Abstract
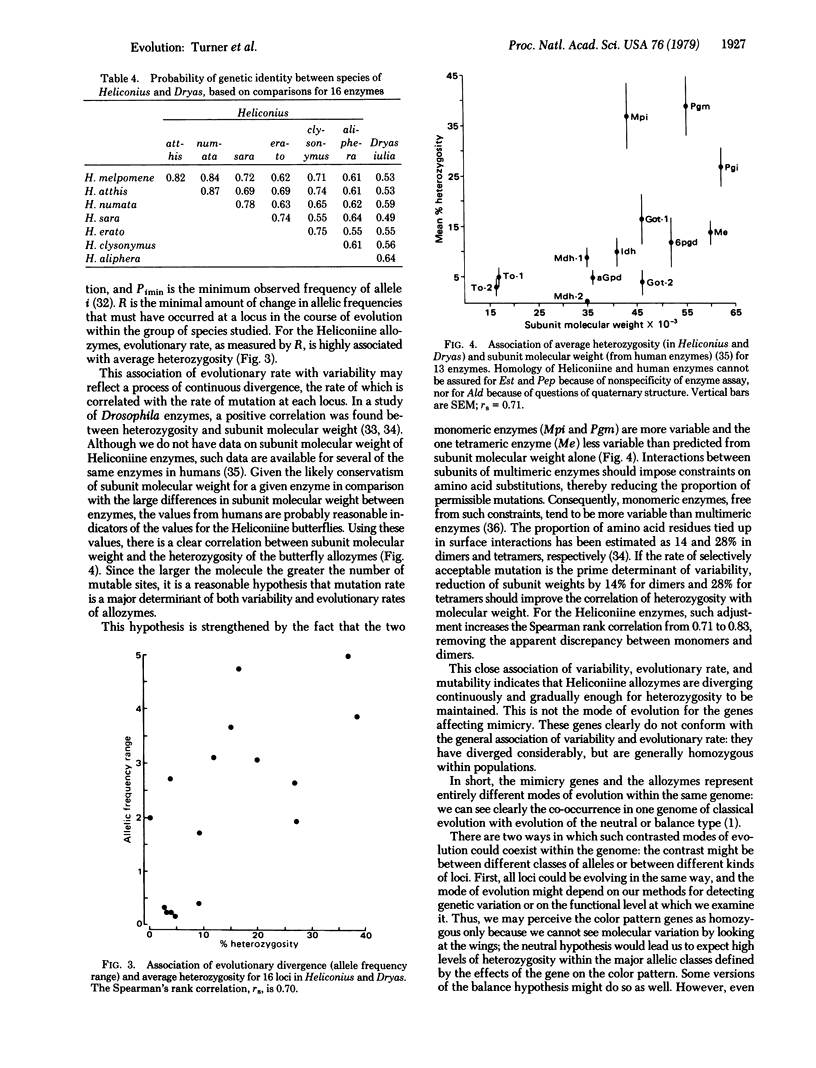
Butterflies in the South American genus Heliconius have undergone a spectacular adaptive radiation (with convergent evolution between some lines) in their color patterns; this has been produced by natural selection for muellerian mimicry. The genetic basis of this radiation, shown by crossing highly differentiated races within two of the species, is homozygosity for alternative alleles at some half dozen loci. In complete contrast, allozyme loci in these butterflies are strongly heterozygous and show only frequency differences (never amounting to homozygosity of alternative alleles) between races; the amount of allozyme divergence is the same between races of H. erato and H. sara, although in color pattern the first forms marked races and the other does not. For the allozymes, there is a strong correlation over loci for rate of divergence between species and average heterozygosity. This is not true of the genes controlling color pattern. Heterozygosity of the enzymes is correlated with subunit molecular weight. Thus, different parts of the genome can evolve in different ways simultaneously; genes controlling color pattern in the "classical" mode, and allozymes in a different mode in which the rate of evolution is related to their heterozygosity (a "balance" or "neutral" mode).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson W. W. Natural Selection for Miillerian Mimicry in Heliconius erato in Costa Rica. Science. 1972 May 26;176(4037):936–939. doi: 10.1126/science.176.4037.936. [DOI] [PubMed] [Google Scholar]
- Cherty L. M., Case S. M., Wilson A. C. Frog perspective on the morphological difference between humans and chimpanzees. Science. 1978 Apr 14;200(4338):209–211. doi: 10.1126/science.635583. [DOI] [PubMed] [Google Scholar]
- Coyne J. A. Lack of genic similarity between two sibling species of drosophila as revealed by varied techniques. Genetics. 1976 Nov;84(3):593–607. doi: 10.1093/genetics/84.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire A. L. The properties of a carboxylesterase from the peach-potato aphid, Myzus persicae (Sulz.), and its role in conferring insecticide resistance. Biochem J. 1977 Dec 1;167(3):675–683. doi: 10.1042/bj1670675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson D. A., Edwards Y. H., Harris H. The distributions of subunit numbers and subunit sizes of enzymes: a study of the products of 100 human gene loci. Ann Hum Genet. 1976 May;39(4):383–411. doi: 10.1111/j.1469-1809.1976.tb00144.x. [DOI] [PubMed] [Google Scholar]
- King M. C., Wilson A. C. Evolution at two levels in humans and chimpanzees. Science. 1975 Apr 11;188(4184):107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Koehn R. K., Eanes W. F. Subunit size and genetic variation of enzymes in natural populations of Drosophila. Theor Popul Biol. 1977 Jun;11(3):330–341. doi: 10.1016/0040-5809(77)90016-8. [DOI] [PubMed] [Google Scholar]
- Maxson L. R., Wilson A. C. Convergent morphological evolution detected by studying proteins of tree frogs in the Hyla eximia species group. Science. 1974 Jul 5;185(4145):66–68. doi: 10.1126/science.185.4145.66. [DOI] [PubMed] [Google Scholar]
- McDonald J. F., Chambers G. K., David J., Ayala F. J. Adaptive response due to changes in gene regulation: a study with Drosophila. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4562–4566. doi: 10.1073/pnas.74.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols E. A., Chapman V. M., Ruddle F. H. Polymorphism and linkage for mannosephosphate isomerase in Mus musculus. Biochem Genet. 1973 Jan;8(1):47–53. doi: 10.1007/BF00485556. [DOI] [PubMed] [Google Scholar]
- Sene F. M., Carson H. L. Genetic variation in Hawaiian Drosophila. IV. Allozymic similarity between D. silvestris and D. heteroneura from the island of Hawaii. Genetics. 1977 May;86(1):187–198. doi: 10.1093/genetics/86.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward R. D. Relationship between enzyme heterozygosity and quaternary structure. Biochem Genet. 1977 Feb;15(1-2):123–135. doi: 10.1007/BF00484555. [DOI] [PubMed] [Google Scholar]
- Whitt G. S. Development genetics of the lactate dehydrogenase isozymes of fish. J Exp Zool. 1970 Sep;175(1):1–35. doi: 10.1002/jez.1401750102. [DOI] [PubMed] [Google Scholar]



