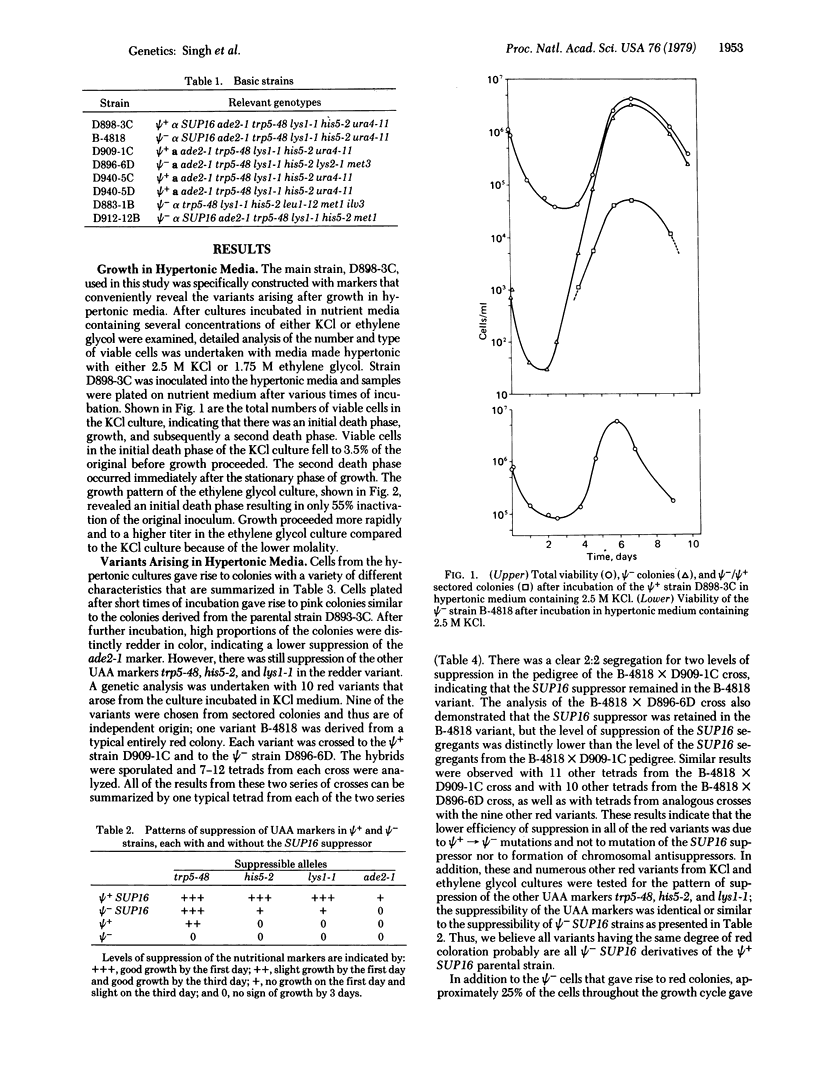
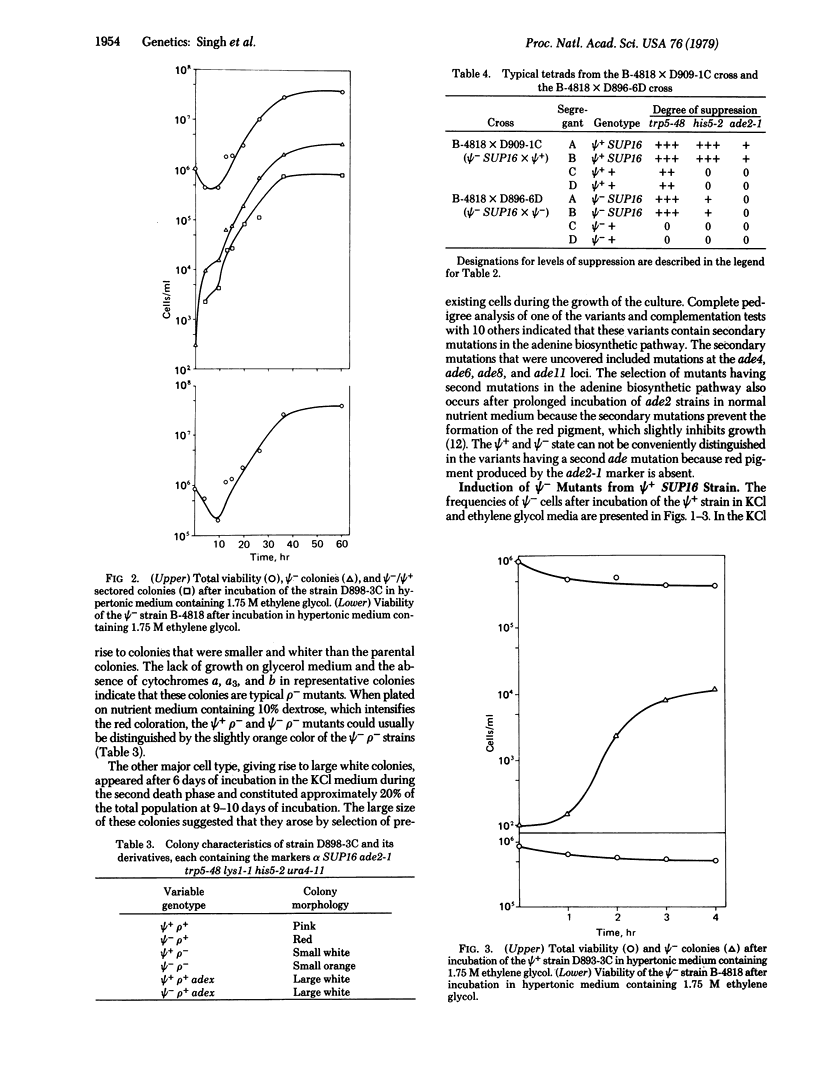
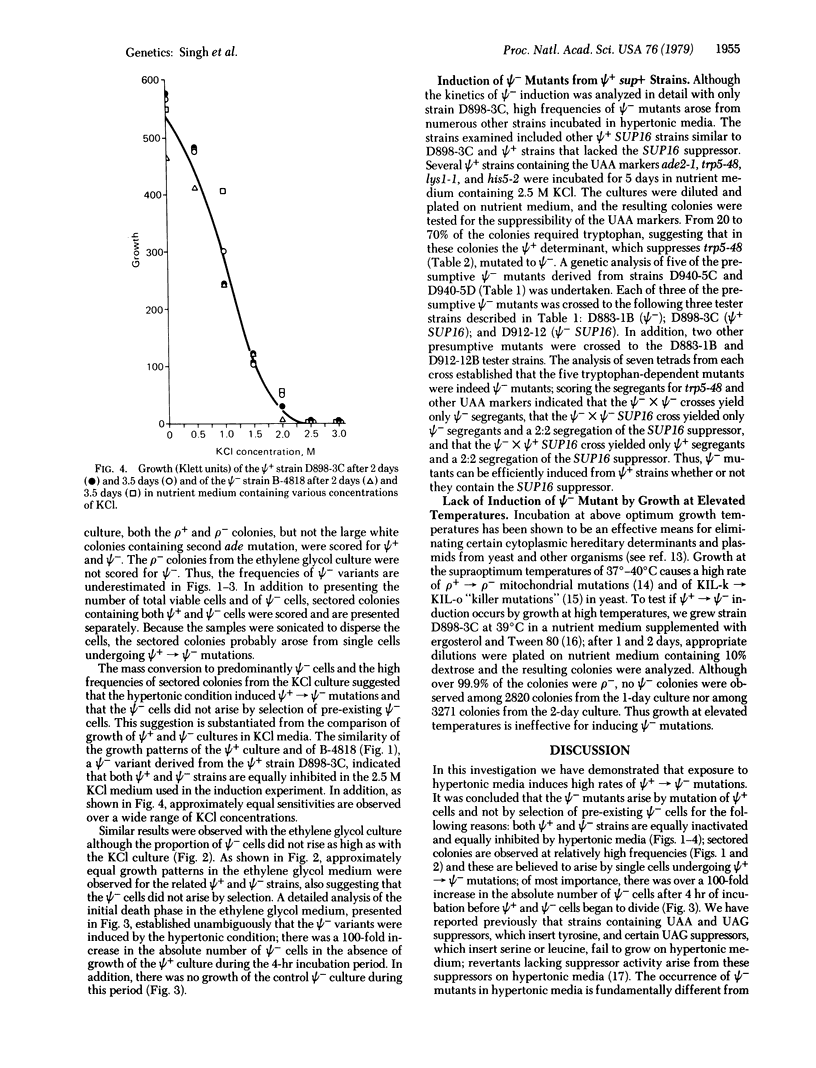
Abstract
The Ψ+ extrachromosomal determinant in the yeast Saccharomyces cerevisiae suppresses certain UAA markers and increases the efficiency of suppression of UAA suppressors and certain frameshift suppressors. Although the exact nature of Ψ+ determinant is unknown, it is believed to be a self-replicating cytoplasmic factor affecting some component of the translational machinery. In this report we describe growth conditions for efficient mutation or elimination of the Ψ+ determinant. Incubation of Ψ+ cultures in hypertonic nutrient medium resulted in rapid conversion to a culture containing predominantly Ψ- cells during the growth cycle. The kinetics of Ψ+ to Ψ- conversion established that the occurrence of Ψ- cells was due to induction and not to selection of pre-existing Ψ- cells. The results suggest that the replication of the Ψ+ determinant is sensitive to hypertonic conditions.
Keywords: nonsense suppression, Saccharomyces cerevisiae, cytoplasmic inheritance
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cox B. S. A recessive lethal super-suppressor mutation in yeast and other psi phenomena. Heredity (Edinb) 1971 Apr;26(2):211–232. doi: 10.1038/hdy.1971.28. [DOI] [PubMed] [Google Scholar]
- Culbertson M. R., Charnas L., Johnson M. T., Fink G. R. Frameshifts and frameshift suppressors in Saccharomyces cerevisiae. Genetics. 1977 Aug;86(4):745–764. doi: 10.1093/genetics/86.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Styles C. A. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R. A. Super-suppressors in Saccharomyces cerevisiae. Genetics. 1967 Aug;56(4):641–658. doi: 10.1093/genetics/56.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E. S., Grossman L. I., Krupnick D., Cryer D. R., Marmur J. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J Mol Biol. 1970 Sep 14;52(2):323–335. doi: 10.1016/0022-2836(70)90033-1. [DOI] [PubMed] [Google Scholar]
- LEDERBERG J. Cell genetics and hereditary symbiosis. Physiol Rev. 1952 Oct;32(4):403–430. doi: 10.1152/physrev.1952.32.4.403. [DOI] [PubMed] [Google Scholar]
- Leibowitz M. J., Wickner R. B. Pet18: a chromosomal gene required for cell growth and for the maintenance of mitochondrial DNA and the killer plasmid of yeast. Mol Gen Genet. 1978 Oct 4;165(2):115–121. doi: 10.1007/BF00269899. [DOI] [PubMed] [Google Scholar]
- Liebman S. W., Stewart J. W., Sherman F. Serine substitutions caused by an ochre suppressor in yeast. J Mol Biol. 1975 Jun 5;94(4):595–610. doi: 10.1016/0022-2836(75)90324-1. [DOI] [PubMed] [Google Scholar]
- McCready S. J., Cox B. S., McLaughlin C. S. The extrachromosomal control of nonsense suppression in yeast: an analysis of the elimination of [psi+] in the presence of a nuclear gene PNM. Mol Gen Genet. 1977 Feb 15;150(3):265–270. doi: 10.1007/BF00268125. [DOI] [PubMed] [Google Scholar]
- SHERMAN F. Respiration-deficient mutants of yeast. I. Genetics. Genetics. 1963 Mar;48:375–385. doi: 10.1093/genetics/48.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F. The effects of elevated temperatures on yeast. II. Induction of respiratory-deficient mutants. J Cell Comp Physiol. 1959 Aug;54:37–52. doi: 10.1002/jcp.1030540106. [DOI] [PubMed] [Google Scholar]
- STARR P. R., PARKS L. W. Effect of temperature on sterol metabolism in yeast. J Cell Comp Physiol. 1962 Apr;59:107–110. doi: 10.1002/jcp.1030590203. [DOI] [PubMed] [Google Scholar]
- Singh A. Nonsense suppressors of yeast cause osmotic-sensitive growth. Proc Natl Acad Sci U S A. 1977 Jan;74(1):305–309. doi: 10.1073/pnas.74.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAVLITZKI J. Sur les conditions de la formation de pigment chez une levure rouge. Rev Can Biol. 1951 Mar;10(1):48–59. [PubMed] [Google Scholar]
- Vodkin M. H., Fink G. R. A nucleic acid associated with a killer strain of yeast. Proc Natl Acad Sci U S A. 1973 Apr;70(4):1069–1072. doi: 10.1073/pnas.70.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. "Killer character" of Saccharomyces cerevisiae: curing by growth at elevated temperature. J Bacteriol. 1974 Mar;117(3):1356–1357. doi: 10.1128/jb.117.3.1356-1357.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Killer of Saccharomyces cerevisiae: a double-stranded ribonucleic acid plasmid. Bacteriol Rev. 1976 Sep;40(3):757–773. doi: 10.1128/br.40.3.757-773.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Twenty-six chromosomal genes needed to maintain the killer double-stranded RNA plasmid of Saccharomyces cerevisiae. Genetics. 1978 Mar;88(3):419–425. doi: 10.1093/genetics/88.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C. S., Cox B. S. Extrachromosomal elements in a super-suppression system of yeast. II. Relations with other extrachromosomal elements. Heredity (Edinb) 1972 Apr;28(2):189–199. doi: 10.1038/hdy.1972.24. [DOI] [PubMed] [Google Scholar]
- Young C. S., Cox B. S. Extrachromosomal elements in a super-suppression system of yeast. III. Enhanced detection of weak suppressors in certain non-suppressed psi-strains. Heredity (Edinb) 1975 Feb;34(1):83–93. doi: 10.1038/hdy.1975.8. [DOI] [PubMed] [Google Scholar]


