Abstract
Piperlongumine (PL) is a naturally occurring small molecule previously shown to induce cell death preferentially in cancer cells relative to non-cancer cells. An initial effort to synthesize analogs highlighted the reactivities of both of piperlongumine's α,β-unsaturated imide functionalities as key features determining PL's cellular effects. In this study, a second-generation of analogs was synthesized and evaluated in cells to gain further insight into how the reactivity, number, and orientation of PL's reactive olefins contribute to its ability to alter the physiology of cells.
Keywords: Michael acceptors, reactive oxygen species, piperlongumine, toxicity
Introduction
In nature, nucleophiles present in proteins and nucleic acids encounter a wealth of biologically active electrophilic small molecules.1 Several drug discovery efforts have explored the potential of targeted covalent inhibitors – electrophilic small molecules whose reactivity is limited to a small fraction of the proteome – to circumvent the off-target effects and potential toxicity of indiscriminate covalent inhibitors.2,3 Many biologically active electrophilic small molecules contain Michael acceptors that can undergo hetero-conjugate addition with the nucleophilic thiols in cysteine residues in irreversible4 or reversible fashion.5 One such thiol-containing tripeptide, glutathione (γ-l-glutamyl-l-cysteinylglycine), serves as a primary source of cellular reducing equivalents (along with NADPH), protects cells from oxidative stress and xenobiotics, and regulates redox-sensitive signaling pathways.6,7 From a chemical standpoint, it is therefore not surprising that small molecules capable of hetero-conjugate addition with thiols could have an impact on cellular redox homeostasis. In fact, several electrophilic small molecules have been shown to perturb the redox state of cancer cells by increasing levels of reactive oxygen species (ROS), which may be the cause of their cytotoxic effects.8
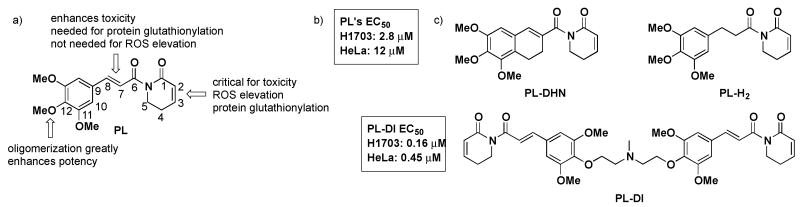
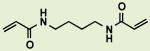
The recent finding that the naturally occurring small molecule piperlongumine (PL, Figure 1)9,10 can selectively kill cancer cells and shrink tumors in xenograft models prompted an in-depth evaluation of the structure/activity relationships and chemical reactivity of this simple molecule.11 This initial study suggested that the electrophilicity of the C2-C3 olefin is critical for many if not all of PL's effects on cells (viability, ROS levels, protein glutathionylation), and demonstrated that this functionality reacts with simple thiols in DMSO solution in vitro. In contrast, modifying or ablating the potential reactivity of the presumably less reactive, C7-C8, olefin (e.g. by hydrogenation, as in PL-H2, or introducing steric hindrance, as in PL-DHN; Figure 1) yielded compounds with reduced effects on viability that in some cases retained the ability to elevate ROS levels, challenging the notion that oxidative stress was sufficient for cell death. In addition, it was found that tethering two PL molecules together through an ether linkage at the C12 alkoxy group enhanced cellular potency with respect to viability by roughly 20-fold compared to PL itself (PL-DI, Figure 1).
Figure 1.
a) Previously established SAR of piperlongumine, PL, with carbon numbering used in this paper; b) EC50 of PL in H1703 and HeLa cells determined from three independent assays; c) Structures of dihydronaphthalene PL analog, PL-DHN; C7-C8 dihydro PL, PL-H2; and tethered PL ‘dimer’, PL-DI.
Building on these observations, we have subsequently designed and synthesized an additional collection of 29 PL analogs – piperlogs – and tested them in assays measuring cell viability, ROS levels, and protein glutathionylation. These analogs vary the nature and the number of electrophilic centers in the PL scaffold and provide new insight into the interdependence of the C2-C3 and C7-C8 olefins, the minimum pharmacophore of PL, and the effect of linker length and its chemical properties on the cellular effects of a series of tethered PL ‘dimers.’ This investigation has demonstrated that subtle differences in the chemical properties of Michael acceptors can have profound implications for their effects on oxidative stress and viability and suggests new testable hypotheses regarding the mechanism of action of these analogs for evaluation in further cellular experiments.
Results and discussion
PL analogs were prepared by acylation of the amide nitrogen of 1 with an appropriate acid chloride (Scheme 1A). Deprotonation of 1 was achieved with n-BuLi, at -78 °C. Subsequent reaction with a variety of acyl chlorides at low temperatures led to imides designed to test hypotheses about PL reactivity. One of the more commonly used amides of type 1, α, (β-unsaturated δ -lactam, 2, was prepared through ring-closing metathesis of homoallyl crotonamide with catalytic amounts of Grubbs' second-generation ruthenium catalyst.12 Notably, this cyclization failed when a homoallyl acrylamide was used as a substrate. PL ‘dimers’ in which two PL units are tethered by an ether linkage were generally made from phenol 39 and various diols by a Mitsunobu dehydrative coupling using azodicarboxylate and triphenylphosphine (Scheme 1B). Alternatively, two PL units could be connected through an ester linkage by acylation of phenol 3 with bis acid chlorides in the presence of triethylamine (Scheme 1C). Non-commercially available mono and bis acid chlorides were prepared from the corresponding acids by using oxalyl chloride and catalytic DMF. The purity of compounds isolated following silica chromatography was evaluated using 1H and 13C NMR; additionally, purity at the time of cellular testing was evaluated using LC-MS analysis. Compounds that had substantively degraded were excluded from further analysis.
Scheme 1.
Synthetic procedures for the preparation of piperlogs.
The resulting collection of small molecules containing multiple Michael acceptors was evaluated in the following cell-based assays: 1) cell viability (ATP coupled luciferase activity assay); 2) ROS levels (measuring conversion of a nonfluorescent dichlorodihydrofluorescein to fluorescent dichlorofluorescein);13 and 3) protein glutathionylation detected by immunofluorescence using a glutathione-recognizing antibody.14 Assay performance of PL and piperlogs can be found in the supplementary content.
PL contains two α,β-unsaturated imide functionalities in a continuous conjugated system, with the C2-C3 olefin more reactive toward thiol addition in vitro.11 To explore the necessity of the imide functionality, a variety of analogs containing multiple α,β-unsaturated amides were prepared (Table 1). Strikingly, these analogs did not diminish cell growth and viability in two cancer cell lines and did not increase ROS or protein glutathionylation levels. Amide analogs retaining or lacking the trimethoxycinnamoyl group found in PL were equally inactive. The diminished reactivity toward hetero-conjugate addition of α,β-unsaturated amides relative to α,β-unsaturated imides appears in this setting sufficient to ablate completely a variety of cellular effects.* Notably, this finding is in accord with previous reports of low background reactivity for acrylamide-containing targeted kinase inhibitors5 and suggests that the limit of reactivity for cellular electrophiles lies between α,β-unsaturated imides and amides.
Table 1.
Bis-α,β-unsaturated amides are inactive in viability, ROS increase, and protein glutathionylation assays.
| ID | ID | ||
|---|---|---|---|
|
BRD2724 |
|
BRD2285 |
|
|
BRD1764 |
|
BRD6008 |
|
BRD1088 |
|
BRD2537 |
|
|
BRD9521 |
BRD2285, an analog bearing an α-cyano amide functionality, obtained through Knoevenagel condensation, had surprisingly little effect on cell viability in spite of its more reactive double bond (Table 1). Curiously, this compound retained the ability to increase ROS levels, suggesting that, as with previous PL analogs PL-H2 and PL-DHN (Figure 1), ROS increase does not necessarily lead to cell death.23 BRD6008, a vinylogous urea analog of BRD2285 presumed to be far less reactive toward hetero-conjugate addition did not show any activity in the performed cellular assays (Table 1).
Previously, the presence of both the C2-C3 and C7-C8 olefin of PL was demonstrated to be essential for its cellular activities. An additional series of analogs was prepared that varied both of these electrophilic functionalities, and the spacing between them, to gain additional perspective into the minimum requirements for the cellular activity of this scaffold. The trimethoxycinnamoyl moiety could be replaced by simpler acryloyl (Table 2, BRD4809,), or methacryloyl substituents (Table 2, BRD3152) to produce analogs with slightly enhanced or diminished potency relative to PL, respectively. Decreased potency as a consequence of α-substitution (fluoro in BRD2847, methyl in BRD2501, phenyl in BRD5545) was also recognized in the previous study.11 Substitution of an ethoxycarbonyl group at C8, which would be predicted to give rise to a more reactive C7-C8 Michael acceptor, led to a less active compound (Table 2, BRD8143) compared to acryloyl analog BRD4809. It is possible that this highly reactive compound undergoes hetero-conjugate addition with other intracellular nucleophiles prior to reacting with the cellular targets that give rise to cell death following treatment with PL or BRD4809. The C1-C5 ring could also be simplified without loss of activity. Replacing it with an acyclic crotonyl imide had a minimal effect on potency, as long as Michael acceptors at the C2-C3 and C7-C8 positions were present (Table 2, BRD6245). Excising both the lactam and the aromatic ring found in PL led to BRD7991, which although less potent than PL, killed cancer cells (H1703 line) and elevated ROS and protein glutathionylation. The efficacy of this analog, whose molecular weight is less than half that of PL (153 Da vs. 317 Da), further supports the central role of electrophilicity in determining cellular efficacy in this compound collection.
Table 2.
EC50 values of imide-containing piperlogs in H1703 and HeLa cell lines determined from three independent assays.
| ID | EC50 H1703 (μM) |
EC50 HeLa (μM) |
ID | EC50 H1703 (μM) |
EC50 HeLa (μM |
||
|---|---|---|---|---|---|---|---|
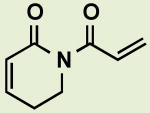
|
BRD4809 | 2.2 | 8.1 |
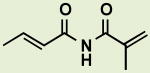
|
BRD7991 | 8.0 | >20 |
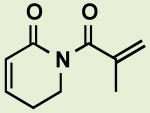
|
BRD3152 | 4.7 | 19 |
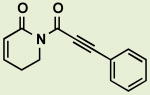
|
BRD5481 | 0.91 | 4.1 |
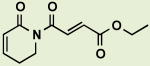
|
BRD8143 | 3.5 | 16 |
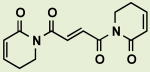
|
BRD6312 | 2.3 | 11 |
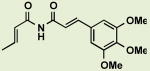
|
BRD6245 | 4.0 | 14 |
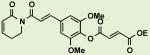
|
BRD8091 | 1.0 | 4.3 |
Replacing the C7-C8 styryl olefin with a phenylacetylene derivative (Table 2, BRD5481) provided a more potent PL analog than des-trimethoxy PL (BRD4524) reported previously (0.91 μM vs. 3.5 μM EC50 in H1703). This outcome may be a function of the enhanced reactivity toward hetero-conjugate addition of such alkynyl Michael acceptors, or may derive from the altered steric or stereoelectronic constraints on reactivity.16 Additionally, although PL was shown to have a variety of protein targets in cell extracts as assessed by quantitative proteomics, the precise range of targets accessed by each analog has not been determined. As such, variation in performance between analogs may reflect either differing affinities for various cellular protein and non-protein targets or differing physicochemical properties, including solubility, permeability, and subcellular localization.
Interestingly, a significant potency increase has been observed when the PL skeleton is duplicated within one molecule (for example, PL-DI, Figure 111). To explore this effect further, an initial set of analogs was synthesized to determine whether the potency of these compounds depends on the length and chemical nature of the linker joining two PL segments.17,18 Changing the chemical linkage from an ether to an ester bond led to a dramatic loss of potency compared to other ‘dimers’, likely due to facile hydrolytic cleavage of phenyl esters in cellular environment.19 Compared to PL, ester-linked dimer BRD1781 was only slightly more active, in line with the expected effective doubling of PL concentration in cells.
A set of analogs with polyethylene glycol ether linkers spanning lengths from 4 to 15 sigma bonds was also prepared (Figure 2). These analogs, as well as PL-DI (EC50 0.16 μM H1703, 0.45 μM HeLa; Figure 1) showed nearly identical EC50 values within each of two cell lines investigated. BRD4547, whose electrophilic functionalities are fixed in close proximity by a phenylene linker, retained the greatly enhanced cellular potency of this set of PL ‘dimers’ without greatly exceeding the molecular weight of PL. Furthermore, few significant differences in potency among these compounds were observed in ROS and glutathionylation assays. Apparently, the gain in efficacy observed from tethering two PL units is independent of the exact spacing between those units.
Figure 2.

EC50 values of tethered ‘dimeric’ imide-containing piperlogs in H1703 and HeLa cell lines. All data are expressed as mean ± standard deviation for three independent experiments.
This observation suggests new hypotheses regarding the enhanced potency of PL ‘dimers’ relative to PL in our assays. Although these ‘dimers’ can likely cross-link distinct sets of cellular nucleophiles as a result of their unique spacing of PL units, their uniform performance in cellular assays suggests a common mode of inducing cell death. Particularly attractive candidates include the cross-linking of DNA or cytoskeletal components.20 These assemblies of repeating alkylatable subunits might be equally reactive with piperlogs of widely varying linker length and are well-established targets for chemotherapeutic agents, including multivalent electrophiles like nitrogen mustards and busulfan.21,22
Additionally, a series of ‘dimers’ was synthesized from PL subunits in which reactivity at C8 was reduced or eliminated (Figure 3). BRD85988 was prepared by tethering two molecules of PL-DHN, an analog previously shown to elevate ROS levels and deplete glutathione, but with diminished effects on cell death relative to PL, using a propanediol linker in direct analogy to BRD4605 (Table 3). An intermediate analog containing one unit of PL and one unit of PL-DHN with the same linker was also prepared. Notably, BRD85988, containing two PL-DHN units, was found to lower cell viability with potency comparable to PL. In contrast, BRD4605, which includes two PL units, displayed 15 to 25-fold greater potency, and BRD5525 displayed intermediate potency. Despite its relatively small effect on cell viability, BRD85988 potently elevated ROS even at nontoxic doses without elevating protein glutathionylation, much like PL-DHN itself.
Figure 3.
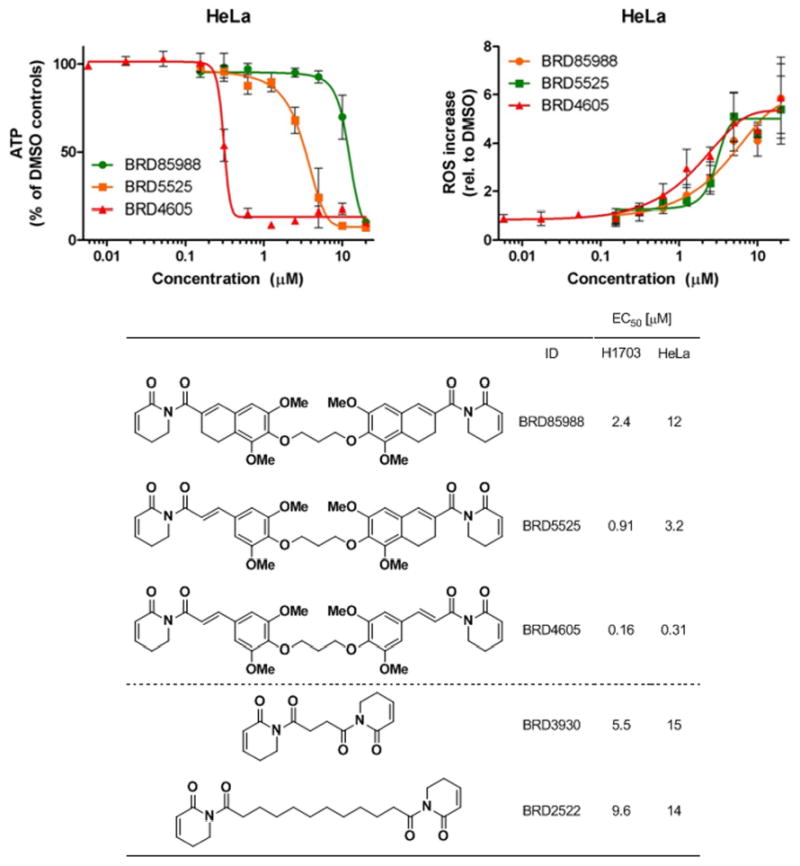
Viability (top left) and ROS increase (top right) of ‘dimeric’ piperlogs with C7-C8 olefin modifications determined from three independent experiments. All data are expressed as mean ± standard deviation for three independent experiments.
Similarly, two analogs were prepared by tethering PL units that both lack a C7-C8 olefin (BRD3930 and BRD2522), a scaffold analogous to PL-H2 (Figure 1). These analogs lack the trimethoxyphenyl moiety of PL, which appears to contribute only modestly to the cellular effects of PL (Figure 3). Use of a variable length aliphatic linker enabled synthesis of ‘dimers’ of substantially reduced molecular weight relative to PL-DI and related structures. In analogy to BRD85988 discussed above, BRD3930 and BRD2522 reduced cell viability less potently than PL despite containing two reactive C2-C3 olefins. Again, linker length did not greatly affect performance in our cellular assays. Owing to their multivalent electrophilicity but strikingly reduced ability to induce cell death relative to other ‘dimers’, these reagents will be helpful control compounds during further exploration of hypotheses regarding cellular mechanisms by which ‘dimers’ induce cell death, including alkylation of DNA or cytoskeletal components as described above.
Like PL-H2, BRD3930 and BRD2522 also elevated ROS at nontoxic doses; however, these analogs also potently elevated protein glutathionylation even at nontoxic doses (Supporting information). These analogs represent the first for which protein glutathionylation has been observed independently of cell death. Together with BRD85988, BRD3930 and BRD2522 reinforce the conclusion that multivalent electrophilicity is insufficient to observe PL's cellular effects; rather, the particular spacing and/or reactivity of the C2-C3 and C7-C8 olefins are critical.
Conclusions
By assembling a second-generation collection of piperlongumine analogs, we have gained new insight into how the structure and chemical reactivity of this scaffold relates to its cellular effects. We note that the α,β-unsaturated imide functionality appears essential for all cellular effects and cannot be recapitulated by α,β-unsaturated amides, presumably due to amides' lower reactivity. Several analogs retain PL-like properties while removing the trimethoxyphenyl ring or simplifying the C1-C5 ring to a crotonyl substituent, further highlighting the key role of the electrophilic functionalities of the PL scaffold in determining its cellular effects. Moreover, a collection of ‘dimeric’ analogs in which two PL units are tethered by linkers of varying length indicates that the high potency of these compounds in cellular assays is largely independent of linker length, an observation that may provide new hypotheses for future cellular studies of the mechanism of action of these compounds. Notably, tethering of PL units lacking a reactive olefin at C7-C8 led to ‘dimeric’ analogs that were no more potent than PL itself and far less potent than various analogs that tether two PL units, further highlighting the critical contribution of the C7-C8 olefin to PL-induced cell death. Together, these observations clarify that not all electrophilic functionalities are created equal and further demonstrate the essential role of the α,β-unsaturated imide electrophiles in determining PL's cellular effects.
Experimental section
General information
Dry solvents were purchased from Sigma-Aldrich. Unless otherwise stated, all reagents were obtained from commercial sources and used without further purification. 1H NMR spectra were recorded on Varian Unity/Inova 500 (500 MHz), or Bruker Ultrashield 300 (300 MHz) spectrometers. 1H NMR data are reported as follows: chemical shift in parts per million relative to CHCl3 (7.27 ppm), multiplicity (s, singlet; d, doublet; t, triplet; q, quartet; quin, quintet; m, multiplet; br, broadened), coupling constant (Hz), and integration. 13C NMR spectra were recorded on Varian Unity/Inova 500 (125 MHz) or Bruker Ultrashield 300 (75 MHz) spectrometers. 13C NMR chemical shifts are reported in parts per million relative to solvent. All 13C NMR spectra were determined with broadband decoupling. High-resolution mass spectra (HRMS) were obtained through the Broad Institute Chemical Biology Analytical Chemistry facility. All reactions were magnetically stirred and monitored by thin-layer chromatography (TLC) using E. Merck silica gel 60 F254 precoated plates (0.25 mm). Flash chromatography was performed using a CombiFlash companion system (Teledyne ISCO, Inc.) with pre-packed FLASH silica gel columns (Biotage, Inc.).
Compound synthesis and characterization
Acylation of amides
To solution of amide in THF (0.2 M, 1.1 equiv for mono acyl chlorides, 2.2 equiv for bis acyl chlorides) at -78 °C was slowly added cold solution of n-BuLi in hexanes (2.5 m, 1 equiv) and stirred for 15 minutes. Appropriate acyl chloride (1 equiv) was added dropwise and reaction was stirred for 30 minutes then warmed up to room temperature slowly. Reaction mixture was diluted with ethyl acetate, partitioned with NH4Cl(aq, sat), dried with brine and Na2SO4 and purified by column chromatography.
Acylation of phenol
To solution of phenol 3 (2 equiv) and triethylamine (6 equiv) in dichloromethane (0.1 m) was added bis acid chloride (1 equiv) dropwise. Reaction was stirred at room temperature for 24 hours. Reaction mixture was evaporated in vacuo and purified by silica gel column chromatography using gradient of methanol (0-5%) in DCM.
Etherification of phenol
To solution of phenol 3 (2.2 equiv) in a 1:1 mixture of toluene and THF (0.023 m) was added diol (1 equiv), triphenylphosphine (5 equiv) and diisopropyldiazadicarboxylate (5 equiv). The mixture was stirred for 24 h, volatiles evaporated and residue was separated by silica gel column chromatography with hexanes/EtOAc 50:50 to 10:90 gradient.
Acylation of amine
To solution of diamine (1 equiv), triethylamine (2.2 equiv), and DMAP (0.1 equiv) in THF (0.215 m) was added acid chloride (2.2 equiv) and the reaction was stirred over night. Volatiles were evaporated in vacuo and residue was fractionated by silica gel column chromatography using methanol (0-20%) in DCM.
Synthesis of BRD6245
To a suspension of (E)-but-2-enamide (59.9 mg, 0.70 mmol, 1.0 equiv) in 3.5 mL THF was added n-BuLi in hexanes (2.5 M, 0.34 mL, 0.85 mmol, 1.2 equiv) slowly at -78 °C. The reaction mixture was warmed up to room temperature and stirred for 8 h before a solution of (E)-3-(3,4,5-trimethoxyphenyl)acryloyl chloride (361 mg, 1.41 mmol, 2.0 equiv) in 2 mL of THF was added drop wise at -78 °C. The reaction mixture was allowed to warm up to rt overnight. The crude reaction mixture was diluted with ethyl acetate, and quenched with cold NH4Cl(aq, sat), and the organic layer separated. The aqueous layer was extracted twice with ethyl acetate. The combined organic layers were washed with brine and dried over MgSO4. After filtration and evaporation under reduced pressure, the residue was purified by column chromatography to yield the pure compound. BRD7991 was prepared in a similar procedure using methacryloyl chloride.
Compound characterization
BRD1088: (2E,2′E)-N,N′-(ethane-1,2-diyl)bis(3-(3,4,5-trimethoxyphenyl)acrylamide). 1H NMR (300 MHz, CHLOROFORM-d) δ 7.42 (d, J = 15.5 Hz, 2H), 6.69 (s, 4H), 6.34 (d, J = 15.6 Hz, 2H), 3.82 (s, 12H), 3.79 (s, 6H), 3.46-3.39 (m, 2H), 3.35-3.24 (m, 4H). Exact mass (M+H)+ calc'd: 501.2237, found 501.2223.
BRD1764: (2E,2′E)-N,N′-(Butane-1,4-diyl)bis(but-2-enamide). 1H NMR (500 MHz, DMSO-d6) δ 7.67 (br. s., 2H), 6.58 (dq, J=15.1, 6.8 Hz, 2H), 5.88 (dd, J=15.4, 1.2 Hz, 1 H), 3.05 - 3.14 (m, 4H), 1.78 (dd, J=6.8, 1.0 Hz, 6H), 1.36-1.46 (m, 4H); 13C NMR (126 MHz, DMSO-d6) δ 16.8, 26.5, 37.9, 125.9, 136.7, 164.6. Exact mass (M+Na)+ calc'd: 247.1422; found 247.1414.
BRD1781: bis(2,6-Dimethoxy-4-((E)-3-oxo-3-(2-oxo-5,6-dihydropyridin-1(2H)-yl)prop-1-en-1-yl)phenyl) dodecanedioate. 1H NMR (500 MHz, CHLOROFORM-d) δ ppm 7.69 (d, J=15.1 Hz, 2H), 7.45 (d, J=15.6 Hz, 2H), 6.96 (dt, J=9.5, 4.0 Hz, 2H), 6.82 (s, 4H), 6.06 (dt, J=9.6, 1.8 Hz, 2H), 4.02-4.08 (m, 4H), 3.85 (s, 12H), 2.62 (t, J=7.6 Hz, 4H), 2.46 -2.53 (m, 4H), 1.78 (quin, J=7.4 Hz, 4H), 1.40 - 1.49 (m, 4H), 1.34 (br, 8H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.8, 25.1, 29.0, 29.3, 29.5, 33.9, 41.6, 56.2, 104.9, 121.9, 125.8, 133.2, 143.5, 145.6, 152.3, 165.8, 168.8, 171.4. Exact mass (M+H)+ calc'd: 801.3599; found 801.3581.
BRD2285: (E)-3-((E)-2-cyano-3-(4-(trifluoromethyl)phenyl)acrylamido)propyl 3-(3,4,5-trimethoxyphenyl)acrylate. 1H NMR (500 MHz, CHLOROFORM-d) d ppm 8.39 (s, 1H), 8.01 (d, J=7.8 Hz, 2H), 7.75 (d, J=8.3 Hz, 2H), 7.66 (d, J=15.6 Hz, 1H), 6.96 - 7.03 (m, 1H), 6.82 (s, 2H), 6.56 (d, J=15.1 Hz, 1H), 4.39 (t, J=5.6 Hz, 2H), 3.90 (d, J=1.0 Hz, 9H), 3.63 (q, J=6.0 Hz, 2H), 2.06 (quin, J=5.9 Hz, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 28.3, 38.8, 56.1 (q, J = 2.6 Hz), 61.0 (q, J = 2.9 Hz), 63.1, 105.5, 116.4, 116.7, 126.1 (q, J = 3.8 Hz), 129.7, 130.5, 135.0 (d, J = 1.4 Hz), 145.7, 151.1, 153.4, 159.4, 167.0. Exact mass (M+Na)+ calc'd: 541.1562, found 541.1552.
BRD2522: 1,12-bis(2-oxo-5,6-dihydropyridin-1(2H)-yl)dodecane-1,12-dione. 1H NMR (500 MHz, CHLOROFORM-d) d ppm 6.89 (dt, J=9.8, 3.9 Hz, 2H), 6.00 (dt, J=9.8, 2.0 Hz, 2H), 3.98 (t, J=6.3 Hz, 4H), 2.92 (t, J=7.3 Hz, 4H), 2.38 - 2.45 (m, 4H), 1.56-1.70 (m, 4 H), 1.16-1.38 (m, 12H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.7, 25.0, 29.2, 29.4, 29.5, 39.3, 40.9, 126.0, 145.0, 165.4, 176.6. Exact mass (M+Na)+ calc'd: 411.2260; found 411.2265.
BRD2537: (2E,2′E)-N,N′-(ethane-1,2-diyl)bis(N-methyl-3-(3,4,5-trimethoxyphenyl)acrylamide). 1H NMR (300 MHz, CHLOROFORM-d) δ 7.42 (d, J = 15.3 Hz, 2H), 7.10 (d, J = 15.4 Hz, 2H), 6.71 (s, 4H), 3.85 (s, 12H), 3.79 (s, 6H), 3.33-3.23 (m, 4H), 3.17 (s, 6H). Exact mass (M+H)+ calc'd: 529.2550, found 529.2549.
BRD2724: N,N′-(Butane-1,4-diyl)diacrylamide. 1H NMR (500 MHz, DMSO-d6) δ 1.42 (br, 4H), 3.12 (d, J = 5.4 Hz, 4H), 5.56 (d, J = 10.3 Hz, 2H), 6.07 (s, 2H), 6.13-6.24 (m, 2H), 8.10 (br, 2H); 13C NMR (126 MHz, DMSO-d6) δ 26.7, 38.3, 125.0, 131.9, 164.6. Exact mass (M+Na)+ calc'd: 219.1109; found 219.1111.
BRD3152: 1-Methacryloyl-5,6-dihydropyridin-2(1H)-one. 1H NMR (300 MHz, CHLOROFORM-d) δ 2.01 (t, J = 1.2 Hz, 3H), 2.50 (tdd, J = 6.4, 6.4, 4.3, 1.9 Hz, 2H), 3.89 (t, J = 6.5 Hz, 2H), 5.24 (quin, J = 1.4 Hz, 1H), 5.29 (t, J = 0.9 Hz, 1H), 5.98 (dt, J = 9.7, 1.9 Hz, 1H), 6.93 (dt, J = 9.7, 4.1 Hz, 1H); 13C NMR (75 MHz, CHLOROFORM-d) δ 18.9, 24.7, 42.3, 117.6, 125.2, 142.6, 145.3, 165.1, 175.0. Exact mass (M+H)+ calc'd: 166.0868; found: 166.0862.
BRD3930: 1,4-bis(2-oxo-5,6-dihydropyridin-1(2H)-yl)butane-1,4-dione. 1H NMR (500 MHz, CHLOROFORM-d) δ ppm 2.41 (dtd, J = 8.5, 4.3, 4.3, 2.0 Hz, 4H), 3.26 (s, 4H), 3.95 (t, J = 6.6 Hz, 4H), 5.98 (dt, J = 9.8, 2.0 Hz, 2H), 6.76-6.95 (m, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.5, 34.3, 40.9, 125.8, 145.1, 165.2, 175.3. Exact mass (M+Na)+ calc'd: 299.1008; found 299.1013.
BRD4547: 1,1′-((2E,2′E)-3,3′-(1,4-phenylene)bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (500 MHz, CHLOROFORM-d) δ 2.49 (tdd, J=6.5, 6.5, 4.4, 1.7 Hz, 4H), 4.05 (t, J=6.3 Hz, 4H), 6.06 (dt, J=9.6, 1.8 Hz, 2H), 6.95 (dt, J=9.3, 4.4 Hz, 2H), 7.54 (d, J=15.6 Hz, 2H), 7.59 (s, 4H), 7.73 (d, J=15.6 Hz, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.8, 41.6, 122.7, 125.8, 128.7, 136.6, 142.5, 145.5, 165.7, 168.8. Exact mass (M+Na)+ calc'd: 399.1321; found 399.1319.
BRD4605: 1,1′-((2E,2′E)-3,3′-((propane-1,3-diylbis(oxy))bis(3,5-dimethoxy-4,1-phenylene))bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 7.68 (d, J = 15.6 Hz, 2H), 7.41 (d, J = 15.5 Hz, 2H), 7.06-6.88 (m, 2H), 6.78 (s, 4H), 6.05 (d, J = 9.7 Hz, 2H), 4.25 (t, J = 6.3 Hz, 4H), 4.04 (t, J = 6.4 Hz, 4H), 3.82 (s, 12H), 2.57-2.41 (m, 4H), 2.18 (quin, J = 6.2 Hz, 2H); 13C NMR (75 MHz, CHLOROFORM-d) δ 169.1, 166.0, 153.7, 145.6, 144.1, 139.7, 130.6, 126.1, 121.1, 105.8, 71.1, 56.3, 41.8, 31.2, 25.0. Exact mass (M+Na)+ calc'd: 669.2424, found 669.2413.
BRD4691: 1,1′-((2E,2′E)-3,3′-((((ethane-1,2-diylbis(oxy))bis(ethane-2,1-diyl))bis(oxy))bis(3,5-dimethoxy-4,1-phenylene))bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 7.67 (d, J = 15.6 Hz, 2H), 7.40 (d, J = 15.5 Hz, 2H), 7.00-6.89 (m, 2H), 6.78 (s, 4H), 6.04 (d, J = 9.7 Hz, 2H), 4.17 (t, J = 5.1 Hz, 4H), 4.04 (t, J = 6.5 Hz, 4H), 3.86 (s, 12H), 3.80 (t, J = 5.1 Hz, 4H), 3.73 (s, 4H), 2.53-2.42 (m, 4H); 13C NMR (75 MHz, CHLOROFORM-d) δ 169.1, 166.0, 153.7, 145.7, 144.0, 139.3, 130.8, 126.0, 121.2, 105.7, 72.5, 70.9, 70.6, 56.4, 41.8, 25.0. Exact mass (M+Na)+ calc'd: 743.2792, found 743.2786.
BRD4698: Bis(2,6-dimethoxy-4-((E)-3-oxo-3-(2-oxo-5,6-dihydropyridin-1(2H)-yl)prop-1-en-1-yl)phenyl) fumarate. 1H NMR (300 MHz, PYRIDINE-d5) δ 8.02 (d, J = 15.6 Hz, 2H), 7.83 (d, J = 15.6 Hz, 2H), 7.61 (s, 2H), 7.11 (s, 4H), 6.85 – 6.73 (m, 2H), 6.08 (d, J = 9.7 Hz, 2H), 4.01 (t, J = 6.4 Hz, 4H), 3.76 (s, 12H), 2.29 – 2.19 (m, 4H); 13C NMR (75 MHz, PYRIDINE-d5) δ 169.2, 166.3, 163.0, 153.3, 146.8, 142.7, 135.2, 135.1, 134.8, 130.6, 125.9, 105.8, 56.7, 42.4, 25.3. Exact mass (M+Na)+ calc'd: 709.2009, found 709.1967.
BRD4809: 1-Acryloyl-5,6-dihydropyridin-2(1H)-one. 1H NMR (500 MHz, CHLOROFORM-d) δ 2.47 (tdd, J = 6.3, 6.3, 4.4, 2.0 Hz, 2H), 4.00 (t, J = 6.6 Hz, 2H), 5.76 (dd, J = 10.3, 1.5 Hz, 1H), 6.02 (dt, J = 9.8, 1.7 Hz, 1H), 6.40 (dd, J = 17.1, 2.0 Hz, 1H), 6.87-6.98 (m, 1H), 7.06 (dd, J = 16.8, 10.5 Hz, 1H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.7, 41.4, 125.6, 128.5, 131.6, 145.7, 165.5, 168.8. Exact mass (M+H)+ calc'd: 152.0712; found: 152.0699
BRD5481: 1-(3-Phenylpropioloyl)-5,6-dihydropyridin-2(1H)-one. 1H NMR (500 MHz, CHLOROFORM-d) δ 2.43-2.53 (m, 2H), 4.06 (t, J = 6.3 Hz, 2H), 6.07 (dt, J = 9.8, 1.5 Hz, 1H), 6.93-6.99 (m, 1H), 7.33-7.40 (m, 2H), 7.41-7.46 (m, 2H), 7.65 (d, J = 7.8 Hz, 1H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.4, 40.8, 83.7, 93.3, 120.5, 125.1, 128.4, 130.4, 133.0, 145.6, 153.0, 163.7. Exact mass (M+Na)+ calc'd: 248.0687; found 248.0685.
BRD5525: (E)-1-(6-(3-(2,6-dimethoxy-4-(3-oxo-3-(2-oxo-5,6-dihydropyridin-1(2H)-yl)prop-1-en-1-yl)phenoxy)propoxy)-5,7-dimethoxy-3,4-dihydronaphthalene-2-carbonyl)-5,6-dihydropyridin-2(1H)-one. 1H NMR (300 MHz, CHLOROFORM-d) δ 2.19 (quin, J = 6.4 Hz, 2H), 2.41-2.60 (m, 6H), 2.76-2.91 (m, 2H), 3.78 (s, 3H), 3.78-3.80 (m, 3H), 3.84 (s, 6H), 3.86-3.93 (m, 2H), 4.05 (t, J = 6.4 Hz, 2H), 4.18-4.32 (m, 4H), 6.00 (d, J = 9.8 Hz, 1H), 6.06 (d, J = 10.0 Hz, 1H), 6.53 (s, 1H), 6.79 (s, 2H), 6.88-7.03 (m, 3H), 7.42 (d, J = 15.3 Hz, 1H), 7.68 (d, J = 15.8 Hz, 1H); 13C NMR (75 MHz, CHLOROFORM-d) δ 174.5, 169.1, 166.1, 165.6, 153.8, 152.1, 151.1, 145.6, 145.3, 144.2, 142.7, 139.7, 135.0, 133.5, 130.7, 128.7, 126.1, 125.6, 123.1, 121.1, 108.5, 105.9, 71.1, 71.1, 60.9, 56.4, 56.3, 43.4, 41.9, 31.3, 29.9, 25.0, 23.9, 20.9. Exact mass (M+Na)+ calc'd: 695.2581; found 695.2571.
BRD6008: (E)-3-((E)-2-cyano-3-(dimethylamino)acrylamido)propyl 3-(3,4,5-trimethoxyphenyl)acrylate. 1H NMR (500 MHz, CHLOROFORM-d) δ 7.79 (s, 1H), 7.61 (d, J = 15.9 Hz, 1H), 6.82 (s, 2H), 6.54 (d, J = 15.9 Hz, 1H), 6.37-6.30 (m, 1H), 4.30 (t, J = 5.8 Hz, 2H), 3.88 (s, 6H), 3.87 (s, 3H), 3.53-3.45 (m, 2H), 3.30 (s, 3H), 3.17 (s, 3H), 1.99-1.90 (m, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 167.2, 165.1, 156.4, 153.5, 145.3, 140.1, 130.2, 120.2, 117.4, 105.5, 63.4, 61.1, 56.3, 47.5, 38.5, 38.1, 28.8. Exact mass (M+H)+ calc'd: 418.1978, found 418.1983.
BRD6245: (E)-N-((E)-3-(3,4,5-trimethoxyphenyl)acryloyl)but-2-enamide. 1H NMR (300 MHz, CHLOROFORM-d) δ 8.93 (s, 1H), 7.76 (d, J = 15.6 Hz, 1H), 7.33-7.09 (m, 2H), 6.82 (s, 2H), 6.51 (d, J = 15.3 Hz, 1H), 3.90 (s, 6H), 3.89 (s, 3H), 1.98 (dd, J = 6.9, 1.4 Hz, 3H); 13C NMR (75 MHz, CHLOROFORM-d) δ 166.9, 166.0, 153.7, 146.3, 146.2, 140.9, 130.1, 124.8, 119.1, 106.0, 61.2, 56.4, 18.5. Exact mass (M+H)+ calc'd: 306.1341, found 306.1347.
BRD6312: (E)-1,4-bis(2-oxo-5,6-dihydropyridin-1(2H)-yl)but-2-ene-1,4-dione. 1H NMR (500 MHz, CHLOROFORM-d + few drops of METHANOL-d4) δ 7.56 (s, 2H), 6.97-6.87 (m, 2H), 5.96 (d, J = 9.8 Hz, 2H), 3.94 (t, J = 6.5 Hz, 4H), 2.48-2.38 (m, 4H); 13C NMR (126 MHz, CHLOROFORM-d) δ 168.1, 165.6, 146.6, 134.1, 125.1, 41.6, 24.7. Exact mass (M+H)+ calc'd: 275.1032, found 275.1039.
BRD6335: bis(2,6-Dimethoxy-4-((E)-3-oxo-3-(2-oxo-5,6-dihydropyridin-1(2H)-yl)prop-1-en-1-yl)phenyl) succinate. 1H NMR (500 MHz, CHLOROFORM-d) δ 2.50 (dt, J=6.3, 4.4 Hz, 4H), 3.12 (s, 4H), 3.85 (s, 12H), 4.06 (t, J=6.3 Hz, 4H), 6.06 (dt, J=9.5, 1.6 Hz, 2H), 6.82 (s, 4H), 6.94-7.01 (m, 2H), 7.45 (d, J=15.6 Hz, 2H), 7.69 (d, J=15.1 Hz, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 24.8, 29.0, 41.6, 56.2, 104.9, 122.0, 125.7, 130.1, 133.4, 143.4, 145.6, 152.3, 165.8, 168.7, 169.6. Exact mass (M+Na)+ calc'd: 711.2166; found 711.2135.
BRD7782: 1,1′-((2E,2′E)-3,3′-(((((oxybis(ethane-2,1-diyl))bis(oxy))bis(ethane-2,1-diyl))bis(oxy))bis(3,5-dimethoxy-4,1-phenylene))bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 2.40-2.54 (m, 4H), 3.70 (br, 8H), 3.75-3.82 (m, 4H), 3.86 (s, 12H), 4.04 (t, J = 6.5 Hz, 4H), 4.16 (t, J = 4.9 Hz, 4H), 6.04 (d, J = 9.8 Hz, 2H), 6.76 (br, 4H), 6.95 (dt, J = 9.5, 4.3 Hz, 2H), 7.40 (d, J = 15.4 Hz, 2H), 7.66 (d, J = 15.4 Hz, 2H); 13C NMR (75 MHz, CHLOROFORM-d) δ 24.8, 41.6, 56.1, 70.3, 70.6, 72.3, 77.2, 105.4, 121.0, 125.8, 130.6, 143.8, 145.4, 153.4, 165.8, 168.8. Exact mass (M+Na)+ calc'd: 787.3054; found 787.3047.
BRD7991: (E)-N-methacryloylbut-2-enamide. 1H NMR (300 MHz, CHLOROFORM-d) δ 8.35 (s, 1H), 7.25-6.90 (m, 2H), 5.79 (d, J = 0.6 Hz, 1H), 5.56 (dd, J = 3.0, 1.4 Hz, 1H), 1.94 (s, 3H), 1.89 (dd, J = 6.5, 1.2 Hz, 3H); 13C NMR (75 MHz, CHLOROFORM-d) δ 167.4, 166.8, 147.1, 140.1, 124.3, 122.7, 18.7, 18.6. Exact mass (M+Na)+ calc'd: 176.0687, found 176.0690.
BRD8091: 2,6-dimethoxy-4-((E)-3-oxo-3-(2-oxo-5,6-dihydropyridin-1(2H)-yl)prop-1-en-1-yl)phenyl ethyl fumarate. 1H NMR (500 MHz, CHLOROFORM-d) δ 7.68 (d, J = 15.5 Hz, 1H), 7.46 (d, J = 15.5 Hz, 1H), 7.14-7.00 (m, 2H), 7.00-6.90 (m, 1H), 6.83 (s, 2H), 6.05 (d, J = 9.7 Hz, 1H), 4.29 (q, J = 7.1 Hz, 2H), 4.05 (t, J = 6.4 Hz, 2H), 3.85 (s, 6H), 2.55-2.40 (m, 2H), 1.34 (t, J = 7.1 Hz, 3H); 13C NMR (126 MHz, CHLOROFORM-d) δ 168.9, 166.0, 164.9, 162.7, 152.4, 145.9, 143.4, 135.6, 134.0, 132.4, 129.8, 125.9, 122.5, 105.0, 61.7, 56.4, 41.9, 25.0, 14.3. Exact mass (M+H)+ calc'd: 430.1502, found 430.1503.
BRD8143: (E)-ethyl 4-oxo-4-(2-oxo-5,6-dihydropyridin-1(2H)-yl)but-2-enoate. 1H NMR (500 MHz, CHLOROFORM-d) δ 7.66 (d, J = 15.4 Hz, 1H), 7.01-6.94 (m, 1H), 6.71 (d, J = 15.4 Hz, 1H), 6.02 (d, J = 9.8 Hz, 1H), 4.24 (q, J = 7.1 Hz, 2H), 3.99 (t, J = 6.5 Hz, 2H), 2.53-2.42 (m, 2H), 1.30 (t, J = 7.1 Hz, 3H); 13C NMR (126 MHz, CHLOROFORM-d) δ 167.7, 165.6, 165.5, 146.5, 137.2, 130.5, 125.2, 61.3, 41.6, 24.8, 14.3. Exact mass (M+H)+ calc'd: 224.0923, found 224.0916.
BRD85988: 1,1′-(6,6′-(propane-1,3-diylbis(oxy))bis(5,7-dimethoxy-3,4-dihydronaphthalene-2,2′-carbonyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 2.20 (quin, J = 6.0 Hz, 2H), 2.48 (m, J = 7.9, 7.9 Hz, 4H), 2.54 (d, J = 4.9 Hz, 4H), 2.85 (t, J = 8.5 Hz, 4H), 3.79 (s, 6H), 3.80 (s, 6H), 3.89 (t, J = 6.6 Hz, 4H), 4.26 (t, J = 6.2 Hz, 4H), 6.00 (d, J = 9.8 Hz, 2H), 6.54 (s, 2H), 6.87-7.01 (m, 4H); 13C NMR (75 MHz, CHLOROFORM-d) δ 174.5, 165.6, 152.1, 151.1, 145.3, 142.7, 135.1, 133.5, 128.7, 125.6, 123.1, 108.5, 71.1, 61.0, 56.3, 43.5, 29.9, 25.1, 23.9, 20.9. Exact mass (M+Na)+ calc'd: 721.2737; found 721.2726.
BRD9080: 1,1′-((2E,2′E)-3,3′-(((Oxybis(ethane-2,1-diyl))bis(oxy))bis(3,5-dimethoxy-4,1-phenylene))bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 2.44-2.53 (m, 4H), 3.74-3.94 (m, 16H), 4.04 (t, J = 6.4 Hz, 4H), 4.19 (t, J = 5.1 Hz, 4H), 6.05 (d, J = 9.6 Hz, 2H), 6.78 (s, 4H), 6.95 (dt, J = 9.4, 4.3 Hz, 2H), 7.41 (d, J = 15.4 Hz, 2H), 7.67 (d, J = 15.6 Hz, 2H); 13C NMR (75 MHz, CHLOROFORM-d) d 24.8, 41.6, 56.1, 70.3, 72.3, 105.5, 121.0, 125.8, 130.6, 139.1, 143.8, 145.4, 153.5, 165.8, 168.9. Exact mass (M+Na)+ calc'd: 699.2530; found 699.2524.
BRD9427: 1,1′-((2E,2′E)-3,3′-((3,6,9,12-tetraoxatetradecane-1,14-diylbis(oxy))bis(3,5-dimethoxy-4,1-phenylene))bis(acryloyl))bis(5,6-dihydropyridin-2(1H)-one). 1H NMR (300 MHz, CHLOROFORM-d) δ 7.67 (d, J = 15.6 Hz, 2H), 7.40 (d, J = 15.5 Hz, 2H), 7.00-6.88 (m, 2H), 6.78 (s, 4H), 6.04 (d, J = 9.7 Hz, 2H), 4.16 (t, J = 5.1 Hz, 4H), 4.03 (t, J = 6.4 Hz, 4H), 3.86 (s, 12H), 3.79 (t, J = 5.0 Hz, 4H), 3.75-3.57 (m, 12H), 2.56-2.40 (m, 4H); 13C NMR (75 MHz, CHLOROFORM-d) δ 169.1, 166.0, 153.7, 145.7, 144.0, 139.3, 130.8, 126.0, 121.2, 105.7, 72.5, 70.8, 70.8, 70.6, 56.4, 41.8, 25.0. Exact mass (M+Na)+ calc'd: 831.3316, found 831.3306.
BRD9521: (2E,2′E)-N,N′-(propane-1,3-diyl)bis(3-(3,4,5-trimethoxyphenyl)acrylamide). 1H NMR (500 MHz, CHLOROFORM-d + few drops of METHANOL-d4) δ 7.45 (d, J = 15.6 Hz, 2H), 6.71 (s, 4H), 6.39 (d, J = 15.6 Hz, 2H), 3.82 (s, 12H), 3.80 (s, 6H), 3.40-3.31 (m, 6H), 1.71 (dt, J = 12.2, 6.3 Hz, 2H); 13C NMR (126 MHz, CHLOROFORM-d) δ 167.3, 153.4, 141.0, 139.5, 130.6, 120.0, 105.1, 61.0, 56.1, 36.4, 29.3. Exact mass (M+H)+ calc'd: 515.2393, found 515.2394.
Cell culture
HeLa and H1703 were obtained from ATCC and cultured in recommended media in a 37 °C incubator (5% CO 2).
ROS assay
Cells were plated at 4,000 cells/well in black 384-well plates (Corning) and allowed to attach overnight. The next day (ca. 90% confluency), dilutions of compounds in DMSO were added by pin transfer (CyBio Vario, 100 nL per well), and incubated for 90 minutes. Media was changed using a Thermo Multidrop Combi liquid handler to phenol red-free DMEM containing 10 μM CM-H2DCF-DA and 10 μg/mL Hoechst 33342. Following incubation for 15-30 minutes, cells were washed twice with PBS. Images were obtained using an IX_Micro automated fluorescence microscope (Molecular Devices). Quantitation of pixel intensity was performed using MetaXpress software and signal intensity was calculated relative to wells in the same plate treated with DMSO.
ATP assay
Cells were plated at 1,000 per well in white 384-well plates and allowed to attach overnight. After addition of compounds by pin transfer, plates were incubated 48 h. At that time, media was removed and replaced with a solution of CellTiter-Glo reagent (Promega) in PBS. After ten minutes, luminescence was read using an EnVision multilabel plate reader (Perkin-Elmer) and signal intensity was calculated relative to in-plate DMSO control wells.
Immunofluorescence detection of glutathionylated proteins
Cells were plated at 3,000 (HeLa) per well and allowed to attach overnight. After addition of compounds by pin transfer, plates were incubated for 6 h. Cells were then fixed with 2% paraformaldehyde in PBS (20 minutes), permeabilized 30 minutes with PBS+0.1% TritonX-100 (“PBST”), and blocked 30 minutes with PBST+2%BSA. Primary antibody (Ms anti-glutathione, Abcam Ab19534) was added (1:1250 in PBST+2%BSA) and incubated at 4 °C overnight. Following two wash es with PBST, cells were incubated at RT 1 h in the dark with secondary antibody solution (AlexaFluor 488-conjugated goat anti-mouse, Jackson Immunochemicals, 1:750, plus Hoechst 33342, 10 μg/ml in PBST+2%BSA). Following two washes with PBST, images were collected using an IX_Micro automated fluorescence microscope (Molecular Devices). Quantitation of pixel intensity was performed as above.
Supplementary Material
Acknowledgments
We thank Weicheng Zhang for synthesizing compounds BRD2724 and BRD1764. This research was supported by the National Institute of Health (NIGMS-38627). S.L.S is a Howard Hughes Medical Institute Investigator.
Footnotes
Superior ability of imides (compared to amides) to stabilize developing negative charge on α-carbon in conjugate addition reactions was used in a number of developed synthetic organic methods.15
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Gersch M, Kreuzer J, Sieber SA. Nat Prod Rep. 2012;29:659. doi: 10.1039/c2np20012k. [DOI] [PubMed] [Google Scholar]
- 2.Singh J, Petter RC, Baillie TA, Whitty A. Nat Rev Drug Discov. 2011;10:307. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 3.Potashman MH, Duggan ME. J Med Chem. 2009;52:1231. doi: 10.1021/jm8008597. [DOI] [PubMed] [Google Scholar]
- 4.Hagel M, Niu D, St Martin T, Sheets MP, Qiao L, Bernard H, Karp RM, Zhu Z, Labenski MT, Chaturvedi P, Nacht M, Westlin WF, Petter RC, Singh J. Nat Chem Biol. 2011;7:22. doi: 10.1038/nchembio.492. [DOI] [PubMed] [Google Scholar]
- 5.Serafimova IM, Pufall MA, Krishnan S, Duda K, Cohen MS, Maglathlin RL, McFarland JM, Miller RM, Frodin M, Taunton J. Nat Chem Biol. 2012;8:471. doi: 10.1038/nchembio.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant CM, MacIver FH, Dawes IW. Curr Genet. 1996;29:511. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Trends Biochem Sci. 2009;34:85. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Wondrak GT. Antioxid Redox Signal. 2009;11:3013. doi: 10.1089/ars.2009.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X, Tolliday NJ, Golub TR, Carr SA, Shamji AF, Stern AM, Mandinova A, Schreiber SL, Lee SW. Nature. 2011;475:231. doi: 10.1038/nature10167. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Bezerra DP, Pessoa C, de Moraes MO, Saker-Neto N, Silveira ER, Costa-Lotufo LV. Eur J Pharm Sci. 2012;48:453. doi: 10.1016/j.ejps.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Adams DJ, Dai M, Pellegrino G, Wagner BK, Stern AM, Shamji AF, Schreiber SL. Proc Natl Acad Sci U S A. 2012;109:15115. doi: 10.1073/pnas.1212802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vougioukalakis GC, Grubbs RH. Chemical Reviews. 2010;110:1746. doi: 10.1021/cr9002424. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Joseph JA. Free Radic Biol Med. 1999;27:612. doi: 10.1016/s0891-5849(99)00107-0. [DOI] [PubMed] [Google Scholar]
- 14.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. Free Radic Biol Med. 2007;43:883. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Sammis GM, Jacobsen EN. J Am Chem Soc. 2003;125:4442. doi: 10.1021/ja034635k. [DOI] [PubMed] [Google Scholar]
- 16.Schwobel JA, Wondrousch D, Koleva YK, Madden JC, Cronin MT, Schuurmann G. Chem Res Toxicol. 2010;23:1576. doi: 10.1021/tx100172x. [DOI] [PubMed] [Google Scholar]
- 17.Reinke AA, Gestwicki JE. Chem Biol Drug Des. 2007;70:206. doi: 10.1111/j.1747-0285.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 18.Zheng FF, Kuduk SD, Chiosis G, Munster PN, Sepp-Lorenzino L, Danishefsky SJ, Rosen N. Cancer Res. 2000;60:2090. [PubMed] [Google Scholar]
- 19.McCracken NW, Blain PG, Williams FM. Biochem Pharmacol. 1993;45:31. doi: 10.1016/0006-2952(93)90373-5. [DOI] [PubMed] [Google Scholar]
- 20.Bezerra DP, Vasconcellos MC, Machado MS, Villela IV, Rosa RM, Moura DJ, Pessoa C, Moraes MO, Silveira ER, Lima MA, Aquino NC, Henriques JA, Saffi J, Costa-Lotufo LV. Mutat Res. 2009;677:8. doi: 10.1016/j.mrgentox.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Brookes P, Lawley PD. Biochem J. 1961;80:496. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco DY, Stratton NK, Gibson NW. Cancer Res. 1989;49:5108. [PubMed] [Google Scholar]
- 23.Adams DJ, Boskovic ZV, Theriault JR, Wang AJ, Stern AM, Wagner BK, Shamji AF, Schreiber SL. ACS Chem Biol. 2013;8:923. doi: 10.1021/cb300653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.