Abstract
Vinculin is a key regulator of the attachment of the actin cytoskeleton to the cell membrane at cellular adhesion sites that is crucial for processes like cell motility and migration, development, survival, and wound healing. Vinculin loss results in embryonic lethality, cardiovascular diseases, and cancer. Its tail domain, Vt, is crucial for vinculin activation and focal adhesion turnover and binds to the actin cytoskeleton and acidic phospholipids upon which it unfurls. The RNA binding protein raver1 regulates the assembly of focal adhesions transcriptionally by binding to vinculin. The muscle-specific splice form, metavinculin, is characterized by a 68 residue insert in the tail domain (MVt) and correlates with hereditary idiopathic dilated cardiomyopathy. Here we report that metavinculin can bind to raver1 in its inactive state. Our crystal structure explains this permissivity, where an extended coil unique to MVt is unfurled in the MVtΔ954:raver1 complex structure. Our binding assays show that raver1 forms a ternary complex with MVt and vinculin mRNA. These findings suggest that the metavinculin:raver1:RNA complex is constitutively recruited to adhesion complexes.
Keywords: adherens junction, cardiomyopathy, focal adhesion, RRM domain, RNA binding
Introduction
Vinculin is a highly conserved 117 kDa helix-bundle protein that provides essential links of the actin cytoskeleton to cell adhesion complexes. Vinculin and its alternatively spliced isoform coined metavinculin are co-expressed in muscle cells where they are required for proper formation of costameres and intercalated discs in cardiomyocytes. For example, targeted deletion of vinculin in mice leads to embryonic heart failure and vinculin+/- mice develop cardiomyopathies. In addition, mechanical load triggers increases in metavinculin expression, whereas there are marked reductions in metavinculin and vinculin levels in cardiomyopathies. Finally, sporadic and familial missense mutations in the metavinculin insert (A934V, ΔL954, and R975W) are associated with cardiomyopathies.
Vinculin is comprised of five helix bundle domains; three N-terminal seven-helix bundles (Vh1-Vh3) and one four-helix bundle form its head (VH) domain, which is connected to a C-terminal five-helix bundle tail (Vt) domain via a proline-rich loop.12 In its inactive, closed conformation vinculin binding to most of its partners is masked by intramolecular hydrophobic interactions of Vh1 with Vt. However, these interactions are severed by the binding of activators like talin to Vh1, leaving Vt free to binds its partners such as F-actin and raver1, a RNA binding protein harboring three RNA recognition motifs (RRM1-3) that appears to deliver mRNA cargo to focal adhesions when bound to vinculin. Further, the Vt domain also directs the formation of higher-order oligomers and this is facilitated by acidic phospholipids that are thought to unfurl regions of Vt.
We have shown that the 68-residue insert in the metavinculin tail (MVt) domain harbors a unique α-helix (H1′) and a preceding N-terminal coil that displaces the H1 α-helix and the preceding N-terminal coil specific to vinculin, forming a unique five-helix bundle.31 This helix swap imparts unique functions to metavinculin, as it directs its actin bundling and oligomerization properties.32 Surprisingly, here we report that metavinculin, unlike vinculin, can bind to raver1 in its closed conformation and that this interaction is permissive for raver1 to bind to vinculin RNA. The crystal structure of the metavinculin:raver1 complex explains this permissivity, which directs constitutive metavinculin functions.
Results
Metavinculin binding to raver1 is independent of its activation state
Activation of vinculin and metavinculin, for example following binding by the vinculin binding sites (VBS) of talin to the Vh1 domain, severs their head-tail interactions. This event is necessary for binding of the Vt domain of vinculin to F-actin and raver1. However, given the unique structure of MVt we tested if metavinculin could bind to tail-interacting partners in its inactive conformation. We thus performed F-actin co-sedimentation and native gel shift assays (Fig. 1). As shown for vinculin previously (Fig. 1b),15 native metavinculin failed to pellet F-actin in co-sedimentation assays, whereas talin-VBS3-activated metavinculin interacted with F-actin (Fig. 1c). Surprisingly, however, native gel shift analyses established that inactive full-length metavinculin bound to the RRM1-3 domains of raver1 while full-length vinculin does not (Fig. 1d). Raver1 alone was not a trigger for metavinculin activation because raver1-bound metavinculin does not bind to F-actin (Fig. 1c); thus, raver1 binding does not sever the metavinculin head-tail interaction. Finally, talin-VBS3-activated, raver1-bound metavinculin bound to F-actin. Therefore, raver1 can bind to inactive metavinculin; raver1 is not a metavinculin activator; and raver1, activated metavinculin, and F-actin can form a ternary complex.
Fig. 1.
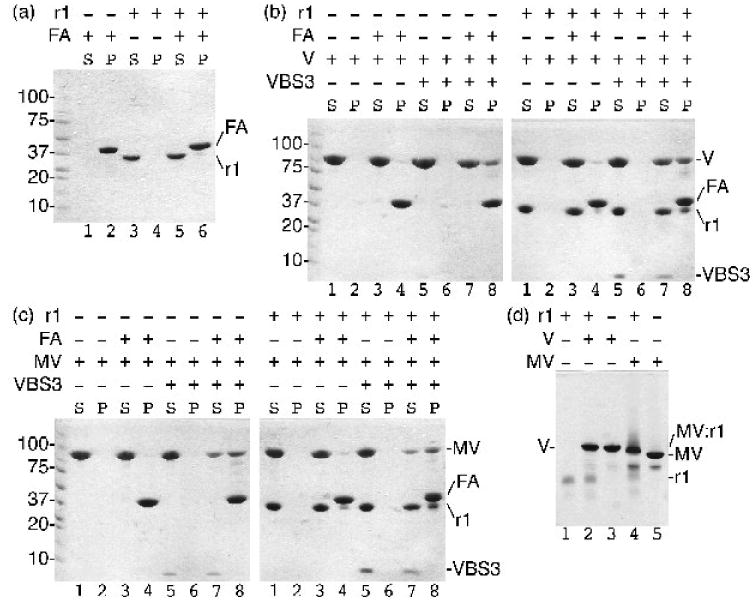
Raver1 binds to inactive metavinculin but not to inactive vinculin. Actin co-sedimentation assays as analyzed on a 8-25% gradient SDS PAGE gel established that:
(a) raver1 (residues 39-321) does not bind to F-actin (FA; raver1, r1, remains in the supernatant, S, while F-actin pellets, P); (b) raver1 binding is not sufficient to activate the latent F-actin binding properties of vinculin (left gel: vinculin, V, remains in the supernatant, lanes 1-2; inactive vinculin does not bind to F-actin, FA, lanes 3-4; vinculin activated by talin VBS3, VBS3, remains in the supernatant, lanes 5-6; the vinculin:VBS3 complex binds to F-actin, lanes 7-8. Right gel: vinculin and raver1, r1, are soluble, lanes 1-2; vinculin and raver1 do not bind to F-actin, lanes 3-4; vinculin, raver1, and VBS3 remain soluble, lanes 5-6; vinculin activated by VBS3 pellets with F-actin, lanes 7-8); and (c) raver1 binding is not sufficient to activate the latent F-actin binding properties of metavinculin (left gel: metavinculin, MV, remains in the supernatant, lanes 1-2; inactive MV does not bind to F-actin, lanes 3-4; MV activated by VBS3 remains in the supernatant, lanes 5-6; the MV:VBS3 complex binds to F-actin, lanes 7-8. Right gel: MV and raver1 are soluble, lanes 1-2; MV and raver1 do not bind to F-actin, lanes 3-4; MV, raver1, and VBS3 remain soluble, lanes 5-6; MV activated by VBS3 pellets with F-actin, lanes 7-8)
(d) Native gel shift mobility assay of raver1 alone (lane 1), vinculin, V, alone (lane3), metavinculin, MV, alone (lane 5), vinculin incubated with raver1 (lane 2), and metavinculin incubated with raver1 (lane 4) shows that a new band is formed corresponding to MV:raver1 complex formation under physiological conditions without pre-activation of MV by a VBS. In contrast, no vinculin:raver1 complex is formed (lane 2).
The metavinculin extended coil is unfurled in the MVtΔ954:raver1 structure
To define the molecular basis of how metavinculin interacts with raver1 in its closed conformation we solved the crystal structures of MVtΔ954 (residues 945-1133) in complex with raver1 (residues 39-321) to 2.5 Å resolution (Fig. 2; Table I). We also obtained an electron density map for the native MVt:RRM1-3 structure to 3.2 Å resolution and while one of the unit cell dimensions cannot be determined with absolute confidence the lower resolution MVt:RRM1-3 structure is essentially the same as the higher resolution MVtΔ954:RRM1-3 structure reported here. The cardiomyopathy associated deletion mutant, Leu954, is disordered in our structure where the final model comprises raver1 and metavinculin residues 39-319 and 955-1133, respectively. Crystal contacts generate another large metavinculin-raver1 interface (Fig. 3) but previous mutagenesis and binding studies unambiguously identified the interface in solution.28
Fig. 2.
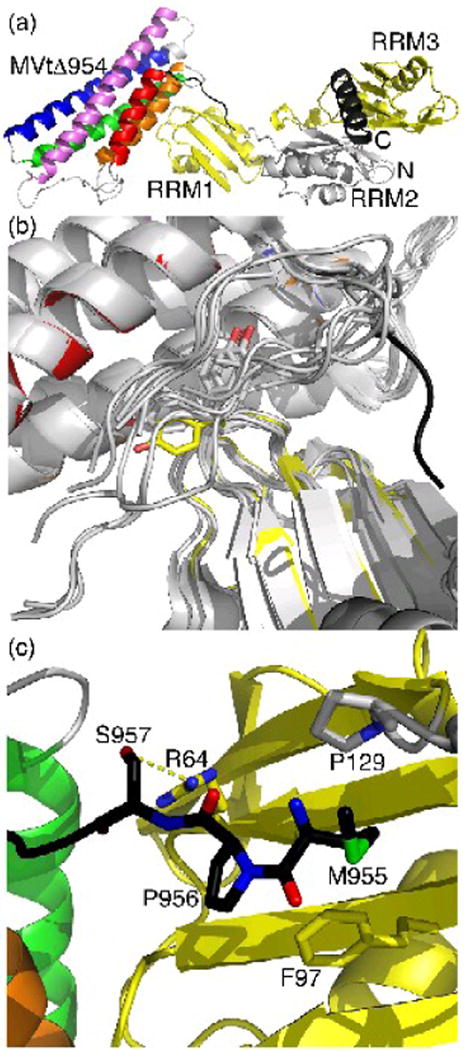
Unfurling of MVtΔ954 leads to distinct interactions with raver1.
(a) Cartoon drawing of the RRM1-3 domains of raver1 (RRM1 and RRM3 in yellow, RRM2 in grey, N- and C-terminal α-helices in dark grey), which induces the unfurling of the metavinculin-specific extended coil region that precedes the H1′ α-helix (black, residues 950-963). The MVtΔ954 α-helices are colored according to the rainbow colors (red, α-helix H1′, residues 964-979; orange, H2, residues 986-1,006; green, H3, residues 1,011-1,040; blue, H4, residues 1,043-1,072; and violet, H5, residues 1,081-1,114). The termini of MVtΔ954, and the MVtΔ954 and RRM1-3 domains are labeled.
(b) The unfurled extended coil of MVtΔ954 interacts with the hydrophobic face of the RRM1 domain of raver1. Superposition of the two five-helix bundle meta/vinculin molecules in the asymmetric unit of full-length human metavinculin, full-length human metavinculinΔ954, full-length human vinculin, Vt bound to RRM1-3, and the four Vt bound to RRM1 (only the tail domain is shown, grey) onto MVtΔ954:RRM1-3 shows that the extended coil (black) is in a distinct conformation in the MVtΔ954:raver1 structure (same color coding as in Figure 2a). In its metavinculin-bound state, raver1 (yellow) residue Tyr92, labeled and shown in sticks presentation, rotates almost 135° into the Vt extended coil-binding site. A similar movement is only seen in the raver1 apo structure, which is superimposed as well (grey).
(c) Novel MVtΔ954-raver1 interactions of the metavinculin extended coil. Ser957 engages in a hydrogen bond with Arg64, Pro956 binds to Phe97, and Met955 binds to P/ro129.
Table I. MVtΔ954:raver1 X-ray data reduction and crystallographic refinement statistics.
| (A) X-ray data reduction statistics | |
| Space group | P6522 |
| Unit cell dimensions | |
| a=b, c | 164.9 Å, 102.6 Å |
| Resolution (last shell) | 35.7 Å – 2.536 Å (2.545 Å – 2.536 Å) |
| Total measurements (last shell) | 595,557 (3,069) |
| Number of unique reflections (last shell) | 27,719 (256) |
| Wavelength | 1 Å |
| R-merge* (last shell) | 0.096 (0.458) |
| Average I/σ(I) (last shell) | 31.9 (6) |
| Completeness (last shell) | 1 (1) |
| Multiplicity (last shell) | 21.5 (12) |
| (B) Crystallographic refinement statistics | |
| Resolution (last shell) | 35.7 Å – 2.54 Å (2.64 Å – 2.54 Å) |
| No. of reflections working set (last shell) | 26,220 (2,703) |
| No. of reflections test set (last shell) | 1,390 (152) |
| R-factor‡ working set (last shell) | 0.169 (0.18) |
| R-free§ (last shell) | 0.204 (0.245) |
| No. of amino acid residues | 467 |
| No. of protein atoms | 3,628 |
| No. of solvent molecules | 232 |
| No. of glycerol residues | 13 |
| No. of Hepes residues | 1 |
| Average B-factor | |
| Protein | 44.5 Å2 |
| Solvent | 47.2 Å2 |
| Glycerol | 69.4 Å2 |
| Hepes | 133 Å2 |
| R.m.s.d from ideal geometry | |
| Bond lengths | 0.01 Å |
| Bond angles | 1.07° |
where < | Fcalc | > denotes the expectation of | Fcalc(hkl)| used in defining the likelihood refinement target
The free R-factor is a cross-validation residual calculated by using about 5% reflections, which were randomly chosen and excluded from the refinement
Fig. 3.
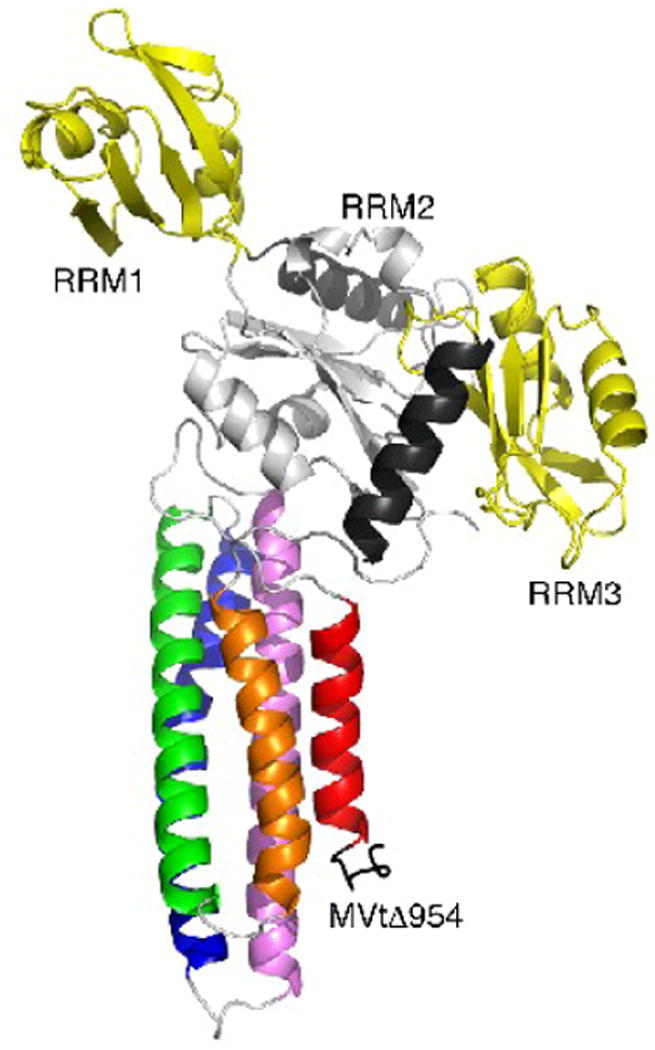
Crystal contacts of MVtΔ954 that do not involve the raver1 RRM1 domain.
Cartoon drawing (same color coding as in Fig. 2a) of the crystal contacts seen in both MVtΔ954:RRM1-3 and Vt:RRM1-3 structures (PDB entry 3h2u) involving mainly the RRM2 domain (residues 130-220) and the C-terminal α-helix (residues 303-319) with the metavinculin C-terminus (residues 1114-1133): hydrophobic interactions are manifest for MVtΔ954 residue Trp979 with RRM2 residue Pro197, Ile1113 with Leu151 and Leu198, and Trp 1131 with Ala313 and Ala317, while polar interactions involve Lys982, Thr1040, Ser1113, Lys1114, Ile1115, Ala1119, Gly1120, Thr1122, Leu1123, Arg1124, Arg1127, and Lys1128 with Glu150, Arg153, Ser191, Arg188, Asp192, Lys196, and Thr202. RRM3 residues Phe264 and Arg227 make further interactions with MVtΔ954 Tyr1132 and Gln1133, respectively. However, in solution, the RRM2-RRM3 domains alone (residues 129-321) do not bind to the vinculin tail domain and the RRM1 E120K and R121E mutations disrupt the binding of mutated raver1 RRM1-3 (residues 39-321) to the vinculin tail domain.28
As seen for the Vt:raver1 structures,28 the three RRM domains of raver1 in complex with metavinculin have a canonical RRM structure, with five-stranded anti-parallel β-strands and two α-helices (Fig. 2a). Further, as seen in the native structure of MVt,31 raver1-bound MVtΔ954 is a five-helix bundle domain (α-helices H1′ and H2-H5) where the Vt α-helix H1 and its preceding extended coil region are disordered and replaced with a highly homologous α-helix H1′ and an unrelated N-terminal extended coil region of the metavinculin-specific insert. Importantly, unlike in the Vt:raver1 structure,28 the N-terminal extended coil that precedes the H1′ α-helix is unfurled in the metavinculin-raver1 interaction (Fig. 2). Furthermore, superposition of Vt or MVt in complex with raver1 and full-length vinculin or metavinculin structures shows that the extended coil is only unfurled in the MVtΔ954:RRM1-3 structure (Fig. 2b). Thus, raver1 binding unfurls a portion of the tail domain yet this does not activate metavinculin (Fig. 1c).
Further, the binding mode of the unfurled coil of metavinculin with raver1 is unique, where the C-terminus of the unfurled coil binds to the hydrophobic β-sheet RNA binding domain of the RRM1 (residues Leu62 and Phe97; Fig. 2c; Table II) through the agency of hydrophobic metavinculin residues 955-MPS-957. By contrast, in vinculin the structurally equivalent residues 888-QKA-890 do not interact with raver1.28 Finally, in the Vt:raver1 structure, Tyr92 of the RRM1 domain of raver1 engages in hydrophobic interactions with Phe885 and Pro886 of the N-terminal strand that precedes the H1 α-helix,28 whereas when bound to metavinculin Tyr92 of raver1 is rotated 135° (Fig. 2b) allowing the Lys93 side chain to occupy its position and clashes with superimposed vinculin residue Glu883. Superposition of the structures of apo raver1 or when bound to vinculin and metavinculin shows that Tyr92 only has the conformation seen in our MVtΔ954:RRM1-3 structure when not bound by vinculin. Thus, vinculin binding, but not metavinculin binding induces this conformational change.
Table II. MVtΔ954 residues in contact with raver1 residues.
| (A) Hydrophobic inter actions | ||
|---|---|---|
| MVtΔ954 residues | raver1 residues (RRM1 location) | distance |
| Met955 CB | Pro129 CG (C-terminus) | 4.4 Å |
| Pro956 CG | Phe97 CZ (β3) | 4 Å |
| Arg992 CD | Asp69 CB (β1-α1 loop) | 4.3 Å |
| Leu995 CD2 | Tyr92 CD1 (β2-β3 loop) | 3.7 Å |
| Leu996 CD2 | Asp69 CB (β1-α1 loop) | 4.2 Å |
| Gln1016 CB | Glu120 CB (β4-β5 loop) | 3.6 Å |
| (B) Polar interactions | ||
|---|---|---|
| MVtΔ954 residues | raver1 residues (RRM1 location) | distance |
| Ser957 OG | Arg64 NH2 (β1) | 3.1 Å |
| His973 NE2 | Tyr92 OH (β2-β3 loop) | 3.4 Å |
| Glu999 OE1 | Arg121 NH2 (β4-β5 loop) | 2.8 Å |
| Glu999 OE2 | Arg121 NE (β4-β5 loop) | 2.8 Å |
| Arg1002 NH1 | Tyr92 O (β2-β3 loop) | 2.8 Å |
| Ser1008 OG | Glu122 OE1 (β5) | 2.9 Å |
| Gly1009 O | Arg117 NH1 (β4) | 3.1 Å |
| Arg1012 NH2 | Glu120 OD2 (β4-β5 loop) | 2.9 Å |
| Arg1012 NE | Glu120 OE1 (β4-β5 loop) | 2.8 Å |
| Asp1020 OD2 | Arg121 NH2 (β4-β5 loop) | 2.9 Å |
| Asp1020 OD2 | Arg119 NH2 (β4-β5 loop) | 3.2 Å |
Movements in raver1 accommodate the unfurled coil of metavinculin
In the Vt:raver1 structure the N-terminal extended coil of Vt interacts with its α-helix H1 to generate a hydrophobic core (Phe885, Pro886, and Met899) that binds to Tyr92 of raver1. Additionally, two electrostatic interactions (raver1 Arg91 with vinculin Asp907 and Arg910 with Glu884) and one hydrogen bond (side chain of Arg903 and the main chain carboxyl group of Pro886) stabilize the specific loop conformation seen in Vt:raver1 complex.28 However, in the metavinculin-raver1 interaction the N-terminus of the unfurled coil interacts with a hydrophobic surface of the RRM1 domain of raver1 (Fig. 2).
The RRM1 domain in the apo raver1 structure is rotated 35° relative to the RRM2-RRM3 domains. This rotation was thought to be induced by a bound sulfate anion, which binds to Arg59 and Glu159, disrupting their electrostatic interaction, and which appears to mimic an RNA binding site.36 In the MVtΔ954:RRM1-3 structure, a glycerol is found within 3.6 Å of the sulfate anion binding site (Fig. 4), again supporting the notion that this is the RNA binding site. Additionally, even though the side-chains of this binding site in the MVtΔ954:RRM1-3 structure have a conformation akin to that of the Vt-bound raver1 structure, the relative RRM1 domain movement resembles that of the apo raver1 structure.
Fig. 4.
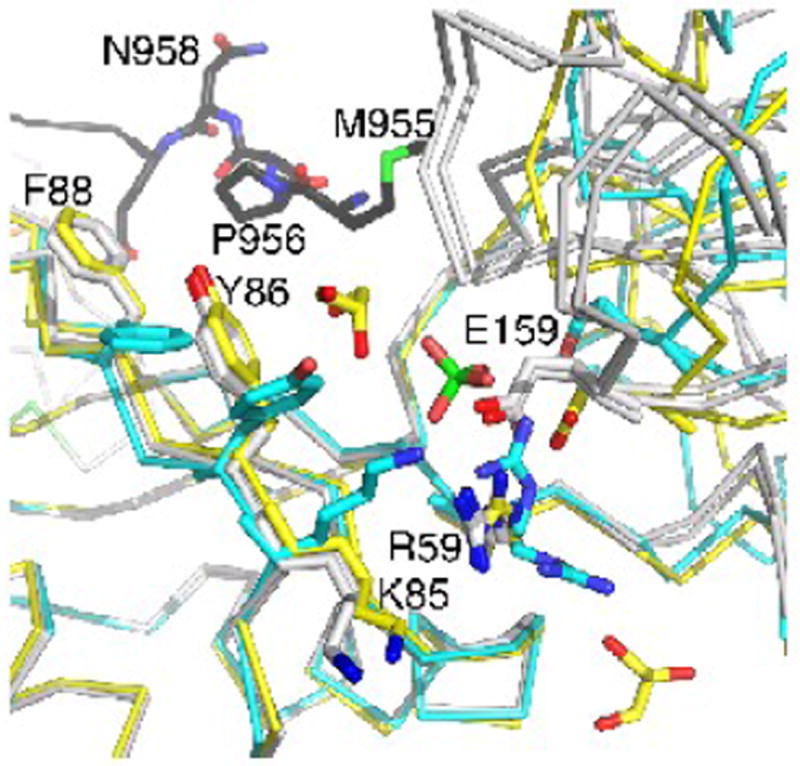
The RRM2-RRM3 domains are rotated when bound by metavinculin.
Superposition of the apo raver1 structure (cyan) onto raver 1 bound to MVtD954 (yellow) or to Vt (grey) showing the relative RRM2-RRM3 movement. The unfurled extended coil of MVtΔ954 binds in the cleft between RRM1 and the RRM2-RRM3 domains. Key residues, the sulfate in the apo raver1 structure, and two of the bound glycerols in the MVtΔ954:raver1 structure are shown in ball-and-stick representation.
The MVt:raver1 complex is permissive for binding to RNA
Since the C-terminus of the extended coil of MVtΔ954 interacts with the hydrophobic β-sheet of the RRM1, we reasoned that this might disrupt raver1 RNA binding. To test this we performed electrophoretic mobility shift assays (EMSA) using biotin-labeled vinculin RNA (3089-UCAUGCAGUCUG-3100), a preferred raver1 cargo.28 Notably, vinculin RNA bound as avidly to the MVt:RRM1-3 complex as to the raver1 RRM1-3 domains alone while it did not bind to the RRM2-RRM3 domains (Fig. 5). Mutation of raver1 residue Lys93, which is located in the canonical RNP1 motif of RRM1, to glutamate impaired vinculin RNA binding by the MVt:RRM1-3 complex (Fig. 5). Thus, the MVt:raver1 complex can bind to RNA and raver1 residue Lys93 is important for RNA interaction.
Fig. 5.
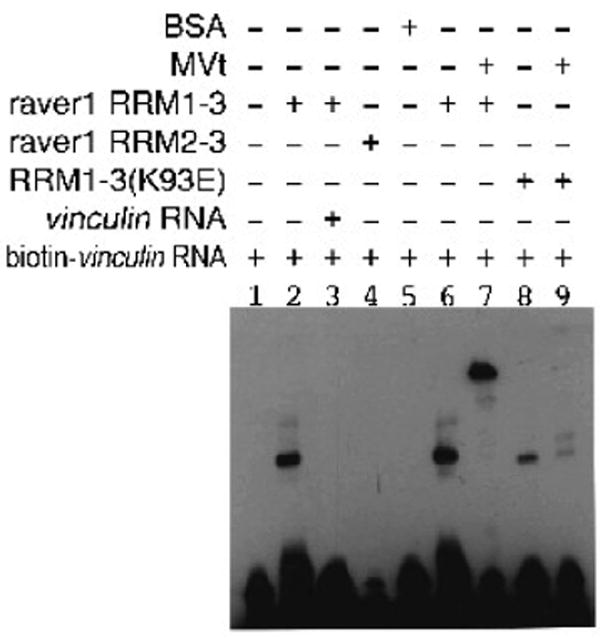
MVt-raver1 interactions are permissive for binding to RNA.
EMSA of biotin-labeled vinculin RNA and raver1 proteins in complex with MVt establishes that the metavinculin:raver1 complex can bind to this RNA. Note that RRM2-RRM3 protein failed to bind vinculin RNA (lane 4) and that vinculin RNA binding was impaired by the K93E raver1 mutant (lanes 8, 9).
Discussion
Severing the intramolecular head-tail interaction of both vinculin and metavinculin has long thought to be necessary for their binding to key partners, which allows their respective tail domains bind to F-actin, the acidic phospholipid PIP2 and raver1. Indeed, severing this intramolecular clamp is essential for the interactions of both vinculin and metavinculin with F-actin and PIP2, and for vinculin to bind to raver1. Thus, it was very surprising that our structure and biochemical studies demonstrated that metavinculin in its native, inactive, and closed-clamp conformation was indeed capable of binding to raver1. These findings underscore the unique effects of the helix swap on the structure and function of metavinculin and they strongly support the notion that even in its closed conformation metavinculin is not a passive molecule, where the N-terminal extended coil that precedes its unique α-helix H1′ is unfurled when bound to raver1, and where the conformation of the metavinculin tail domain when bound to raver1 differs from its native, unbound state. Our crystal structure fully explains these metavinculin-raver1 interactions. However, these alterations in metavinculin structure are not sufficient to activate metavinculin or affect it's binding to F-actin, nor do they affect the binding of RNA cargo to raver1. Collectively, these findings support a model where in muscle cells even native metavinculin is constitutively bound to RNA-bearing raver1, a scenario that would allow for local and rapid de novo synthesis of components of adhesion complexes and intercalated discs.
Materials and Methods
Cloning, protein expression, and purification
Human full-length metavinculin, MVt (residues 856-1134), MVtΔ954 (residues 856-1133), RRM1-3 (residues 39-321), and RRM2-RRM3 domains (residues 129-321) were generated as described. The K93E raver1 mutant was generated by site-directed mutagenesis. For crystallization, the MVtΔ954:RRM1-3 complex was obtained by co-purification using a Superdex 200 size exclusion chromatography column.
MVtΔ954:raver1 crystallization and X-ray data collection
Complex crystals containing MVtΔ954 and RRM1-3 were grown by sitting drop vapor diffusion at 23 °C using a 1:1 ratio of protein (24.5 mg/ml) and reservoir solution [0.1 M Hepes-NaOH pH 7, 5% (v/v) tacsimate, and 10% (w/v) PEG MME 5K]. Prior to data collection, crystals were briefly transferred to mother liquor contianing 15% glycerol and flash cooled in liquid nitrogen. Data were collected at the APS SER-CAT 22BM beam line and processed using autoPROC39 using XDS,40 POINTLESS and SCALA.41 The X-ray diffraction data statistics are shown in Table I.
Structure determination of the MVtΔ954:raver1 complex
The structure was solved by molecular replacement using the program MOLREP and the search model consisted of MVt (residues 961-1129 of chain A in PDB entry 3myi) and RRM1-3 (residues 39-294 in chain D of PDB entry 3h2u). This gave a clear solution in spacegroup P6522 (other screw axes in P6x22 as well as P6x were tested) which was refined in BUSTER42 to an initial crystallographic and free R-factor of 0.292 and 0.328, respectively. ARP/wARP43 was then used to allow rebuilding from that starting model. This initial 447-residue model was subjected to several cycles of BUSTER refinement and model-building in Coot44 resulted in the model described in Table I.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed using purified proteins and 5′-biotin end-labeled 12-mer vinculin RNA (UCAUGCAGUCUG). The binding mixtures contained 0.4 μM biotinylated RNA, purified 26 μM raver1 protein, 10 mM Tris-HCl pH 7.5, 50 mM KCl, and 1mM DTT. After incubating for 20 min at room temperature electrophoresis was performed on 8% polyacrylamide gel at 100 V and the gel was transferred to nylon membrane by electroblotting at 380 mA for 30 min. Transferred RNA was cross-linked for 1 min using UV cross-linker. Biotin labeled RNA was detected using a LightShift Chemiluminescent EMSA kit and X-ray film.
Native gel electrophoresis
The native gel electrophoresis of RRM1-3 (30 μM) bound to metavinculin (30 μM) was performed using the Phast system (Amersham). Proteins were incubated for 15 min at 23 °C and loaded onto a 10-15% gradient polyacrylamide gel with native buffer strips. Complexes were visualized by Coomassie Blue staining.
Actin co-sedimentation assays
F-actin co-sedimentation assays were performed as previously described21 in PBS containing 25 μM raver1, 12 μM metavinculin, and 40 μg of polymerized F-actin. Actin was purchased from Cytoskeleton Inc., BSA from Sigma, and talin-VBS3 (residues 1944-1969) was synthesized. Samples were incubated for 20 min at room temperature +/- 10-fold molar excess of talin-VBS3 and centrifuged at 95,000 × g at 25 °C for 15 min. The pellets were suspended in SDS sample buffer and equal amounts of supernatants and resuspended pellets analyzed on a 8-25% gradient polyacrylamide PhastGels using SDS buffer strips. Proteins were stained with Coomassie Blue.
Research Highlights.
Metavinculin binds to raver1 at muscle cell adhesion complexes via its tail domain
Raver1 forms a ternary complex with metavinculin and vinculin mRNA
Metavinculin binds to raver1 in both its inactive and activated states
Crystal structure of the metavinculin:raver1 complex explain this permissivity
The metavinculin:raver1:RNA complex is constitutively recruited to adhesion complexes
Acknowledgments
We are grateful to our colleagues at Scripps Florida, in particular to John Cleveland for critical review and editing of the manuscript, to Zhen Wu and Philippe Bois for sequencing, Philippe Bois for very helpful discussions, and Krishna Chinthalapudi for providing metavinculin and vinculin proteins for the actin pull-down assays. We are also indebted to the staff of the Advanced Photon Source, SBC-CAT, for synchrotron support. TI is supported by grants from the National Institutes of Health (NIGMS) and by start-up funds provided to Scripps Florida from the State of Florida. JHL was supported by a fellowship from the American Heart Association. This is publication no. 20695 from The Scripps Research Institute.
Abbreviations
- Vt
vinculin tail domain
- MV
full-length human metavinculin
- MVt
metavinculin tail domain
- VH
vinculin head domain
- RRM
RNA recognition motif
- VBS
vinculin binding site
- RMSD
root mean square deviation
- EMSA
electrophoretic mobility shift assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Geiger B. A 130K protein from chicken gizzard: its localization at the termini of microfilament bundles in cultured chicken cells. Cell. 1979;18:193–205. doi: 10.1016/0092-8674(79)90368-4. [DOI] [PubMed] [Google Scholar]
- 2.Jockusch BM, Isenberg G. Interaction of α-actinin and vinculin with actin: opposite effects on filament network formation. Proc Natl Acad Sci U S A. 1981;78:3005–9. doi: 10.1073/pnas.78.5.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkins JA, Lin S. High-affinity interaction of vinculin with actin filaments in vitro. Cell. 1982;28:83–90. doi: 10.1016/0092-8674(82)90377-4. [DOI] [PubMed] [Google Scholar]
- 4.Pardo JV, Siliciano JD, Craig SW. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements (“costameres”) mark sites of attachment between myofibrils and sarcolemma. Proc Natl Acad Sci U S A. 1983;80:1008–12. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkin AM, Ornatsky OI, Glukhova MA, Koteliansky VE. Immunolocalization of meta-vinculin in human smooth and cardiac muscles. J Cell Biol. 1988;107:545–53. doi: 10.1083/jcb.107.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gimona M, Small JV, Moeremans M, Van Damme J, Puype M, Vandekerckhove J. Porcine vinculin and metavinculin differ by a 68-residue insert located close to the carboxy-terminal part of the molecule. Embo J. 1988;7:2329–34. doi: 10.1002/j.1460-2075.1988.tb03076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witt S, Zieseniss A, Fock U, Jockusch BM, Illenberger S. Comparative biochemical analysis suggests that vinculin and metavinculin cooperate in muscular adhesion sites. J Biol Chem. 2004;279:31533–43. doi: 10.1074/jbc.M314245200. [DOI] [PubMed] [Google Scholar]
- 8.Maeda M, Holder E, Lowes B, Valent S, Bies RD. Dilated cardiomyopathy associated with deficiency of the cytoskeletal protein metavinculin. Circulation. 1997;95:17–20. doi: 10.1161/01.cir.95.1.17. [DOI] [PubMed] [Google Scholar]
- 9.Zemljic-Harpf AE, Ponrartana S, Avalos RT, Jordan MC, Roos KP, Dalton ND, Phan VQ, Adamson ED, Ross RS. Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am J Pathol. 2004;165:1033–44. doi: 10.1016/S0002-9440(10)63364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87:169–74. doi: 10.1016/j.ymgme.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Olson TM, Illenberger S, Kishimoto NY, Huttelmaier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–7. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 12.Borgon RA, Vonrhein C, Bricogne G, Bois PR, Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–97. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Johnson RP, Craig SW. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J Biol Chem. 1994;269:12611–9. [PubMed] [Google Scholar]
- 14.Wachsstock DH, Wilkins JA, Lin S. Specific interaction of vinculin with α-actinin. Biochem Biophys Res Commun. 1987;146:554–60. doi: 10.1016/0006-291x(87)90564-x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RP, Craig SW. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 1995;373:261–4. doi: 10.1038/373261a0. [DOI] [PubMed] [Google Scholar]
- 16.Brindle NP, Holt MR, Davies JE, Price CJ, Critchley DR. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem J. 1996;318(Pt 3):753–7. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, Vermeulen S, van Roy F, Adamson ED, Takeichi M. α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol. 1998;142:847–57. doi: 10.1083/jcb.142.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, Yamada KM, Aota S. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izard T, Vonrhein C. Structural basis for amplifying vinculin activation by talin. J Biol Chem. 2004;279:27667–78. doi: 10.1074/jbc.M403076200. [DOI] [PubMed] [Google Scholar]
- 20.Bois PR, Borgon RA, Vonrhein C, Izard T. Structural dynamics of α-actinin-vinculin interactions. Mol Cell Biol. 2005;25:6112–22. doi: 10.1128/MCB.25.14.6112-6122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Bois PR, O'Hara BP, Nietlispach D, Kirkpatrick J, Izard T. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J Biol Chem. 2006;281:7228–36. doi: 10.1074/jbc.M510397200. [DOI] [PubMed] [Google Scholar]
- 22.Izard T, Tran Van Nhieu G, Bois PR. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J Cell Biol. 2006;175:465–75. doi: 10.1083/jcb.200605091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park H, Lee JH, Cossart P, Gouin E, Izard T. The Rickettsia surface cell antigen 4 applies mimicry to bind to and activate vinculin. J Biol Chem. 2011;286:35096–35103. doi: 10.1074/jbc.M111.263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park H, Valencia-Gallardo C, Sharff A, Tran Van Nhieu G, Izard T. A novel Vinculin binding site of the IpaA invasin of Shigella. J Biol Chem. 2011;286:23214–23224. doi: 10.1074/jbc.M110.184283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yogesha SD, Sharff A, Bricogne G, Izard T. Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin. Protein Science. 2011;20:1471–6. doi: 10.1002/pro.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yogesha SD, Rangarajan ES, Vonrhein C, Bricogne G, Izard T. Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin. Protein Science. 2012;21:583–8. doi: 10.1002/pro.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–86. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Rangarajan ES, Yogesha SD, Izard T. Raver1 interactions with Vinculin and RNA Suggest a Feed-Forward Pathway in Directing mRNA to Focal Adhesions. Structure. 2009;17:833–842. doi: 10.1016/j.str.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson RP, Craig SW. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem Biophys Res Commun. 1995;210:159–64. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 30.Palmer SM, Playford MP, Craig SW, Schaller MD, Campbell SL. Lipid Binding to the Tail Domain of Vinculin: SPECIFICITY AND THE ROLE OF THE N AND C TERMINI. J Biol Chem. 2009;284:7223–31. doi: 10.1074/jbc.M807842200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangarajan ES, Lee JH, Yogesha SD, Izard T. A helix replacement mechanism directs metavinculin functions. PLoS ONE. 2010;5:e10679. doi: 10.1371/journal.pone.0010679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudiger M, Korneeva N, Schwienbacher C, Weiss EE, Jockusch BM. Differential actin organization by vinculin isoforms: implications for cell type-specific microfilament anchorage. FEBS Lett. 1998;431:49–54. doi: 10.1016/s0014-5793(98)00723-6. [DOI] [PubMed] [Google Scholar]
- 33.Madl T, Sattler M. Adhesion dance with raver. Structure. 2009;17:781–3. doi: 10.1016/j.str.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 34.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–41. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Izard T, Evans G, Borgon RA, Rush CL, Bricogne G, Bois PR. Vinculin activation by talin through helical bundle conversion. Nature. 2004;427:171–5. doi: 10.1038/nature02281. [DOI] [PubMed] [Google Scholar]
- 36.Rangarajan ES, Lee JH, Izard T. Apo raver1 structure reveals distinct RRM domain orientations. Protein Science. 2011;20:1464–70. doi: 10.1002/pro.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huttelmaier S, Bubeck P, Rudiger M, Jockusch BM. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur J Biochem. 1997;247:1136–42. doi: 10.1111/j.1432-1033.1997.01136.x. [DOI] [PubMed] [Google Scholar]
- 38.Sun N, Critchley DR, Paulin D, Li Z, Robson RM. Human α-synemin interacts directly with vinculin and metavinculin. Biochem J. 2008;409:657–67. doi: 10.1042/BJ20071188. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 42.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek P, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.9. Cambridge: United Kingdom: Global Phasing Ltd.; 2011. [Google Scholar]
- 43.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature Protocols. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]


