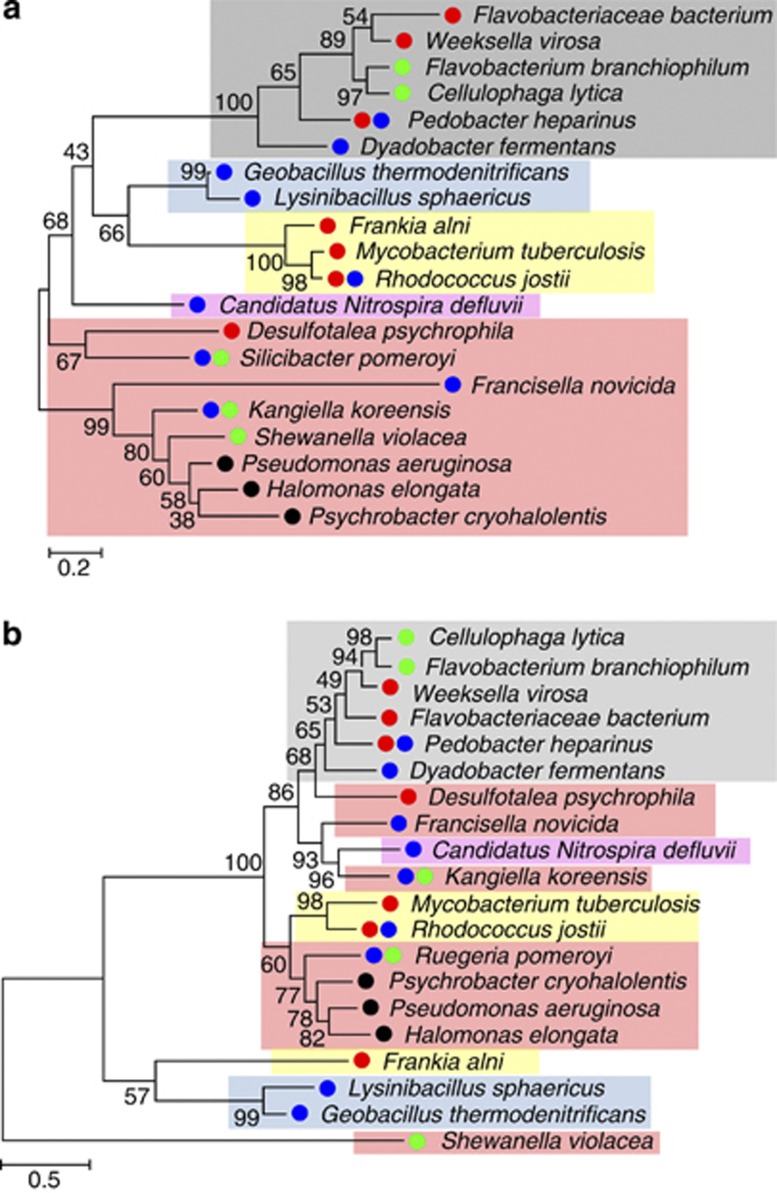
Lateral gene transfer may explain the presence of lysine-to-AASA pathways in distantly related taxa. To test for Lateral gene transfer we first reconstructed the reference species' phylogenetic tree and then reconciled it with the Lateral gene transfer for a candidate gene tree. Because all four pathways have AASADH in common, we used this enzyme to analyze the phylogenetic relationships among these organisms. We built a tree using the RNA polymerase subunit-α (rpoA), a gene commonly used for phylogenetic analysis (Zeigler, 2003), to trace the phylogenetic relationships among bacteria containing LKR, SDH, LYSDH, LAT or PIPOX. Next, we created a phylogenetic tree using AASADH for the same organisms. The phylogenetic tree constructed with RpoA is consistent with the phylogenetic relationships established for bacteria (Figure 2a), but the phylogenic relationships among the AASADH sequences are not congruent with the taxonomic results from the topology of the RpoA tree (Figure 2b). This result suggests that the lysine-to-AASA pathways may have been acquired via lateral gene transfer in some taxa (for example, Proteobacteria and Actinobacteria). Interestingly, some phyla, such as bacteroidetes, contain organisms that have genes for the LYSDH, LAT or saccharopine pathway, suggesting that gene transfer may have occurred several times. Taken together, these observations suggest that the lysine-to-AASA pathways have been widely spread among bacteria living in different ecosystems, and the prevalence of these genes among bacteria living in hyperosmotic environments points to a potential role in adaptive advantage in such habitats.
Figure 2.
Phylogenetic tree based on the maximum likelihood method. (a) RpoA and (b) AASADH amino-acid sequences from organisms that contain LKR and SDH (●), LYSDH (●), LAT (●) and PipOX (●). The shadowed boxes indicate bacterial phyla: (gray) Bacteroidetes; (blue) Firmicutes; (yellow) Actinobacteria; (pink) Nitrospirae; (red) Proteobacteria.
Another interesting finding is that S. pomeroyi DSS-3, Silicibacter sp. TM1040 and Kangiella koreensis, all moderately halophytic seawater bacteria (Lima et al., 2009), have both the saccharopine and LYSDH pathways. Other species, such as the soil and sea bacterium Pedobacter heparinus, bacteria from the genus Rhodococcus and the extremely halophytic bacteria Salinibacter ruber, contain both LYSDH and LAT. Some Rhodococcus species have more than one copy of the aasadh gene. Coincidently, all of these organisms live or are able to live in hyperosmotic stress environments. Salinibacter species are among the most halophilic bacteria known. S. ruber, for example, cannot grow in salt concentrations below 15% and is very successful at colonizing saltern ponds (Anton et al., 2002).
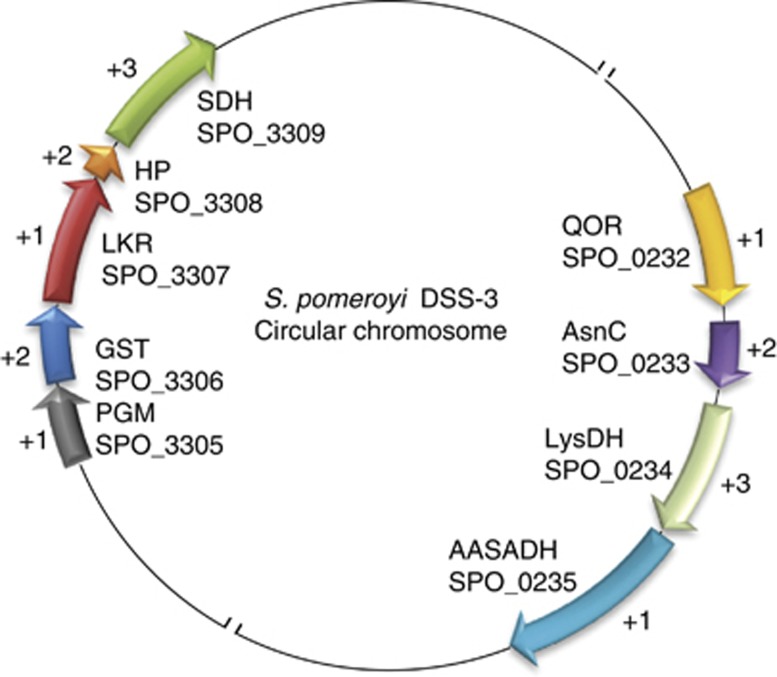
S. pomeroyi has two lysine-to-AASA pathways. The lkr and sdh genes colocalize with GST and the glycolysis gene encoding phosphoglycerate mutase (PGM). The lysdh gene colocalizes with aasadh, the transcriptional regulator asnc and the oxidative stress-associated gene encoding quinone oxidoreductase (QOR; Figure 3). To probe the functionality of the operons pgm–gst–lkr–sdh and qor–asnc–lysdh–aasadh, they were amplified from the complementary DNA synthesized from total RNA isolated from S. pomeroyi grown in MB medium. Primers designed to amplify individual as well as concatenate genes produced amplicons, supporting the hypothesis that these genes are produced as polycistronic mRNA. This evidence supports the idea that the genes encoding lysine catabolic enzymes are functionally linked in operons together with stress responsive genes (Supplementary Figure S3).
Figure 3.
Genomic organization of the genes comprising the lkr–sdh and lysdh–aasadh operons in S. pomeroyi. The (+) signal preceding the numbers 1, 2 and 3 indicates the transcribed frame. SPO followed by 4 digits indicates the NCBI gene identifier. The genes of the LKR/SDH operon encode: PGM, phosphoglycerate mutase; GST, glutathione-S-transferase; LKR, lysine-ketoglutarate reductase; HP, hypothetical protein; SDH, saccharopine dehydrogenase. The genes for the lysdh–aasadh operon encode: QOR, quinone oxidorreductase, AsnC, AsnC transcriptional regulator family; LysDH, lysine dehydrogenase, AASADH, α-aminoadipic-δ-semialdehyde dehydrogenase.
Lysine catabolism genes are upregulated in high-salt conditions in a lysine-dependent manner
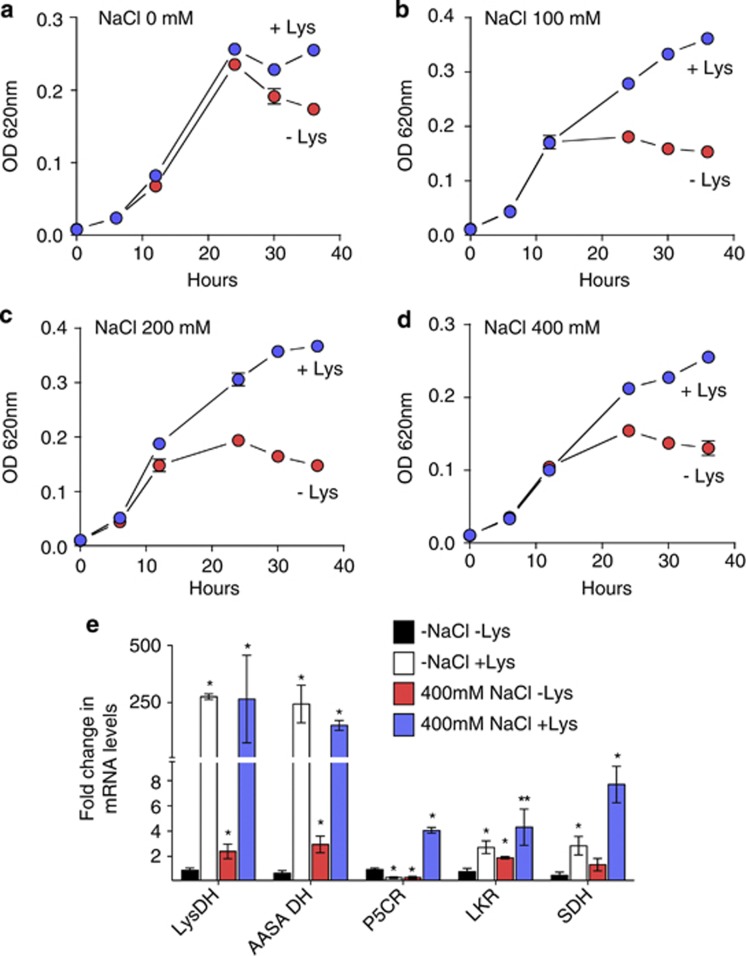
To verify whether the lkr, sdh, lysdh and aasadh genes are induced by salt stress, S. pomeroyi was grown in 12.5% MB medium supplemented with increasing concentrations of NaCl in the presence or absence of lysine (Figure 4). The addition of 5 mM lysine to the MB medium in the absence of salt had no significant effect on the bacterial growth rate (Figure 4a), but addition of the amino acid to MB medium containing 100, 200 or 400 mM NaCl significantly increased the bacterial growth rate and extended the exponential growth phase. (Figure 4b–d). These results suggest that S. pomeroyi can use lysine to adapt to high-salt environments.
Figure 4.
Effect of lysine supplementation on the S. pomeroyi growth rate in high-salt medium. Bacterial cultures were grown for 36 h in 12.5% MB medium containing the indicated salt concentrations: (a) no salt; (b) 100 mM NaCl, (c) 200 mM NaCl, (d) 400 mM NaCl, in the presence or absence of 5 mM lysine. Growth curves were analyzed for statistic significance based on comparison of log phases slopes using covariance analysis. (a) not significant (P=0.43); (b) significant (P=0.008); (c) significant (P=0.0007). (d) significant (P=0.0008). (e) Relative mRNA levels of lysdh, aasadh, lkr, sdh and p5cr, as determined by quantitative real-time PCR, in bacterial cells grown for 24 h in high salt. The fold changes in mRNA levels were estimated using the ΔΔCT method based on the CT of glyceraldehydes-3-phosphate dehydrogenase as the endogenous control. Changes in the relative mRNA levels were compared using Student's t-test. One and two asterisks represent significant differences at P<0.05 and P<0.1, respectively.
We then asked whether the lkr–sdh and lysdh–aasadh operons are induced in S. pomeroyi grown under salt stress. Samples were collected from S. pomeroyi grown for 24 h in MB medium containing 400 mM NaCl in the presence or absence of 5 mM lysine. In addition, we verified the relative mRNA level of the p5cr gene, which encodes the enzyme δ1-pyrroline-5-carboxylate reductase (P5CR). P5CR uses δ1-piperideine-6-carboxylate (P6C), the cyclic form of AASA, as a substrate to produce pipecolate (Fujii et al., 2002; Eduard and Jakobs, 2010). P5CR also catalyzes the formation of L-proline using δ1-pyrroline-5-carboxylate as a substrate (Peisach and Strecker, 1962). Quantitative real-time PCR analyses revealed that both the lkr–sdh and lysdh–aasadh operons are induced by lysine and salt stress, but the lysdh–aasadh pathway is 100-fold more responsive to lysine than the lkr–sdh pathway, indicating that this pathway is preferentially expressed under salt stress (Figure 4e and Supplementary Figure S4). Interestingly, the p5cr gene is also upregulated by high salt, but only in a lysine-dependent manner (Figure 4e). The upregulation of the p5cr gene might indicate that lysine and salt can trigger pipecolate production under salt stress.
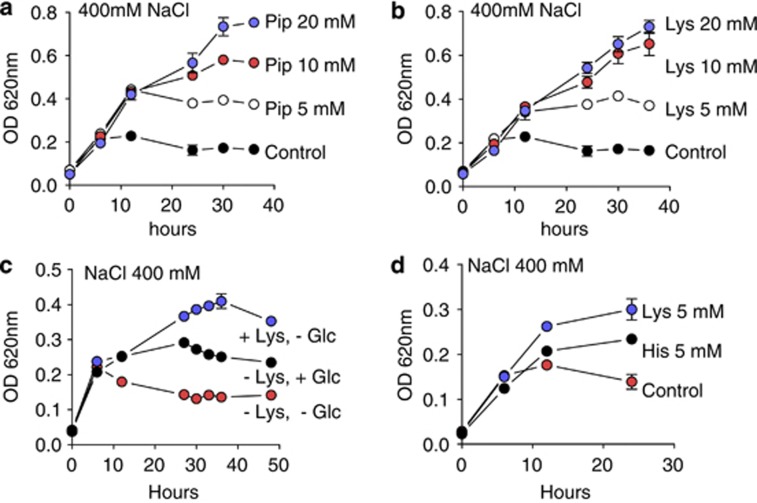
If pipecolate or its derivatives act as an osmoprotectant,the addition of this amino acid to the salt stress medium might improve adaptation to saline environments. The addition of 5, 10 or 20 mM pipecolate to 12.5% MB medium containing 400 mM NaCl improved the growth of S. pomeroyi (Figure 5a) at a rate similar to that observed for lysine supplementation (Figure 5b). The presence of pipecolate inside the bacterial cells was determined by mass spectrometry. We could not detect pipecolate accumulation in the bacteria grown in 5 mM pipecolate or 5 mM lysine, but it accumulated at high levels inside the bacterial cells at high concentrations of pipecolate (10 and 20 mM).
Figure 5.
Effects of pipecolate, lysine, glucose or histidine on the S. pomeroyi growth rate in high-salt medium. Bacterial cultures were grown in 12.5% MB medium containing 400 mM NaCl supplemented with: (a) 0, 5, 10 or 20 mM pipecolate; (b) 0, 5, 10 or 20 mM lysine; (c) 5 mM glucose or 5 mM lysine; (d) 5 mM glucose was replaced by 5 mM histidine.
To address the question of whether lysine is simply being used as a carbon source for bacterial growth, S. pomeroyi was grown in 12.5% MB medium supplemented with high salt and either glucose or lysine (Figure 5). The bacteria grew more densely with lysine than with glucose in the high-salt medium (Figure 5c). We also assayed whether the addition of other positively charged amino acids to the high-salt MB medium provided the same growth advantages as lysine. When histidine was added to the high-salt MB medium, the bacteria did not grow as densely as they did with the lysine supplementation (Figure 5d).
Expression of lysdh–aasadh operon in E. coli increases high-salt tolerance
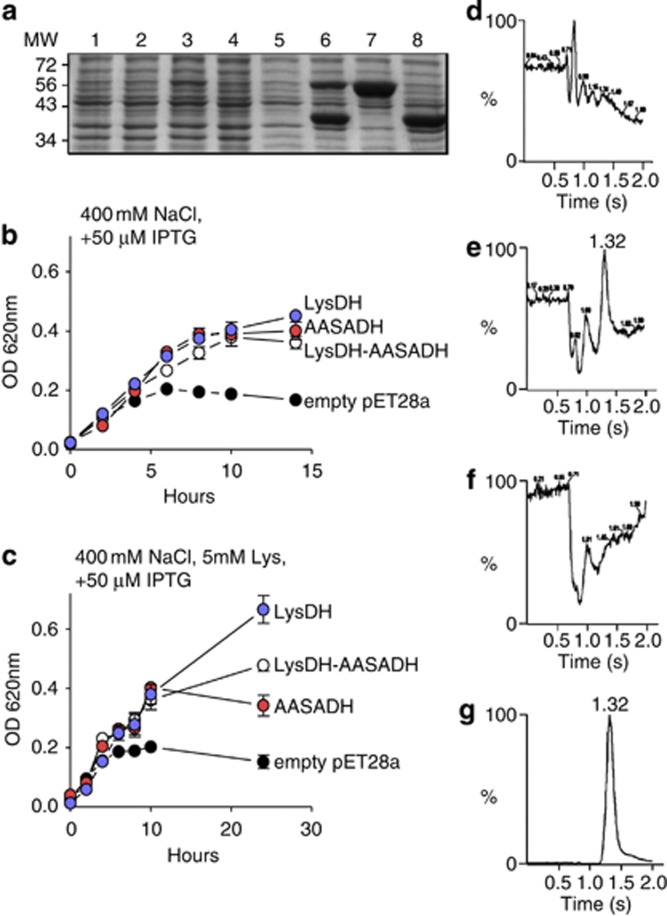
The S. pomeroyi lysdh and aasadh genes were cloned into pET28a under the control of the lac promoter and transformed into the E. coli strain BL21 DE3 pRIL. The two genes were cloned both individually and together in operon configuration. Individual E. coli lines harboring the empty pET28a plasmid or one of the three recombinant plasmids had similar growth rates in LB medium without high salt (Supplementary Figure S5A). When induced with 50 μM IPTG, the E. coli expressing lysdh, aasadh or lysdh–aasadh grew similarly to those without IPTG induction, but those transformed with the empty pET28a plasmid had a significantly reduced growth rate (Supplementary Figure S5B). The higher growth rate observed in the E. coli harboring the lysdh, aasadh or lysdh–aasadh genes may indicate that these genes alleviate the stress caused by IPTG (Yee and Blanch, 1993). The E. coli strains harboring the lysdh, aasadh or lysdh–aasadh constructs correctly expressed the recombinant 40 kDa LYSDH protein and/or the recombinant 52 kDa AASADH protein when induced with 50 μM IPTG (Figure 6a). Under high-salt conditions, the three strains harboring the recombinant constructs showed increased growth rates compared with the control strain harboring the empty pET28a plasmid (Figure 6b). The addition of 5 mM lysine to the high-salt medium further increased the growth rate of the E. coli strains harboring the recombinant lysdh and lysdh–aasadh constructs as compared with the strain harboring the empty pET28a plasmid or that containing solely aasadh (Figure 6c). The LB medium used in this assay has lysine originated from hydrolyzed tryptone and yeast extract. This lysine-containing medium may help the strains grow in high salt for the first 10 h. After this period, the medium supplemented with additional lysine maintained the high growth rate of the strain harboring the lysdh construct compared with the strains harboring the other constructs or the empty pET28a plasmid (Figure 6c).
Figure 6.
Effect of salt stress on the growth rate of E. coli BL21 DE3 harboring constructs expressing the lysdh–aasadh operon components. (a) SDS-PAGE of proteins extracted from bacterial cells grown for 8 h in non-induced and IPTG-induced cultures. Lanes 1, 2, 3 and 4 correspond to the protein extracts of bacterial cells harboring the pET28a empty plasmid, lysdh–aasadh, lysdh and aasadh, respectively, grown in non-induced LB medium. Lanes 5, 6, 7 and 8 correspond to the protein extracts of bacterial cells harboring the pET28a empty plasmid, lysdh–aasadh, lysdh and aasadh, respectively, grown in LB medium containing 50 μM IPTG. (b) Growth rates of E. coli harboring the pET28a empty plasmid, lysdh–aasadh, lysdh and aasadh constructs in high-salt LB medium containing 50 μM IPTG. (c) Growth rates of E. coli harboring the the pET28a empty plasmid, lysdh–aasadh, lysdh and aasadh constructs in high-salt LB medium containing 50 μM IPTG in the presence or absence 5 mM lysine. (d–g) Elution pattern of bacterial cell extracts from E. coli harboring empty pET28a plasmid (d) lysdh–aasadh construct (e) aasadh construct (f) or lysdh construct (g). Peaks with the expected retention time for L-pipecolic acid (1.32 min) were found for lysdh–aasadh and lysdh constructs but not for the aasadh construct or the empty pET28a.
Samples of the E. coli BL21 DE3 pRIL harboring one of the recombinant constructs or the empty pET28a plasmid grown in high-salt medium were analyzed for pipecolate accumulation. The pipecolate standard peaked at m/z=130.2, corresponding to C6H11NO2. The base peak was at m/z∼84 (Supplementary Figure S5C). Nearly identical peak patterns were found for bacteria harboring lysdh and lysdh–aasadh, confirming the presence of pipecolate in the samples (Supplementary Figure S5D). The strains harboring the aasadh-only construct or the empty pET28a plasmid did not show any pipecolate accumulation (Figure 6d and f). The bacteria harboring the lysdh-only or the lysdh–aasadh construct accumulated pipecolate to approximately 611 and 32.9 nmol mg−1 protein, respectively (Figure 6e and g). The accumulation of high amounts of pipecolate indicates that the lysine-to-AASA pathway derives compatible solutes, which can improve bacterial performance under salt stress, and suggests that LYSDH may have a major role in salt stress tolerance.
Discussion
The saccharopine pathway is involved in the regulation of lysine levels and the stress response in plants and animals (Arruda and Neshich, 2012). In this pathway, LKR and SDH catalyze, in two reaction steps, the conversion of lysine to AASA and the formation of glutamate. AASA is a toxic semialdehyde, and AASADH, the third enzyme of the pathway, may keep the AASA concentration at very low levels by converting it to AAA (Figure 1). The cyclic isomer of AASA, P6C, can be used as a P5CR substrate to produce pipecolate (Fujii et al., 2002; Eduard and Jakobs, 2010), a physiological osmoprotectant (Moulin et al., 2006) (Figure 1). The saccharopine pathway was recently found in the seawater bacteria S. pomeroyi (Serrano et al., 2012). Interestingly, the S. pomeroyi genes encoding LKR and SDH are localized in an operon together with GST, suggesting that this pathway has a significant role in the stress responses of diverse organisms.
The LKR and SDH proteins were found in only a few bacterial species, whereas AASADH was identified in a much larger number of organisms (Table 1). The existence of other enzymatic pathways that produce AASA from lysine (Figure 1) indicates that this lysine catabolic derivative is an important metabolite that is used by bacteria for growth and adaptation to stress. This view is supported by the fact that lysdh, lat and pipox genes are located in operons together with aasadh in diverse organisms (Supplementary Figure S2). LYSDH and LAT occur more frequently and are distributed among distantly related organisms, including Archaea. There is no apparent clade specificity for each pathway, although most of the LYSDH-containing organisms belong to the proteobacteria phylum and the majority of the LAT-containing organisms are actinobacteria. Nearly all phyla represented have LYSDH-containing organisms (Table 1), and phylogenetic analyses suggest that horizontal gene transfer may have contributed to the spread of these pathways to distinct taxa. In the four lysine-to-AASA pathways identified, the genes are frequently organized in the form of lkr–sdh, lysdh–aasadh, lat–aasadh and pipox–aasadh operons that also comprise genes encoding oxidative stress enzymes. This suggests a connection among lysine catabolism and stress adaptation.
The LYSDH and LAT pathways are more widely distributed among the analyzed prokaryotes than the LKR–SDH and PIPOx–AASADH pathways. We hypothesize that the LYSDH and LAT pathways produce the osmoprotectant pipecolate more efficiently than the other two pathways. The efficient production of pipecolate from P6C quickly relieves osmotic stress allowing for rapid response of the bacteria to stress. For example, pipecolate accumulates at high levels and protects against osmotic stress in Corynebacterium ammoniagenes (Gouesbet et al., 1992) and has been described as a genuine osmoprotectant for E. coli grown in high salt (Gouesbet et al., 1994). Pipecolate was also shown to accumulate in rapeseed leaf tissues subjected to osmotic stress (Moulin et al., 2006) and in halophytic and sand dune plants subjected to permanent water stress (Goas et al., 1975; Jefferies et al., 1979). Furthermore, pipecolate co-accumulates with proline in water-stressed soybeans (Fukutoku and Yamada, 1981). Proline could also be a derivative of the LAT and saccharopine pathways, as it can be synthesized from the glutamate formed in the first transamination step (Figure 1). In plants, glutamate is a substrate for proline synthesis; the enzyme δ1-pyrroline-5-carboxylate synthetase converts it to δ1-pyrroline-5-carboxylate, which is reduced to proline by P5CR (Delauney et al., 1993), the same enzyme that converts P6C to pipecolate (Fujii et al., 2002; Eduard and Jakobs, 2010). Thus, in the presence of AASA and glutamate, the protective osmolytes pipecolate and proline could be formed.
Another possibility is that these pathways produce AASA, which may act as a signaling molecule. In yeast for example, AASA modulates the transcription factor Lys14, which regulates the transcription of LYS genes (Feller et al., 1994).
In S. pomeroyi, the LYSDH pathway is preferentially expressed, as compared to the saccharopine pathway, upon salt stress and lysine induction (Figure 4). The fact that the gene encoding P5CR is also upregulated in high-salt medium in a lysine-dependent manner (Figure 4) corroborates with the notion that lysine may fuel pipecolate and its derivatives in bacteria subjected to stress. In plants, the lysine derived from osmo-induced proteolysis increases under stress and stimulates the LKR/SDH expression channeling lysine to AASA, which can be used to form either Acetyl-CoA or pipecolate (Moulin et al., 2000, 2006). The accumulation of pipecolate reached remarkable concentrations in the E. coli expressing lysdh alone and, to a lesser extent, in the bacteria expressing both lysdh and aasadh, but not in the bacteria expressing aasadh alone. These results indicate that pipecolate is accumulated rather than being metabolized in the E. coli harboring lysdh and, thus, potentially acting as a genuine osmoprotectant. Bacteria containing lysdh–aasadh showed a reduced accumulation of pipecolate due to the conversion of AASA to AAA by AASADH (Figure 6e). Although no pipecolate accumulation was observed in E. coli expressing aasadh alone, these bacteria performed better in high-salt medium than the E. coli harboring the empty pET 28a plasmid. This suggests that salinity, in addition to its specific ionic and osmotic stress, may increase oxidative stress and the accumulation of toxic aldehydes that can be degraded by AASADH. This physiological AASADH function has already been demonstrated in plants and animals (Rodrigues et al., 2006; Brocker et al., 2010). However, in the case of the lysine-to-AASA pathways, as demonstrated in S. pomeroyi, the colocalization of lysdh and aasadh in an operon suggests that AASADH has a role in maintaining AASA at very low levels, whereas its cyclic isomer P6C can be used by P5CR to produce pipecolate (Figure 4e). Thus, the lysine-to-AASA pathways could be used to alleviate hyperosmotic stress by producing compatible solutes and preventing the accumulation of toxic aldehydes. In addition, AASADH could have a role in oxidizing aldehyde precursors to produce other compatible solutes, such as the conversion of betaine aldehyde to the osmoprotectant betaine (Brocker et al., 2010).
Acknowledgments
We thank Professor Paulo Mazzafera and Dr Alexandra Sawaya for their help with the mass spectrometry. This study was funded by FAPESP – 10/50114-4. Izabella Pena Neshich received PhD fellowship from FAPESP – 12/00235-5. Paulo Arruda is a CNPq productivity research fellow. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alexander DC, Anders CL, Lee L, Jensen SE. Pcd mutants of Streptomyces clavuligerus still produce cephamycin. J Bacteriol. 2007;189:5867–5874. doi: 10.1128/JB.00712-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- Arruda P, Kemper EL, Papes F, Leite A. Regulation of lysine catabolism in higher plants. Trends Plant Sci. 2000;5:324–330. doi: 10.1016/s1360-1385(00)01688-5. [DOI] [PubMed] [Google Scholar]
- Arruda P, Neshich IP. Nutritional rich and stress tolerant crops by saccharopine pathway manipulation. Food Energy Sec. 2012;2:1–7. [Google Scholar]
- Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- Brocker C, Lassen N, Estey T, Pappa A, Cantore M, Orlova V, et al. Aldehyde Dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J Biol Chem. 2010;285:18452–18463. doi: 10.1074/jbc.M109.077925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, et al. Sorghum bicolor's transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol. 2005;58:699–720. doi: 10.1007/s11103-005-7876-2. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMOL molecular graphics system. Delano Scientific: San Carlos, CA, USA; 2002. [Google Scholar]
- Delauney AJ, Hu CA, Kishor PB, Verma DP. Cloning of ornithine delta-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J Biol Chem. 1993;268:18673–18678. [PubMed] [Google Scholar]
- Deleu C, Cousrtaut M, Niogret M-F, Larher F. Three new osmotic stress-regulated cDNAs identified by differential display polymerase chain reaction in rapeseed leaf discs. Plant Cell Environ. 1999;22:979–988. [Google Scholar]
- Eduard AS, Jakobs C. Metabolism of lysine in α-aminoadipic semialdehyde dehydrogenase-deficient fibroblasts: Evidence for an alternative pathway of pipecolic acid formation. FEBS Lett. 2010;584:181–186. doi: 10.1016/j.febslet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- Feller A, Dubois E, Ramos F, Piérard A. Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol Cell Biol. 1994;14:6411–6418. doi: 10.1128/mcb.14.10.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Mukaihara M, Agematu H, Tsunekawa H. Biotransformation of L-lysine to L-pipecolic acid catalyzed by L-lysine 6-aminotransferase and pyrroline-5-carboxylate reductase. Biosci Biotechnol Biochem. 2002;66:622–627. doi: 10.1271/bbb.66.622. [DOI] [PubMed] [Google Scholar]
- Fujii T, Narita T, Agematu H, Agata N, Isshiki K. Characterization of L-lysine 6-aminotransferase and its structural gene from Flavobacterium lutescens IFO3084. J Biochem. 2000;128:391–397. doi: 10.1093/oxfordjournals.jbchem.a022766. [DOI] [PubMed] [Google Scholar]
- Fukutoku F, Yamada Y. Sources of Proline-nitrogen in water-stressed soybean (Glycine max L.) I. Protein metabolism and proline accumulation. Plant Cell Physiol. 1981;22:1397–1404. [Google Scholar]
- Gebhard S, Hümpel A, McLellan AD, Cook GM. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology. 2008;154:2786–2795. doi: 10.1099/mic.0.2008/018044-0. [DOI] [PubMed] [Google Scholar]
- Goas G, Goas M, Larher F. Formation de l'acide pipecolique chez Triglochin maritime. Can. J. Bot. 1975;54:1221–1227. [Google Scholar]
- Gouesbet G, Blanco C, Hamelin J, Bernard T. Osmotic adjustment in Brevibacterium ammoniagenes: pipecolic acid accumulation at elevated osmolalities. Microbiology. 1992;138:959–965. [Google Scholar]
- Gouesbet G, Jebbar M, Talibart R, Bernard T, Blanco C. Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiology. 1994;140:2415–2422. doi: 10.1099/13500872-140-9-2415. [DOI] [PubMed] [Google Scholar]
- Gratão PL, Polle A, Lea PJ, Azevedo RA. Making the life of heavy metal-stressed plants a little easier. Func Plant Biol. 2005;32:481–494. doi: 10.1071/FP05016. [DOI] [PubMed] [Google Scholar]
- Guerrero FD, Jones JT, Mullet JE. Turgor-responsive gene transcription and RNA levels increase rapidly when pea shoots are wilted. Sequence and expression of three inducible genes. Plant Mol Biol. 1990;15:11–26. doi: 10.1007/BF00017720. [DOI] [PubMed] [Google Scholar]
- Jefferies RL, Rudmik T, Dillon EM. Responses of halophytes to high salinities and low water potentials. Plant Physiol. 1979;64:989–994. doi: 10.1104/pp.64.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Karlsson JO, Ostwald K, Kåbjörn C, Andersson M. A method for protein assay in Laemmli buffer. Anal Biochem. 1994;219:144–146. doi: 10.1006/abio.1994.1243. [DOI] [PubMed] [Google Scholar]
- Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol. 2005;57:315–332. doi: 10.1007/s11103-004-7796-6. [DOI] [PubMed] [Google Scholar]
- Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misono H, Nagasaki S. Occurrence of L-lysine epsilon-dehydrogenase in Agrobacterium tumefaciens. J Bacteriol. 1982;150:398–401. doi: 10.1128/jb.150.1.398-401.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F, Bouchereau A. The lysine-ketoglutarate reductase-saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol Biochem. 2006;44:474–482. doi: 10.1016/j.plaphy.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Moulin M, Deleu C, Larher F. L-Lysine catabolism is osmo-regulated at the level of lysine-ketoglutarate reductase and saccharopine dehydrogenase in rapeseed leaf discs. Plant Physiol Biochem. 2000;38:577–585. doi: 10.1016/j.plaphy.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Papes F, Kemper EL, Cord-Neto G, Langone F, Arruda P. Lysine degradation through the saccharopine pathway in mammals: involvement of both bifunctional and monofunctional lysine-degrading enzymes in mouse. Biochem J. 1999;344:555–563. [PMC free article] [PubMed] [Google Scholar]
- Peisach J, Strecker HJ. The interconversion of glutamic acid and proline. V. The reduction of delta 1-pyrroline-5-carboxylic acid to proline. J Biol Chem. 1962;237:2255–2260. [PubMed] [Google Scholar]
- Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL. Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440. J Bacteriol. 2005;187:7500–7510. doi: 10.1128/JB.187.21.7500-7510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues SM, Andrade MO, Gomes AP, Damatta FM, Baracat-Pereira MC, Fontes EP. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J Exp Bot. 2006;57:1909–1918. doi: 10.1093/jxb/erj132. [DOI] [PubMed] [Google Scholar]
- Serrano GC, Silva Figueira TR, Kiyota E, Zanata N, Arruda P. Lysine degradation through the saccharopine pathway in bacteria: LKR and SDH in bacteria and its relationship to the plant and animal enzymes. FEBS Lett. 2012;586:905–911. doi: 10.1016/j.febslet.2012.02.023. [DOI] [PubMed] [Google Scholar]
- Stroeher VL, Boothe JG, Good AG. Molecular cloning and expression of a turgor-responsive gene in Brassica napus. Plant Mol Biol. 1995;27:541–551. doi: 10.1007/BF00019320. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal EA, Toone WM, Jones N, Morgan BA. Distinct roles for glutathione S-transferases in the oxidative stress response in Schizosaccharomyces pombe. J Biol Chem. 2002;277:35523–35531. doi: 10.1074/jbc.M111548200. [DOI] [PubMed] [Google Scholar]
- Yee L, Blanch HW. Recombinant trypsin production in high cell density fed-batch cultures of Escherichia coli. Biotechnol Bioeng. 1993;41:781–790. doi: 10.1002/bit.260410804. [DOI] [PubMed] [Google Scholar]
- Yoneda K, Fukuda J, Sakuraba H, Ohshima T. First crystal structure of L-lysine 6-dehydrogenase as an NAD-dependent amine dehydrogenase. J Biol Chem. 2010;285:8444–8453. doi: 10.1074/jbc.M109.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol. 2003;53:1893–1900. doi: 10.1099/ijs.0.02713-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.