Abstract
Background
Molecular programs employed by Mycobacterium tuberculosis (Mtb) for the establishment of non-replicating persistence (NRP) are poorly understood. In order to investigate mechanisms regulating entry into NRP, we asked how cell cycle regulation is linked to downstream adaptations that ultimately result in NRP. Based on previous reports and our recent studies, we reason that, in order to establish NRP, cells are halted in the cell cycle at the point of septum formation by coupled regulatory mechanisms.
Results
Using bioinformatic consensus modeling, we identified an alternative cell cycle regulatory element, SojMtb encoded by rv1708. SojMtb coordinates a regulatory mechanism involving cell cycle control at the point of septum formation and elicits the induction of the MazF6 toxin. MazF6 functions as an mRNA interferase leading to bacteriostasis that can be prevented by interaction with its cognate antitoxin, MazE6. Further, MazEF6 acts independently of other Maz family toxin:antitoxin pairs. Notably, soj Mtb and mazEF6 transcripts where identified at 20, 40 and 100 days post-infection in increasing abundance indicating a role in adaption during chronic infection.
Conclusions
Here we present the first evidence of a coupled regulatory system in which cell cycle regulation via SojMtb is linked to downstream adaptations that are facilitated through the activity of the MazEF6 TA pair.
Keywords: Mycobacterium tuberculosis, Cell cycle, MazF, Toxin:antitoxin, SojMtb
Background
A hallmark of Mycobacterium tuberculosis (Mtb) is its ability to establish and maintain a latent infection capable of resuming growth after long periods of time during which no clinical symptoms of disease are evident. This substantial reservoir of latent infections with the potential for relapse to active disease and transmission presents a significant obstacle for successful disease management. Compounding this issue is the limited number of treatment options for chronic tuberculosis infections. Accordingly, there is a considerable need for new anti-tuberculosis therapeutics capable of treating both chronic and acute infections. However, before new drugs with novel modes of action can be developed more information about the coordination of adaptive metabolism associated with latency needs to be obtained.
The transition from rapid growth to the non-replicating persistent state (NRP) that is characteristic of chronic tuberculosis infection involves the downshift of multiple biological processes and a remodeled and reduced metabolism [1]. Previous studies utilizing in vitro models of Mtb NRP indicate the transition into this state occurs at a specific point during the bacterial cell cycle [1-3]. While these experimental observations highlight the required coordination of cell cycle progression with cessation of growth and metabolic shutdown, the precise regulatory mechanisms coupling cell cycle regulation with downstream global metabolic remodeling remain undefined.
Regulation of the cell cycle at the point of septum formation is a key checkpoint in coordinating bacterial stress responses and for transition into NRP [4-6]. The SOS response, which prevents cell cycle progression in response to DNA damage and other stresses, is a classic example of cell division regulation under conditional stress [7-16]. Although proteins that regulate septum formation have been widely characterized in other bacterial species, proteins that carry out this function have not all been identified or thoroughly characterized in Mtb[17]. A previous study from our laboratory identified a septum site determining protein, Ssd, and showed that it is capable of regulating septum formation and inducing alternative metabolic pathways, demonstrating a relationship between the regulation of cell cycle progression and the induction of molecular programs associated with NRP [6].
There is growing evidence that genome encoded toxin:antitoxin (TA) loci have important roles in Mtb biology and they are increasingly being associated with stress responses. TA loci encode an unstable antitoxin immediately upstream of a toxin and these proteins form a complex with one another under optimal growth conditions. This interaction prevents the toxin from exerting its effects on the specific cellular process it targets [18,19]. Several environmental stresses are known to result in the selective degradation of the antitoxin and accumulation of free toxin [20-23]. Of the 88 putative TA systems encoded in the Mtb genome, many have been shown to be responsive to hypoxia, participate in growth transitions, contribute to virulence, and regulate growth in macrophages or in sputum [20,24-29]. Yet, their regulation, coordination and specific contributions to the many stressful states encountered by Mtb are not fully understood [21,30].
Because of the large number of genome encoded TA loci, and lack of information about their regulation and interaction with other members of TA loci, our objective was to identify a cell cycle regulatory component, that was part of an alternative response that included TA loci, and to asses the interaction of the TA loci components. In this study, we identify SojMtb encoded by rv1708 as a cell cycle regulatory protein that governs cell cycle progression at the point of septum formation and elicits a complex adaptive response that includes the MazEF6 TA loci of Mtb. The MazF6 toxin is capable of inducing bacteriostasis that can be inhibited by interaction with its cognate antitoxin, MazE6. The MazEF6 TA pair form a stable protein complex and MazF6 is not capable of interacting with non-cognate antitoxins. This is the first report of a coupled regulatory system consisting of a cell cycle regulator, SojMtb, and a TA locus, that are involved in a global adaptive response ultimately associated with the transition into the NRP state of Mtb.
Results
ORF rv1708 encodes a Soj-like protein in Mtb
Previously, we used transcriptional mapping to assign rv1708, originally annotated as a putative initiation inhibition protein, as a septum formation regulatory protein [4]. To improve the functional assignment of rv1708, a bioinformatics approach using a consensus sequence derived from alignments of proteins with an annotated function of septum inhibition was employed. This substantiated the functional assignment of rv1708 as encoding a septum regulatory protein because of shared similarity with the septum regulatory protein MinD. Septum inhibiting proteins, including MinD and Soj proteins, belong to the same family of P-Loop ATPases [31]. To narrow the functional assignment of Rv1708 we used the Phyre2 protein homology engine, which successfully revealed that Rv1708 shares greater sequence identity to Soj proteins (46% identity with 78% coverage; E value 2e-82) than to true MinD proteins (27% identity with 80% coverage; E value 6e-18). Alignments of the Rv1708 protein with Soj and MinD consensus models, Orthologous Matrix (OMA) groups 95838 and 78690 respectively, revealed that Rv1708 shares Soj-specific DNA-binding residues while lacking the hydrophobic C-terminal aliphatic helical extension that is involved in membrane association and is characteristic of MinD proteins [31,32] (Figure 1). Together, these analyses improve the functional annotation and indicate that rv1708 encodes an alternative Soj protein, referred to here as SojMtb.
Figure 1.

Multialignment of Rv1708 with Soj and MinD consensus models. Blue shading indicates conserved residues while red shading indicates residues involved in non-specific DNA binding by Soj proteins.
Overexpression of soj Mtb regulates cell cycle progression and acts prior to septum formation
To assess whether SojMtb is involved in regulation of cell cycle progression, we performed gene dosage experiments. Overexpression of soj Mtb caused a decrease in growth in Mycobacterium smegmatis and Mtb compared to the vector control strain (Figure 2). To visualize alterations in cellular morphology due to overexpression of soj Mtb , mycobacterial cells were imaged using electron microscopy (Figure 3). These studies revealed that overexpression of soj Mtb in M. smegmatis and Mtb produced similar elongated cell phenotypes with a predominant ultra-structural morphology absent of concentric rings indicative of septa. This elongated cell phenotype is consistent with previous studies performed in M. smegmatis[33]. The absence of septa is characteristic of regulation of FtsZ polymerization prior to Z-ring formation [4,5,16,34,35]. Together, these data indicate that SojMtb reduces the frequency of septum formation and governs cell cycle progression prior to septum formation.
Figure 2.
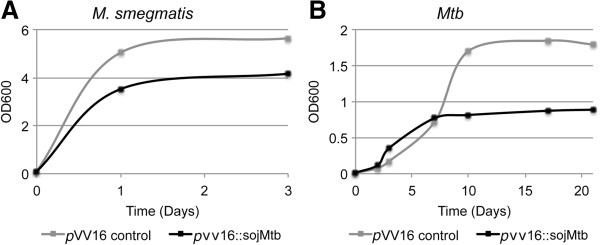
Overexpression ofsojMtbslows growth of mycobacteria. Bacterial growth curves of (A)M. smegmatis containing pVV16::sojMtb or pVV16 alone or (B)Mtb containing pVV16::sojMtb or pVV16 alone. Data include technical replicates and are representative of three independent experiments.
Figure 3.
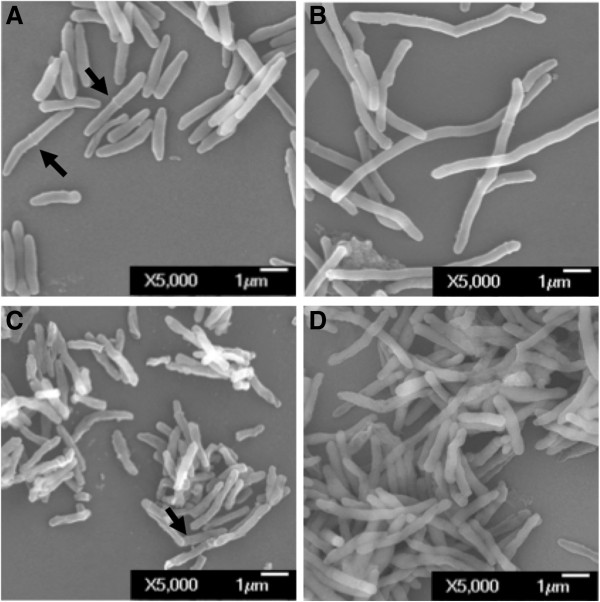
Morphological characterization of mycobacteria overexpressingsojMtb. (A-B)M. smegmatis control and sojMtb merodiploid. (C-D)Mtb control and sojMtb merodiploid.
MazF6 toxin is elicited by SojMtb
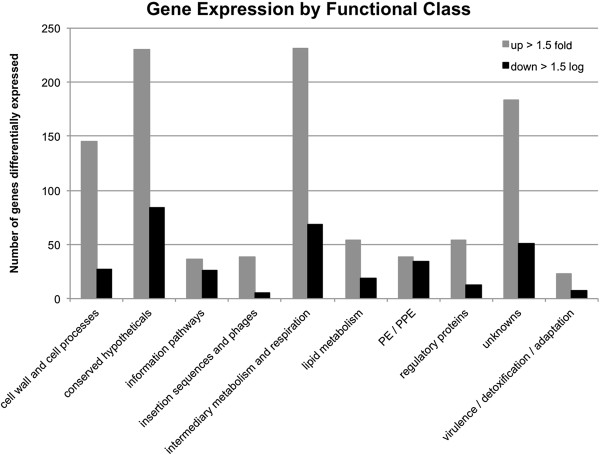
To assess the involvement of SojMtb in eliciting alternative adaptive responses, the global transcriptional response of Mtb upon overexpression of soj Mtb was investigated through whole genome DNA microarray analysis. A total of 1,368 genes from all functional categories displayed a 1.5 fold or greater change in expression (p values < 0.05) in the Mtb merodiploid strain compared to the vector alone control strain (Figure 4). The microarray data analysis revealed that SojMtb is part of a large alternative stress response based on the induction of alternative sigma factors sigH, sigG, sigL, sigM, sigI in response to overexpression of soj Mtb . The pleotropic nature of this transcriptional response is likely the result of the induction of these alternative sigma factors, several of which have been shown to regulate stress response pathways [36]. Notably, rv1991c encoding the MazF6 toxin of the MazEF6 TA loci showed significant change in expression.
Figure 4.
The global transcriptional response ofMtbupon overexpression ofsojMtborganized by functional class. A total of 1,368 genes from all functional categories displaying a 1.5 fold or greater change in expression (p values < 0.05) in the Mtb merodiploid strain compared to the vector alone strain.
The MazF6 toxin is the most well characterized out of nine predicted maz-family TA loci encoded in the Mtb genome [20]. Molecular studies have demonstrated that the MazF6 is an endoribonuclease that cleaves ACA mRNA sequences, and that this activity is inhibited by interaction with the MazE6 antitoxin [37]. MazF6 is thought to induce rapid changes in the transcriptome due to its endoribonuclease activity, allowing coupling of cell cycle inhibition by SojMtb and the transcriptional remodeling associated with the transition into an alternative state such as NRP.
We compared the soj Mtb response with previously described programs associated with NRP in order to determine if there was overlap in the transcriptional responses. Overlap was determined by taking into account the number of genes in the given transcriptional response in relation to the number of genes in the genome (4,124 as indicated from the Broad Institute) to determine the number of genes that would be shared only by chance, as described previously [38]. There was no significant degree of overlap between the up-regulated genes of the soj Mtb response and the dos-response or EHR (Table 1), and in fact, the dos-response was actively down-regulated (Table 2). However, there was significant overlap between the soj Mtb response and the “non-culturable” response, with 14 up-regulated genes in common, the mazEF6 transcript among them.
Table 1.
Comparisons of genes up-regulated upon overexpression of soj Mtb to previously described responses associated with NRP
|
Response associated with NRP |
Soj
Mtb
response (923 up-regulated genes, over 2-log) |
|
|---|---|---|
| Overlapping genes | Overlapping by chance | |
| Dos1 (48 genes) |
4 |
11 |
| Enduring hypoxic response2 (230 genes) |
44 |
52 |
| Nonculturable3 (51 genes significantly up) | 14 | 11 |
Table 2.
Comparisons of genes down-regulated upon overexpression of soj Mtb to previously described responses associated with NRP
|
Response associated with NRP |
Soj
Mtb
response (325 down-regulated genes, over 2-log) |
|
|---|---|---|
| Overlapping genes | Overlapping by chance | |
| Dos1 (48 genes) |
21 |
4 |
| Enduring hypoxic response2 (230 genes) |
15 |
18 |
| Nonculturable3 (51 genes significantly up) | 4 | 4 |
MazE6 and MazF6 functionally and physically interact
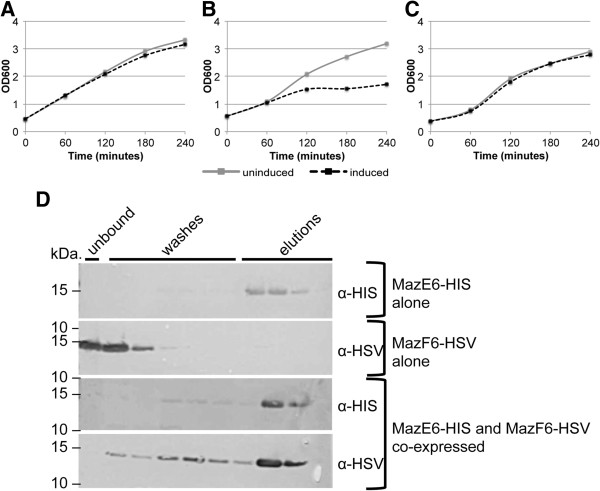
To better understand the functional role of Maz-family TA pairs, we sought to characterize the functionality and interaction of MazE6 and MazF6. When expressed in vivo, the MazF6 toxin induces bacteriostasis, while expression of MazE6 causes no stalling of bacterial growth (Figures 4B and 5A, respectively). Importantly, this bacteriostatic phenotype can be rescued when the toxin and antitoxin are co-expressed thus demonstrating a functional interaction in vivo (Figure 5C). To show a physical interaction between these two recombinant proteins, co-purification was performed and protein identity was confirmed via Western blot and mass spectrometry. When both antitoxin and toxin are produced individually and subject to metal affinity chromatography, MazE6-HIS is present in the elution fraction, while MazF6-HSV is present in the unbound flow and wash fraction, indicating that it is not retained in the affinity column (Figure 5D). In contrast, when co-expressed, MazF6-HSV elutes primarily with MazE6-HIS indicating that they physically interact in vivo and that the bacteriostatic phenotype associated with overproduction of MazF6 can be prevented by its interaction with its cognate antitoxin, MazE6 (Figure 5). Western blot analysis was confirmed by in-gel trypsin digestion of the gel fragment corresponding to the recombinant MazEF6 complex and identification by LTQ mass spectrometry. MazEF6 were among the top hits returned when searched against the NCBInr database for eubacteria. MazE6 was identified with 70% coverage with 12 non-duplicate peptides, and MazF6 was identified with 95% coverage and 8 non-duplicate peptides.
Figure 5.
MazE6-HIS antitoxin and MazF6-HSV toxin functionally and physically interactin vivo.A) Growth of E. coli BL21 (DE3) pLysE containing MazE6-HIS alone, B) MazF6-HSV alone, or C) MazE6-HIS co-produced with MazF6-HSV. Growth arrest due to MazF6-HSV production is recued by co-production of MazE6-HIS antitoxin. D) MazE6-HIS is present in the elution fraction, while MazF6-HSV is present in the unbound flow and wash fraction when each is produced alone. When co-produced, MazF6-HSV elutes primarily with MazE6-HIS, indicating that they physically interact. Data are representative of multiple independent experiments.
To determine if there is any degree of functional redundancy among the numerous MazEF loci in Mtb, we assessed the interactions between MazF6 and non-cognate MazE antitoxins. Functional redundancy was assessed by the ability of non-cognate MazE antitoxins to prevent bacteriostasis induced by MazF6. Growth arrest due to MazF6-HSV production was not rescued by co-production of non-cognate MazE-HIS antitoxins (Table 3). The MazF8 antitoxin encoded by rv2274a was not tested because it is absent in some clinical strains of Mtb[20,39]. Co-purification was also performed and with all pairs tested, MazF6-HSV was only able to physically interact with its cognate antitoxin, MazE6 (Figures 5D and 6). Our data confirm that the MazF6 toxin does not interact with non-cognate MazE antitoxins in vivo indicating that there may be little functional redundancy across MazEF pairs.
Table 3.
Growth kinetics in cultures co-expressing the mazF6 toxin with cognate and non-cognate antitoxins
| MazE | % Reduction in growth | |
|---|---|---|
| MazF6 expressed with non-cognate antitoxins |
1 |
17 |
| 2 |
30 |
|
| 3 |
11 |
|
| 4 |
17 |
|
| 5 |
19 |
|
| 7 |
33 |
|
| 9 |
24 |
|
| MazF6 alone |
- |
46 |
| MazE6 alone |
- |
5 |
| MazF6 | 6 | 4 |
Growth reduction associated with overexpression of the mazF6 toxin can be rescued by co-expression of its cognate antitoxin. Full rescue of the growth defect is not observed when co-expressed with non-cognate antitoxins.
Figure 6.
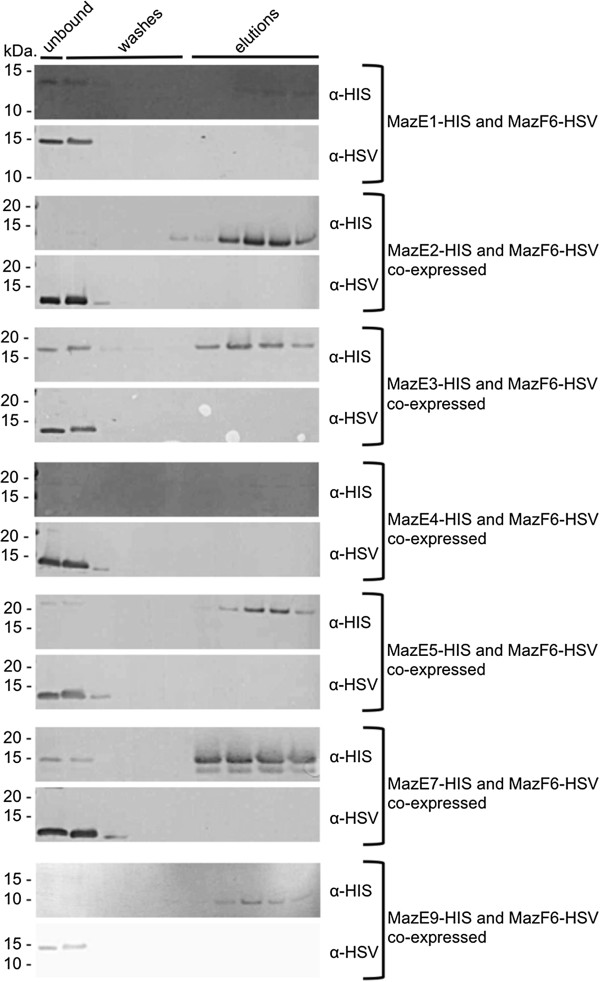
MazF6-HSV toxin does not physically interact with non-cognate MazE-HIS antitoxinsin vivo. With all pairs tested, MazE-HIS antitoxins are present in the elution fractions, while MazF6-HSV is present in the unbound flow and wash fractions only, indicating that MazF6-HSV is only able to physically interact with its cognate antitoxin, MazE6. From top to bottom, antitoxins tested are: MazE1 (Rv0456b), MazE2 (Rv0660c), MazE3 (Rv1103c), MazE4 (Rv1494), MazE5 (Rv1943c), MazE7 (Rv2063), MazE9 (Rv2801a).
soj Mtb and mazEF6 are expressed in bacteria in the lungs and spleen of Mtb infected mice
Adaptations resulting in a NRP state have been associated with the execution of complex and complementary molecular programs designed to allow the cell to survive the stresses experienced within the host [40]. These include programs to slow bacterial growth, induce alternative metabolic pathways, and to withstand acid, hypoxic, nutrient and additional stresses. TA loci have been associated with stress responses in bacteria and are becoming the focus of increasing efforts to elucidate their contributions to alternative states in vivo[18].
In order to determine if SojMtb and MazEF6 play a role during infection, we assessed their transcriptional activity during the infectious processes using the established mouse model of infection [41]. Quantitative-PCR was used to assess soj Mtb and mazEF6 abundance in cDNA obtained from in vitro Mid-Log phase cultures, and the lungs and spleen at 20, 40 and 100 days post-infection (Figure 7AB). The analyses revealed an overall difference in expression patterns and overall abundance of soj Mtb and the mazEF6 loci as compared to in vitro grown bacteria. soj Mtb was abundant at day 20, 40, and 100. The transcriptional response in the lungs was characterized by a significant difference in the transcriptional abundance of mazE6 and mazF6 at day 20, while this difference in the abundance of mazE6 and mazF6 was not noted at days 40 and 100 (Figure 7A). Similarly to the transcriptional pattern observed at day 20 in the lungs, in the spleen soj Mtb was abundant at day 20, 40 and 100, and there was a two log or greater difference in the abundance between mazE6 and mazF6 throughout the course of infection (Figure 7B). The observed transcriptional abundances observed between mazE6 and mazF6 is consistent with observations of TA loci in other systems [42]. Specifically, only the toxin component of several TA loci were identified in Mtb in a human macrophage model of late stage infection [43]. The transcriptional activity of soj Mtb , and mazE6 and mazF6 in bacteria obtained from infected tissues indicate that MazF6 is important throughout the course of infection, and is uniquely temporally functional in facilitating adaptations to the alternative environments encountered during infection.
Figure 7.
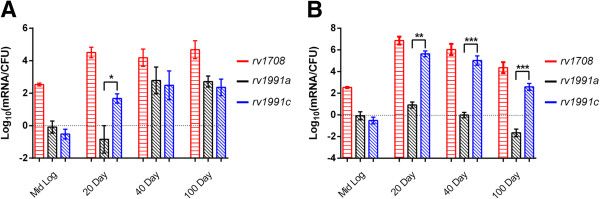
sojMtbandmazEF6are expressed during infection. Quantative-PCR carried out on cDNA generated from RNA pools obtained from Mid-Log phase Mtb and mouse (A) lungs and (B) spleen at day 20, 40, and 100 post-infection. sojMtb(rv1708) transcripts increased in abundance throughout the course of infection in comparison to Mid-Log phase Mtb. mazEF6 (rv1991a, rv1991c) transcripts were identified at day 20, 40 and 100 infection in the lungs and spleen.
Discussion
The establishment of the NRP state requires the coordination of several important cellular processes. These include the halting of cell cycle progression at the point of septum formation, induction of alternative metabolic pathways, and coordination of multiple regulatory mechanisms, including TA loci, in modulating this adaptive response. Previous studies have observed that during macrophage infection, as well as under in vitro models of NRP, the bacteria are observed as multinucleoidal smooth filaments, indicating the transition into NRP occurs after chromosomal replication but prior to formation of the Z-ring septal precursor [1-3]. Although proteins that carry out this function in Mtb have not all been identified or characterized, we predicted that they must be present in the Mtb genome, as this is a critical checkpoint in the cell cycle. Homology searches for cell division regulators known to act by inhibiting septum formation identified rv1708 (soj Mtb ). Septum inhibiting proteins including MinD, ParA and Soj, show extensive similarity while participating in a variety of different cellular processes [31]. Homology models and local sequence alignments confirm that rv1708 encodes a Soj-like, rather than MinD-like protein, as it contains the non-specific DNA-binding residues highly conserved in Soj proteins but lacking in MinD proteins [32]. While Soj and chromosomal ParA proteins are considered to be equivalent in respect to their roles in chromosome segregation, Soj proteins play an important role in the regulation of sporulation-specific gene expression and entry into stationary phase [44]. Previously, studies conducted in M. smegmatis reported that rv1708 encodes a ParA homolog [33]. However, based on genomic context, we do not believe that the protein encoded by rv1708 contributes significantly to chromosomal segregation as ParA.
Together, our bioinformatic analysis and morphological studies indicate a role in regulating cell division and demonstrate that SojMtb performs a role in Mtb that is analogous to Soj proteins described in other bacteria. SojMtb regulates the expression of a subset of genes that act to establish a state of low metabolic activity and reduced growth rate along with increasing adaptive functions required for entry into NRP. Transcriptional analyses of Mtb under a variety of conditions have uncovered several molecular programs that have been extensively linked to the NRP state of Mtb including the dos (dormancy) response, the enduring hypoxic response (EHR) and the “non-culturable” transitional state [45-48]. Our previous studies have demonstrated that septum site determining protein Ssd causes the induction of alternative metabolic pathways, including the dos regulon, further illustrating an intimate coordination of cell cycle progression with induction of molecular programs associated with NRP [6,45].
Upon overexpression of soj Mtb , alternative sigma factors sigH, sigG, sigL, sigM, sigI, and sigB are up-regulated. The pleotropic nature of this transcriptional response is likely the result of the induction of these alternative sigma factors, several of which have been shown to regulate stress response pathways [36]. In addition to the up-regulation of alternative sigma factors, the greatest increase in gene expression is seen for rv1991c, encoding the MazF6 toxin. A 2009 study characterized the transition to NRP by evaluating the “non-culturable” phenotype that has been associated with dormancy in Mtb[47]. Notably, mazF6 was significantly up-regulated in this physiological model. In fact, there is significant overlap in the differential expression patterns between the “non-culturable” response and the response observed upon overexpression of soj Mtb . There was no significant degree of overlap between the up-regulated genes of the soj Mtb response and the dos response or EHR, and in fact, the dos response was actively down-regulated. The dos response contributes to the preparation of Mtb for NRP, while the EHR contributes to the maintenance of NRP [46]. Although the dos response has been shown to aid in the adaptation to low oxygen state associated with NRP, mutants in the response have shown varying degrees of phenotypes and suggest that it does not play as prominent a role in NRP as once thought [38]. This further supports the role of the soj Mtb response in preparing the bacterium for alternative conditions in a manner that is distinct from the preparation elicited by the dos response. Therefore, we posit that MazF6 functions to facilitate the transition into NRP. Likely, the endoribonuclease activity of MazF6 is utilized to erase the existing transcriptome to free cellular machinery to produce transcripts needed for successful entry into NRP.
Currently, there are nine predicted maz-family TA loci encoded in the Mtb genome [20]. Despite a number of reports, the information about the interaction of cognate and non-cognate Maz-family TA pairs is limited [49]. In this study we have demonstrated that the MazF6 toxin is capable of inducing bacteriostasis and that this state can be inhibited by interaction with its cognate antitoxin, MazE6. In addition, we have confirmed a physical interaction between these two proteins by purifying the proteins in complex from live cells. We have also shown that the interaction is specific and that MazF6 is not capable of interacting with non-cognate antitoxins. This suggests that although there are a high number of TA loci in Mtb, cross-talk via interaction is unlikely to occur, at least among this family of TA pairs. The identification of both soj Mtb and mazEF6 transcripts in bacteria isolated from the lungs and spleen of infected mice further supports the role of soj Mtb and this toxin in facilitating the transition to alternative host environments through a coupled regulatory mechanism. The unique differential temporal profile of transcriptional abundance of the mazEF6 polycistron throughout the host infection is concordant with previous observations. Post-transcriptional regulation of TA loci that are co-transcribed is a complex dynamic phenomenon that results in the accumulation of the toxin component of the bicistronic mRNA during times of environmental stress. A study of Mtb during human macrophage infection identified only the toxin component of several TA orthologs during late stage infection [43]. Similarly, a study in E. coli substantiated post-transcriptional modification of endogenous mazEF mRNA in the presence of RelE toxin resulting in the accumulation of mazF toxin mRNA [42]. Together with previous reports, our data indicate that regulation of TA loci is not strictly occurring at the post-translational level but also at the post-transcriptional level.
Conclusions
All environmental conditions or cellular processes that induce the accumulation of the toxin component of TA loci or the specific phenotypes that results have not been fully elucidated. The identification of SojMtb, whose overproduction elicits the induction of this particular toxin loci, links the halting of cell division prior to the point of septum formation to molecular programs associated with alternative responses. MazF6 is strongly up-regulated upon overproduction of SojMtb, suggesting that the regulation of cell division and its eventual halting is facilitated through the action of this toxin. This is the first report of a coupled regulatory system that links cell cycle regulation by SojMtb molecular programs associated with adaptation to stress in Mtb via the activity of MazEF6 TA loci. Importantly, this information provides a foundation for further analysis of how TA loci are coordinated with regards to other essential cellular processes and to define their contribution to the establishment of the NRP state of Mtb.
Methods
Bioinformatic analysis
To identify putative cell division regulators in Mtb, consensus models were derived from septum formation inhibitors from OMA groups 78690, 63437, 84083, 90270, and 245137–40 from a variety of bacterial species using a MAFFT local multialignment tool. Resulting consensus sequences were used to BLAST the Mtb H37Rv genome to identity mycobacterial ORFs that encode septum regulatory proteins. BLAST searches were optimized for percent identity and score. Top hits were analyzed by the Phyre2 server to confirm or refute identification as putative septum formation inhibitors.
Generation of recombinant mycobacterial strains
For all mycobacteria experiments, Mtb strain H37Rv (ATCC 27294) and M. smegmatis mc2155 was cultured at 37°C with shaking in Middlebrook 7H9 liquid medium containing 0.4% glycerol, 10% OADC, and 0.05% Tween80 or on Middlebrook 7H10 agar containing 0.4% glycerol and OADC containing appropriate antibiotics as noted. For generation of recombinant mycobacterial strain overexpressing soj Mtb , the rv1708 gene was PCR amplified from Mtb H37Rv genomic DNA using AccuPrimePfx DNA polymerase (Invitrogen) and cloned into the NdeI and HindIII of the pVV16 constitutive, extrachromosomal mycobacterial vector, which is a derivative of pMV261 [50,51] (Additional file 1). DNA constructs were transformed into One Shot® TOP10 chemically competent E. coli (Invitrogen) and selected by growth on LB agar containing 50 μg ml-1 kanamycin. Sequence confirmed plasmid DNA was transformed into electrocompetent M. smegmatis or Mtb and selection was carried out using kanamycin at 25 μg ml-1.
Microscopy and ultrastructural analysis by SEM
Recombinant M. smegmatis strains containing the bacterial shuttle vector pVV16 as a control and soj Mtb ::pVV16 were grown to an O.D.600nm of 1.0 and collected by centrifugation. Cells were washed three times in PBS, pH 7.4, and fixed with 2.5% glutaraldehyde in Buffer A (0.1 M potassium phosphate (pH 7.4), 1 mM CaCl2 and 1 mM MgCl2) at 4°C for 48 hrs. The fixed bacterial cells were collected by centrifugation, washed three times in Buffer A and treated with 1% OsO4 in Buffer A for 30 minutes at 4°C. Again, cells were washed three times with Buffer A. and prepared for SEM with a graded series of ethanol treatments (20-100%). Ultrastructural examination was performed using a JOEL JEM -100CX electron microscope. Recombinant Mtb strains containing the bacterial shuttle vector pVV16 as a control and pvv16::soj Mtb were prepared as described above. However, the pvv16::soj Mtb culture did not reach an O.D.600nm of 1.0 and was prepared once the growth had plateaued.
Isolation of total RNA for microarray analysis, microarray processing, and data analysis
At Mid-Log growth, Mtb cells were harvested by centrifugation and resuspended in TRIzol (Invitrogen). Cells were lysed by bead beating for one-minute increments for three minutes total, cooling the sample on ice between each step. RNA was separated from other cellular products by the addition of chloroform and centrifugation, according to the TRIzol protocol. The upper aqueous layer was combined with an equal volume of 70% ethanol, and the entire volume was applied to an RNeasy Mini kit (Qiagen) column for RNA cleanup.
The Mtb whole genome microarrays were obtained through the TB Research Materials and Vaccine Testing Contract at Colorado State University. Slides were post-processed using succinic anhydride as described previously [4]. Approximately 8 μg of total RNA was converted to cDNA in the presence of either Cy5- or Cy3-labeled nucleotides as previously described [4,6]. Hybridization was performed at 42°C for 12 hr. Slides were scanned using an Axon Genepix scanner. The final microarray data set (Additional file 2) resulted from combining independent biological replicates. Data reduction and global normalization was performed on the raw fluorescent intensities. The normalized intensity values of treated and control cultures were used to generate ratio and log2 expression values for each gene.
MazF6 interaction analysis
For the Maz6 TA loci interaction studies, mazE and mazF gene fragments were PCR amplified from Mtb H37Rv genomic DNA using Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England BioLabs Inc.). Generally, mazE antitoxins were cloned in the BamHI and XhoI restriction sites of pET28a (Novagen), and mazF6 toxin was cloned into the SphI and NotI restriction sites of pETcoco2 (Novagen) (Additional file 1). DNA constructs were transformed into One Shot® TOP10 chemically competent E. coli (Invitrogen) or Z-Competent E. Coli - Strain DH5α (Zymo Research) and transformants selected by growth on LB agar containing 50 μg ml-1 kanamycin for pET28a and 50 μg ml-1 ampicillin for pETcoco2. Sequence confirmed plasmid DNA was transformed into One Shot® BL21(DE3) pLysE Chemically Competent E. coli (Invitrogen) and selection was carried out overnight by growth in LB broth supplemented with 0.2% glucose, 34 μg ml-1 chloramphenicol, and containing 50 μg ml-1 kanamycin for pET28a selection or 50 μg ml-1 ampicillin for pETcoco2 selection, or both in cases of co-transformation. Glucose was added to the media to maintain low basal expression from the pET vectors as previously described [52]. The overnight outgrowths were used for recombinant protein induction. Briefly, cultures were diluted back 1:50 in fresh media containing antibiotics without glucose. When mazF6::pETcoco2 construct was used, diluted cultures were grown for one hour with shaking at 37°C and L-arabinose was added to a final concentration of 0.01% to amplify plasmid copy number prior to protein induction. Once cultures reached Mid-Log, protein production was induced using 1 mM final concentration of isopropyl-beta-D-thiogalactopyranoside (IPTG)(Fermentas). Cultures were incubated for four hours with shaking at 37°C, with timepoints being taken every hour to monitor growth by O.D.600nm. After four hours, bacterial cell pellets were collected by centrifugation and stored at −80°C.
Purification of MazE-HIS in complex with MazF6-HSV was carried out under native conditions to maintain protein-protein interactions. Cleared lysates of bacterial cell pellets were obtained using BugBuster® including Benzonase® (Novagen) according to manufacturer’s instructions. Generally, cleared lysate was combined with one-fourth the volume of Ni-NTA His-Bind® Resin and mixed gently at 4°C for 1 hour before packing into a column. The column was washed with wash buffer 1 (500 mM NaCl, 20 mM Tris–HCl, 60 mM imidazole, pH 9.0), followed by wash buffer 2 (250 mM NaCl, 20 mM Tris–HCl, 100 mM imidazole, pH 9.0). Recombinant protein or protein complexes were eluted with elution buffer (250 mM NaCl, 20 mM Tris–HCl, 300 mM imidazole, pH 9.0). All fractions were collected and samples were resolved using pre-cast 12% NuPAGE® gels (Invitrogen), followed by transfer to 0.2 micron nitrocellulose membrane (BioRad) for Western blotting. Membranes were blocked in 4% BSA in TBST, incubated with primary Penta-His antibody (Qiagen) or anti-HSV antibody (Novagen), both diluted 1:10,000, followed by goat anti-mouse-alkaline phosphatase (Sigma) secondary antibody diluted 1:10,000. Membranes were developed using SIGMAFAST™ BCIP®/NBT tablet (Sigma) according to manufacturer’s instructions.
Mouse infection, bacterial quantification, and RNA isolation
All use of vertebrate animals took place at Colorado State University, which is AAALAC approved and has an OLAW number of A3572-01. Care was provided by veterinarians in the Laboratory Animal Resources of Colorado State University under the supervision of the University Veterinarian. Six week-old female C57BL/6 mice (Jackson Laboratories) were infected via low dose aerosol with Mtb H37Rv in a Glas-Col Inhalation Exposure System (Glas-Col, Inc.). Exposure was conducted by aerosolizing approximately 2×106 cfuml-1 in a volume of 5 cubic feet over a period of 30 minutes, followed by a 20 minute period of cloud decay. Infected mice were housed in sterile micro-isolator cages in the ABSL-3 facility at Colorado State University and provided water and food ad libitum and monitored for morbidity. Five animals per time point were sacrificed at 20, 40, and 100 days post-infection by CO2 inhalation and lungs and spleens were harvested. Organs were cut into halves, and half of each organ was homogenized in 2 mL sterile saline using an Omni Tissue homogenizer (Omni International). Bacterial burden in the lungs and spleen at each time point were assessed by plating 10-fold serial dilutions of organ homogenates on Middlebrook 7H11 agar containing 10% OADC and carbenicillin (Sigma) and cycloheximide (Sigma) at 50 μg ml-1 and 10 μg ml-1, respectively. Plates were incubated at 37°C for three weeks.
The remaining half of each lung and spleen were homogenized in 5 ml TRIzol by bead beating. Total RNA was isolated by organic phase separation. RNA samples were treated with DNAse (Fermentas) for 1 hour at 37°C, re-extracted in phenol/chloroform/isoamyl alcohol (25:24:1)(Sigma), and precipitated with ammonium acetate.
Reverse transcription and quantitative-PCR
cDNA synthesis from total RNA from the lungs and spleen of infected mice was carried out using a gene-specific reverse primer cocktail with Transcriptor First Strand cDNA Synthesis Kit (Roche). The reverse primer cocktail contained reverse primers specific for mazE6, mazF6, and soj Mtb in addition to random hexamers (Additional file 3). Reactions contained 1 μg total RNA, 2.5 μM gene-specific reverse primers, and 60 μM random hexamers. cDNA synthesis were carried out according to manufacturer instructions along with no reverse transcriptase control and no template control reactions. Quantative-PCR was carried out using 50 ng of cDNA generated from RNA isolated from Mid-Log phase Mtb, and RNA isolated from lungs and spleens of mice at Day 20, 40,and 100 post-infection. Calibration curves were generated using respective mazE6, mazF6, and soj Mtb specific dsDNA amplicons generated from PCR using GoTaq®Green Master Mix (Promega). Calibration curves of respective dsDNA amplicons ranged from 1 ng to 1-8 ng per reaction in 10-fold serial dilutions. Modeled linear fits and primer efficiencies were used to determine concentration of respective gene. Amplicons in experimental samples were converted to total mRNA copy number per mL. Each sample was normalized by taking total mRNA copy number per mL and dividing by the associated cfu mL-1 recovered from each biological sample. No reverse transcriptase and no template controls confirmed no gDNA contamination or non-specific amplification.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MVR carried out the bacterial growth analysis, recombinant protein studies, interaction studies, animal studies, and isolation of total RNA and bacterial transcripts from infected tissues, CCD performed the quantitative-PCR studies from infected tissues, KE carried out the microarrays and morphological studies, and RC performed the bioinformatics. RAS designed the studies, and coordination of the manuscript. All authors participated in drafting, and editing the final manuscript. All authors have read and approved the manuscript.
Supplementary Material
Differential gene expression of Mtb upon overexpression of soj Mtb .
Primer sequences used for plasmid construction.
Primer sequences used for end-point PCR.
Contributor Information
Melissa V Ramirez, Email: vtmelissav@gmail.com.
Clinton C Dawson, Email: clinton.dawson@colostate.edu.
Rebecca Crew, Email: rebecca.crew@colostate.edu.
Kathleen England, Email: englandk@stanford.edu.
Richard A Slayden, Email: richard.slayden@colostate.edu.
Acknowledgments
We acknowledge Laurel Respicio for her work that provided the background for this study, Dr. Susan Knudson for her help with mouse infections, and Dr. Darragh G. Heaslip for assistance with mass spectrometry analysis. We also thank Drs. Peter A. Bowerman, Analise Z. Reeves, and Kevin C. Crosby for their critical reading of the manuscript and helpful feedback. This work was supported by an award from the Research Council of the College of Veterinary Medicine and Biomedical Science at Colorado State University and R01 AI055298 (RAS).
References
- Wayne LG. Dormancy of mycobacterium tuberculosis and latency of disease. Eur J Clin Microbiol Infect Dis. 1994;13(11):908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188(5):1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Knudson DL, Belisle JT. Identification of cell cycle regulators in mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology. 2006;152(Pt 6):1789–1797. doi: 10.1099/mic.0.28762-0. [DOI] [PubMed] [Google Scholar]
- Respicio L, Nair PA, Huang Q, Anil B, Tracz S, Truglio JJ, Kisker C, Raleigh DP, Ojima I, Knudson DL. et al. Characterizing septum inhibition in mycobacterium tuberculosis for novel drug discovery. Tuberculosis (Edinb) 2008;88(5):420–429. doi: 10.1016/j.tube.2008.03.001. [DOI] [PubMed] [Google Scholar]
- England K, Crew R, Slayden RA. Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol. 2011;11:79. doi: 10.1186/1471-2180-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli Is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179(6):1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002;99(26):17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y, Moriya S, Ogasawara N. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol Microbiol. 2003;47(4):1113–1122. doi: 10.1046/j.1365-2958.2003.03360.x. [DOI] [PubMed] [Google Scholar]
- Savijoki K, Ingmer H, Frees D, Vogensen FK, Palva A, Varmanen P. Heat and DNA damage induction of the LexA-like regulator HdiR from lactococcus lactis is mediated by RecA and ClpP. Mol Microbiol. 2003;50(2):609–621. doi: 10.1046/j.1365-2958.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science. 2004;305(5690):1629–1631. doi: 10.1126/science.1101630. [DOI] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Seed PC, Hultgren SJ. Filamentation by escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci U S A. 2006;103(52):19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. Interference of mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62(1):132–147. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- Cirz RT, Jones MB, Gingles NA, Minogue TD, Jarrahi B, Peterson SN, Romesberg FE. Complete and SOS-mediated response of staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. 2007;189(2):531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice SS, Hunstad DA, Cegelski L, Hultgren SJ. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol. 2008;6(2):162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- Slayden RA, Belisle JT. Morphological features and signature gene response elicited by inactivation of FtsI in mycobacterium tuberculosis. J Antimicrob Chemother. 2009;63(3):451–457. doi: 10.1093/jac/dkn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayden RA, Jackson M, Zucker J, Ramirez MV, Dawson CC, Crew R, Sampson NS, Thomas ST, Jamshidi N, Sisk P. et al. Updating and curating metabolic pathways of TB. Tuberculosis. 2013;93(1):47–59. doi: 10.1016/j.tube.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 2009;5(12):e1000767. doi: 10.1371/journal.pgen.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of mycobacterium tuberculosis persisters. MBio. 2011;2(3):e00100–00111. doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- Albrethsen J, Agner J, Piersma SR, Hojrup P, Pham TV, Weldingh K, Jimenez CR, Andersen P, Rosenkrands I. Proteomic profiling of mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol Cell Proteomics. 2013;12(5):1180–1191. doi: 10.1074/mcp.M112.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Sisk P, Stolte C, Weiner B, Koehrsen M, Wymore F, Reddy TB, Zucker JD, Engels R, Gellesch M. et al. TB database 2010: overview and update. Tuberculosis (Edinb) 2010;90(4):225–235. doi: 10.1016/j.tube.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6(7):e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TB, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, Gellesch M, Hubble J, Jen D, Jin H. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37(Database issue):D499–508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta NK, Mehra S, Didier PJ, Roy CJ, Doyle LA, Alvarez X, Ratterree M, Be NA, Lamichhane G, Jain SK. et al. Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis. 2010;201(11):1743–1752. doi: 10.1086/652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste DJ, Espasa M, Bonde B, Kierzek AM, Stewart GR, McFadden J. The genetic requirements for fast and slow growth in mycobacteria. PLoS One. 2009;4(4):e5349. doi: 10.1371/journal.pone.0005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrid AM, Rustad TR, Cangelosi GA, Sherman DR. Characterization of a Clp protease gene regulator and the reaeration response in mycobacterium tuberculosis. PLoS One. 2010;5(7):e11622. doi: 10.1371/journal.pone.0011622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, Woychik NA. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal a site. Proc Natl Acad Sci U S A. 2013;110(21):8501–8506. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. Embo J. 2005;24(2):270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney E, Madiraju M, Rajagopalan M. Overproduction and localization of mycobacterium tuberculosis ParA and ParB proteins. Tuberculosis. 2009;89(Suppl 1):S65–69. doi: 10.1016/S1472-9792(09)70015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Kirikae F, Kirikae T, Pepe A, Amin A, Respicio L, Slayden RA, Tonge PJ, Ojima I. Targeting FtsZ for antituberculosis drug discovery: noncytotoxic taxanes as novel antituberculosis agents. J Med Chem. 2006;49(2):463–466. doi: 10.1021/jm050920y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Levin PA. Assembly dynamics of the bacterial cell division protein FTSZ: poised at the edge of stability. Annu Rev Microbiol. 2003;57:125–154. doi: 10.1146/annurev.micro.57.012903.074300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva P, Misra R, Tyagi AK, Singh Y. The sigma factors of mycobacterium tuberculosis: regulation of the regulators. Febs J. 2010;277(3):605–626. doi: 10.1111/j.1742-4658.2009.07479.x. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from mycobacterium tuberculosis. J Biol Chem. 2006;281(27):18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]
- Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of mycobacterium tuberculosis. PLoS One. 2008;3(1):e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine N, Guglielmini J, Wilbaux M, Van Melderen L. The decay of the chromosomally encoded ccdO157 toxin-antitoxin system in the escherichia coli species. Genetics. 2009;181(4):1557–1566. doi: 10.1534/genetics.108.095190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings CL, Glickman MS. Is mycobacterium tuberculosis stressed out? a critical assessment of the genetic evidence. Microbes Infect. 2010;12(14–15):1091–1101. doi: 10.1016/j.micinf.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Juarrero M, Kingry LC, Ordway DJ, Henao-Tamayo M, Harton M, Basaraba RJ, Hanneman WH, Orme IM, Slayden RA. Immune response to mycobacterium tuberculosis and identification of molecular markers of disease. Am J Respir Cell Mol Biol. 2009;40(4):398–409. doi: 10.1165/rcmb.2008-0248OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasari V, Mets T, Tenson T, Kaldalu N. Transcriptional cross-activation between toxin-antitoxin systems of escherichia coli. BMC Microbiol. 2013;13:45. doi: 10.1186/1471-2180-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch SB, Contreras H, Clark-Curtiss JE. Three mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J Bacteriol. 2009;191(5):1618–1630. doi: 10.1128/JB.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135(1):74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into mycobacterium tuberculosis latency. Cell Microbiol. 2009;11(8):1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- Salina EG, Mollenkopf HJ, Kaufmann SH, Kaprelyants AS. M. Tuberculosis gene expression during transition to the “Non-culturable” state. Acta Nat. 2009;1(2):73–77. [PMC free article] [PubMed] [Google Scholar]
- Chao MC, Rubin EJ. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. Noncognate mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J Biol Chem. 2010;285(51):39732–39738. doi: 10.1074/jbc.M110.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, De la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. et al. New use of BCG for recombinant vaccines. Nature. 1991;351(6326):456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Jeevarajah D, Patterson JH, Taig E, Sargeant T, McConville MJ, Billman-Jacobe H. Methylation of GPLs in mycobacterium smegmatis and mycobacterium avium. J Bacteriol. 2004;186(20):6792–6799. doi: 10.1128/JB.186.20.6792-6799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman TH, Kawasaki ES, Punreddy SR, Osburne MS. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene. 1998;209(1–2):95–103. doi: 10.1016/s0378-1119(98)00020-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential gene expression of Mtb upon overexpression of soj Mtb .
Primer sequences used for plasmid construction.
Primer sequences used for end-point PCR.