Abstract
The ubiquitin proteasome system (UPS) is important in maintaining protein homeostasis. NFE2-related factor 1 (Nrf1), a transcription factor in the cap “n” collar (CNC) basic leucine zipper family, regulates expression of cytoprotective genes. It was previously shown that liver-specific knockout of Nrf1 (Nrf1LKO) leads to hepatic cell death, steatohepatitis and cancer. However, the mechanisms underlying these pathologies are not clear. Here, we report that Nrf1 is critical for proteasome gene expression in the liver. Liver-specific knockout of Nrf1 results in impaired basal and induced expression of proteasome genes, and diminished proteasome activity in hepatocytes. In addition, our findings demonstrated that ER stress signaling pathway was also activated in Nrf1LKO livers. Inhibition of proteasome activity leads to ER stress in Nrf1-deficient hepatocytes, prompting the development of steatosis in the liver. Our results indicate that Nrf1 plays an integral role in the maintenance of proteasome function in hepatocytes and in the prevention of liver steatosis development. Moreover, these results highlight an association between proteasome dysfunction, ER stress and steatosis.
Keywords: Nrf1, proteasome, ER stress, steatohepatitis, transcriptional regulation
Introduction
The Ubiquitin-Proteasome System (UPS) is the major intracellular proteolytic pathway in the cell [1, 2]. The UPS plays a major role in the degradation of mutant proteins, proteins that are terminally misfolded, or damaged by oxidative stress [3, 4]. In addition, the UPS controls the turnover of regulatory molecules involved in gene transcription, cell cycle control, and various signal transduction pathways. It is crucial for cells to maintain adequate proteasomal function, as aberrations in the UPS have been shown to contribute to various pathological conditions in humans [5, 6]. In neurodegenerative disorders, apoptosis of neurons is associated with the accumulation of mutant proteins and proteasome dysfunction [7, 8]. A number of liver diseases, including non-alcoholic steatohepatitis [9], alcoholic cirrhosis [10], and hepatocellular carcinoma [11], show accumulation of ubiquitin-conjugated proteins suggesting that proteasome function is also compromised in these conditions [12].
Proteins destined for proteolysis by the proteasome are tagged by covalent attachment of polyubiquitin chains and subsequently recognized by the 26S proteasome for degradation [13]. The 26S proteasome is a multi-protein complex consisting of a central proteolytic core (20S) particle with regulatory caps (19S) at either end. The core is arranged into two outer and inner rings, each consisting of seven different alpha- and beta-subunits, respectively. Each 19S particle is made of ATPase (Rpt 1-6) and non-ATPase (Rpn 1-14) subunits. The outer rings of the core regulate access of protein substrates to the inner chamber that contains the proteolytic sites. The 19S cap functions to bind, unfold, and regulate entry of polyubiquitinated proteins into the 20S core particle, where they are degraded into small peptides [14, 15].
Nuclear factor erythroid-derived 2-related factor 1 (Nrf1) is a member of the CNC subfamily of basic-leucine zipper transcription factors [16]. CNC factors form heterodimers with small-Maf-proteins and regulate transcriptional activation through the antioxidant response element (ARE) located at the promoter region of various antioxidant genes [17, 18]. Antioxidant genes regulated by Nrf1 include those encoding NAD(P)H:quinone oxidoreductase 1, metallothioneins, glutamate cysteine ligase catalytic and modifier subunits that are involved in glutathione biosynthesis and hemeoxygenase 1 [19-22]. Aside from antioxidant genes, Nrf1 has been shown to regulate genes involved in development and other cellular functions [23]. Osterix, a zinc finger transcription factor that plays an important role in the differentiation of osteoblast and bone formation, has been shown to be regulated by Nrf1 [24]. Nrf1 has also been reported to function as a repressor of transcription. Nrf1 interacts with C/EBP-β to repress expression of the dentin sialophosphoprotein (DSPP) gene in undifferentiated odontoblast [25], and Nrf1 has also been implicated in the negative regulation of iNOS expression [26].
Recent findings indicate that Nrf1 is also involved in regulating proteasome gene expression. Inactivation of Nrf1 in neurons leads to a coordinate down-regulation of Psma and Psmb genes encoding alpha- and beta-subunits of the 20S core, as well as components of the 19S regulatory subcomplex, and neurodegeneration [27]. While these findings indicate that Nrf1 modulates constitutive expression of proteasome genes in neurons, studies in both human and mouse cells demonstrate that induction of proteasome subunit genes in response to proteasome inhibition is also Nrf1-dependent [28]. These studies suggest a regulatory role for Nrf1 beyond oxidative stress response. However, the function of Nrf1 in regulating proteasome activity in other tissue compartments remained to be determined. Previously, we showed that inactivation of Nrf1 in mouse hepatocytes lead to the spontaneous development of steatohepatitis and liver tumors [29]. Here, we demonstrate that loss of Nrf1 in the liver results in proteasome dysfunction in hepatocytes. Nrf1 function is required for both basal and induced expression of proteasome genes in mouse hepatocytes. In turn, Nrf1-deficiency exacerbates ER stress induced liver steatosis. Furthermore, we establish the link of proteasome dysfunction to the ER stress induced hepatic steatosis. Thus, our data suggest an important role for the Nrf1 transcription factor in homeostatic liver maintenance through the regulation of proteasome function.
Results
Nrf1 is required for basal and induced expression of proteasome genes in mouse hepatocytes
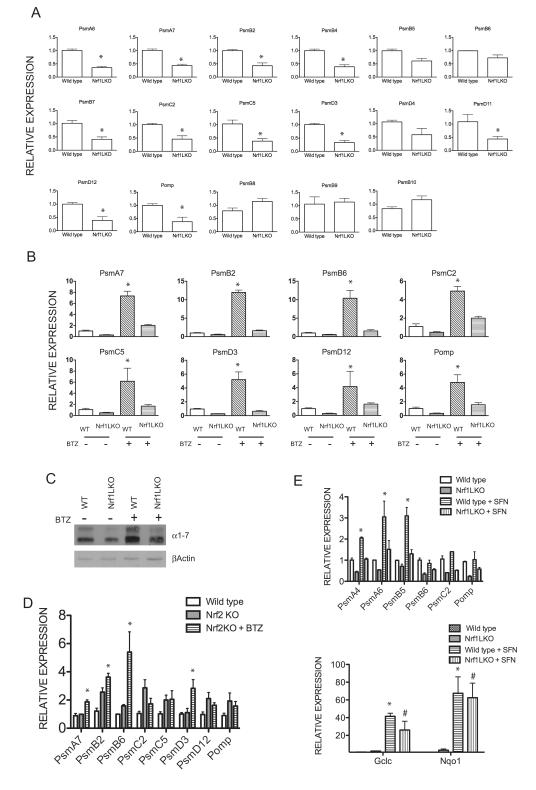
To determine whether Nrf1 mediates the regulation of proteasome gene expression in mouse hepatocytes, RNA was extracted from livers of 1-month old wild type control and Nrf1LKO mice and analyzed by quantitative RT-PCR. Results comparing Nrf1LKO liver samples to controls show a decrease in expression of multiple genes encoding subunits of the 20S and 19S proteasome, and Pomp (proteasome maturation protein), a key component involved in proteasome assembly. However, expression of genes encoding subunits of immunoproteasome; PSMB8, PSMB9, and PSMB10 were unaltered in Nrf1LKO livers (Fig. 1A).
Figure 1. Nrf1 but not Nrf2 is required for both basal and induced proteasome gene expression in hepatocytes.
(A) Comparison of mRNA encoding proteasomal genes in wild type and Nrf1LKO livers by real time RTPCR analysis. Mean values ± SEM of 3-5 mice per group, and significance was assessed by Student’s t test (*P < 0.05). (B) Comparison of mRNA encoding proteasome genes in vehicle-treated or bortezomib-treated wild type and Nrf1LKO livers by real time RTPCR analysis. Mice were intraperitoneally injected with DMSO or bortezomib (2.5 mg/kg), and livers were harvested 24 hr after for RNA extractions. Mean values ± SEM of 3-5 mice per group, and significance was assessed by Student’s t test within genotype between treatments (*P < 0.05). (C) Levels of proteasome alpha-subunits analyzed by Western blotting. (D) Comparison of mRNA encoding proteasome genes in vehicle-treated or bortezomib-treated wild type and Nrf2KO liver by real time RTPCR analysis. Mice were intraperitoneally injected with DMSO or bortezomib (2.5 mg/kg), and livers were harvested 24 hr after for RNA extractions. (E) Comparison of mRNA encoding proteasome genes and oxidative stress genes in vehicle-treated or Sulforaphane-treated wild type and Nrf1LKO livers by real time RTPCR analysis. Mice were intraperitoneally injected with DMSO or sulfurophane (5 mg/kg), and livers were harvested 24 hr after for RNA extractions. Mean values ± SEM of 3 mice per group, and significance was assessed by Student’s t test within genotype between treatments. *P < 0.05 compared to vehicle-treated wild type livers; #P < 0.05 compared to Nrf1LKO vehicle-treated livers.
It has been shown that a decrease in proteasome activity leads to induction of proteasomal genes due to an autoregulatory feedback loop [30]. To determine whether Nrf1 is required for induction of proteasome genes in the liver, quantitative RT-PCR analysis of proteasome gene expression in 1-month old wild type and Nrf1LKO mice were treated with bortezomib, a proteasome inhibitor that also induces proteasome gene expression in cells [31]. The expression of proteasome genes was substantially up-regulated after 24 hours treatment with bortezomib in the livers of wild type mice (Fig. 1B). In contrast, induction of proteasome genes was severely blunted in livers of Nrf1LKO mice after bortezomib treatment (Fig. 1B). To confirm the RT-PCR results, alpha subunits of the 20S proteasome was analyzed by Western blotting. In accord with RNA expression data, both basal and bortezomib-induced expression of α-subunits was decreased in Nrf1 knockout liver (Fig 1C). These results indicate that Nrf1 is required for induction of proteasome in response to bortezomib inhibition.
Nrf2, which regulates oxidative stress response, has been implicated in regulating the expression of proteasome genes [32, 33]. It was shown that expression of various proteasome genes is blunted in Nrf2 knockout livers in response to sulforaphane (SFN) and dithiole-3-thione, which are known Nrf2 activators. To determine the role of Nrf2 in proteasome gene expression in hepatocytes, mRNA from livers of 1-month old vehicle-treated, or bortezomib-treated wild type and Nrf2−/− mice were analyzed. Basal and bortezomib-induced expression of proteasome genes was not affected by loss of Nrf2 in the liver (Fig. 1D). To determine the effects of SFN on proteasome expression in Nrf1LKO livers, wild type and Nrf1LKO were treated with SFN for 24 hours and RNA was extracted for analysis by quantitative RT-PCR. SFN treatment resulted in marked up-regulation of proteasome genes in wild type livers. In contrast, induction of proteasome genes by SFN was blunted in Nrf1LKO livers compared to wild type livers (Fig. 1E, upper panel). However, expression of known Nrf2 target genes, GCLC and NQO1, was up-regulated by SFN treatment in both wild type and Nrf1LKO livers (Fig. 1E, lower panel). Together, these data suggest that proteasome genes are primarily regulated by Nrf1 in mouse liver.
Nrf1 inactivation leads to proteasome insufficiency in hepatocytes
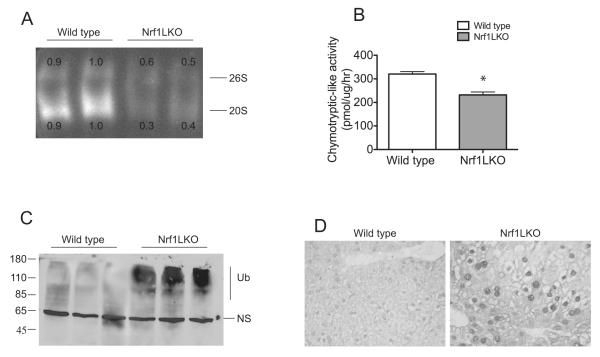
Next, we determined whether diminished expression of proteasome genes in Nrf1LKO livers leads to proteasome insufficiency. Livers were harvested from 1-month old wild type control and Nrf1LKO mice, and proteasome function assessed by an in-gel assay. Activities associated with both 20S and 26S proteasome complexes were markedly diminished in the livers of Nrf1LKO mice compared to controls (Fig. 2A). To confirm these results, an in-tube assay to measure proteasome peptidase activity was performed. The chymotrypsin-like activity was markedly reduced in Nrf1LKO livers compared with controls (Fig. 2B). Consistent with diminished proteasome activity, Western blot analysis of liver lysates from Nrf1LKO mice revealed an accumulation of high molecular weight ubiquitinated proteins (Fig. 2C). In addition, Nrf1LKO liver sections showed increased ubiquitin immunoreactivity (Fig. 2D). Together, these findings indicate that loss of Nrf1 in the liver leads to proteasome dysfunction.
Figure 2. Nrf1LKO livers show accumulation of ubiquitin and decreased proteasome activity.
(A) Measurement of chymotrypsin-like activity by in-gel assay of 1-month old wild type and Nrf1LKO liver homogenates. Fluorescence from free AMC was visualized on a UV transilluminator. Two representative samples from each genotype are shown. The upper band represents the 26S particle and the lower band represents the 20S core. Densitometric quantitations for 26S and 20S levels are shown. (B) Chymotrypsin-like activities in wild type and Nrf1LKO liver homogenates. Mean values ± SEM (n=6 per genotype). *P < 0.05. (C) Western blot analysis of wild type and Nrf1LKO liver lysates with ubiquitin antibody. Note presence of high molecular weight ubiquinated proteins running as a smear in Nrf1LKO samples. (D) Liver sections of 1-month old wild type and Nrf1LKO mice immunostained for ubiquitin.
Deficiency of Nrf1 causes ER stress in liver
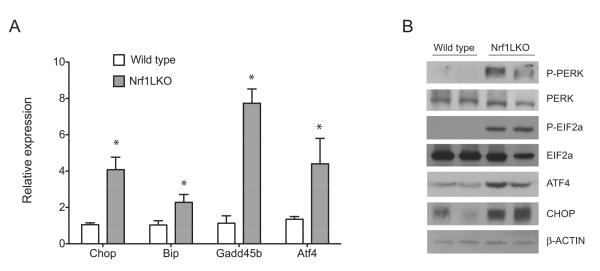
Impairment in proteasomal function can cause endoplasmic reticulum (ER) stress [34-37], and ER stress can lead to hepatic steatosis [38-40]. Thus we sought to determine whether Nrf1LKO livers are characterized by ER stress. Liver lysates from wild type and Nrf1LKO mice were analyzed for markers of ER stress. Quantitative RT-PCR analysis showed increased ATF4, BiP, Gadd45b, and CHOP in Nrf1LKO livers (Fig. 3A). ATF4 is known to activate the expression of BiP, Gadd45b, and CHOP [41]. BiP is an ER-resident molecular chaperone that binds to unfolded proteins and regulates the activation of ER stress transducers, while CHOP plays an important role in ER stress-induced apoptosis [42]. Furthermore, ER stress in mammalian cells induces phosphorylation and activation of PERK and EIF2α [43]. Increased levels of phosphorylated PERK and EIF2α were detected in Nrf1LKO livers (Fig. 3B). Consistent with quantitative RT-PCR results, Nrf1LKO livers showed increased ATF4 and CHOP by Western blotting (Fig. 3B). These data indicate that ER stress is activated in Nrf1LKO livers.
Figure 3. ER stress pathway is activated in Nrf1LKO livers.
(A) Liver lysates from wild type and Nrf1LKO mice were analyzed for PERK and eIF2α phosphorylation, and total levels of PERK, eIF2α, ATF4, CHOP by immunoblotting. Beta-actin levels were used as loading control. (B) Expression of CHOP, BiP, GADD45b and ATF4 mRNA in control and Nrf1LKO livers. Data represents the mean ± SEM of 3 mice per group, and significance was assessed by Student’s t test (*P < 0.05).
Nrf1 heterozygous mice are prone to bortezomib-induced ER stress and steatosis
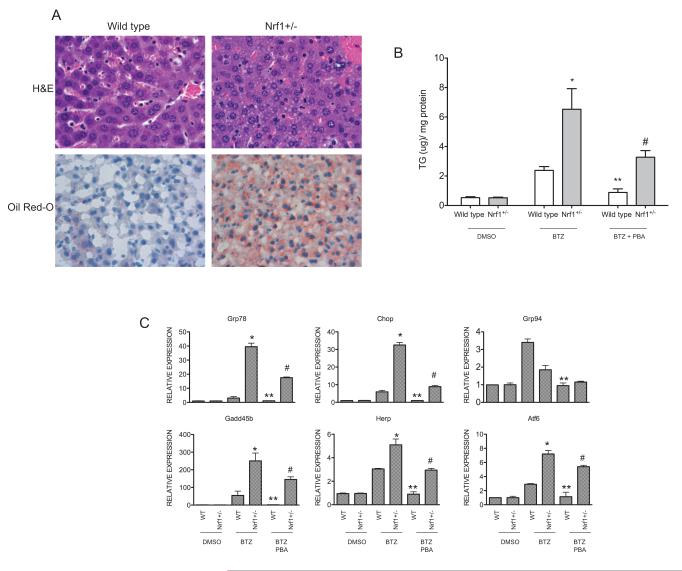
We next examined whether loss of Nrf1 sensitizes hepatocytes to ER stress and steatosis in response to inhibition of proteasome function. Nrf1 heterozygous mice were utilized for these experiments, as they do not show any apparent phenotype under normal conditions. Nrf1+/− and wild type littermate controls were treated with bortezomib, and livers were harvested after 48 hours. H&E and Oil Red O staining demonstrate diffused microvesicular steatosis in Nrf1+/− livers (Fig. 4A), and consistent with the histologic findings, liver triglyceride content in bortezomib treated mice was significantly elevated compared to wild type controls that were similarly treated (Fig. 4B). Next, we sought to examine whether sensitization to steatosis in Nrf1+/− livers is associated with ER stress activation. Quantitative RT-PCR analysis showed elevated levels of ER stress markers including, Grp78, Grp94, Gadd34, Pdi, Atf4, and Chop in Nrf1+/− livers compared to wild type controls after treatment with bortezomib (Fig. 4C). These findings indicate that Nrf1+/− livers are prone to ER stress and steatosis in response to proteasome inhibition. To demonstrate that steatosis in response to inhibition of proteasome function by bortezomib is linked to ER stress, we examined the effects of 4-phenylbutyrate (PBA), a chemical chaperone shown to improve ER folding and reduce ER stress [44, 45]. Bortezomib-induced steatosis, as indicated by triglyceride levels, was attenuated in both wild type and Nrf1+/− by concomitant treatment with PBA (Fig. 4B). In addition, expression of ER stress genes in the liver of wild type and Nrf1LKO with bortezomib treatment was attenuated by PBA treatment (Fig. 4C). These findings are consistent with the idea that hepatic steatosis induced by proteasome inhibition in Nrf1+/− mice is linked to ER stress.
Figure 4. Livers of Nrf1 heterozygous mice exhibit enhanced ER stress and steatosis in response to proteasome inhibition.
(A) H&E and Oil Red O staining of livers of vehicle-treated or bortezomib-treated (2.5 μg/kg) wild type and Nrf1+/− mice. (B) Liver triglyceride (TG) content of wild type and Nrf1+/− mice treated with vehicle or bortezomib (2.5 mg/kg) or bortezomib and PBA (120 mg/kg). Bars represent the mean ± SEM of 3 independent samples in each group, and significance was assessed within genotype by Student’s t test. *P < 0.05 compared to bortezomib-treated wild type livers; **P < 0.05 compared to bortezomib-treated wild type livers; and # P< 0.05 compared to bortezomib-treated Nrf1+/− livers. (C) Expression of ER stress genes in livers of wild type and Nrf1+/− mice treated with bortezomib or bortezomib and PBA. Bars represent the mean ± SEM of 3 different samples in each group, and significance was assessed within genotype by Student’s t test. *P < 0.05 compared bortezomib-treated wild type livers; **P < 0.05 compared to bortezomib-treated wild type livers; and # P< 0.05 compared to bortezomib-treated Nrf1+/− livers. [32]
Discussion
Loss of Nrf1 in hepatocytes results in apoptosis, steatosis, and spontaneous development of tumors in the mouse liver. Nrf1-deficient livers exhibit oxidative stress and lowered expression of genes involved in oxidative stress defense. Hence, these earlier studies suggested that the liver pathology observed in Nrf1LKO mice is mediated by mechanisms involving oxidative stress [20, 29]. Our study here reveals that (1) hepatic deficiency of Nrf1, but not Nrf2, leads to decreased basal and stress-induced proteasome gene expression in the liver; (2) livers of Nrf1LKO mice exhibit proteasome insufficiency and accumulation of ubiquitin conjugates; (3) loss of Nrf1 in hepatocytes leads to ER stress; (4) mice heterozygous for Nrf1 are sensitized to liver ER stress and steatosis in response to drug-induced proteasome inhibition; (5) inhibition of proteasome function leads to ER stress and hepatic steatosis. Together, these findings suggest that impaired proteasome function and attendant ER stress contributes to the liver pathology observed in Nrf1LKO mice.
Both Nrf1 and Nrf2 are members of CNC-bZIP transcription factor family that regulate ARE-dependent genes [16, 18, 46]. Nrf2 has been shown to be the main regulator of oxidative stress genes [18]. Nrf1 shares significant sequence similarity with Nrf2, and Nrf1 has also been shown to regulate oxidative stress genes [19]. However, functions of Nrf1 and Nrf2 are not entirely redundant; Nrf1 has functions that are distinct from Nrf2. Previous studies have shown that Nrf1, but not Nrf2 null mice, die during late gestation [47]. Although Nrf1 and Nrf2 both bind to the ARE, studies suggest that they regulate different subsets of oxidative stress genes. Recently, we have shown that expression of proteasome genes in neurons is Nrf1-dependent [27, 28]. Although Nrf2 is an important regulator of ARE genes, our data from brain-specific Nrf1 knockout mice and Nrf1 knockout MEF cells show that proteasome gene expression is not dependent on Nrf2 [27, 28]. Our results here indicate that Nrf1 is also essential for both basal and proteotoxic-stress inducible expressions of proteasome genes in mouse liver. Transcripts encoding the various subunits that make up the 20S core and 19S regulatory particle of the proteasome, as well as Pomp, a molecular chaperone essential for 20S formation, were significantly decreased in Nrf1LKO livers but not in Nrf2 knockout livers. The marked accumulation of ubiquitinated protein conjugates associated with defects in proteasome and PQC gene expression in Nrf1LKO livers, indicates that Nrf1 is also critical in maintaining protein homeostasis in hepatocytes. Although Nrf2 has also been shown to also regulate proteasome expression in mouse liver [33], our current finding indicates that loss of Nrf2 does not affect basal or proteotoxic-stress inducible proteasome expression. Interestingly, induction of proteasome genes by SFN, a known Nrf2 activator, was attenuated, but not completely abolished, in Nrf1LKO livers. This suggests the possibility is that the transcriptional network required for proteasome gene induction are different depending on the types of cellular stress. Thus, while Nrf1 drives expression when cells undergo proteotoxic stress, Nrf2 may regulate proteasome expression under oxidative stress in hepatocytes. In addition, the mechanism by which Nrf1 function is activated by reduced proteasome inhibition is not known. One possibility is that Nrf1 expression is up-regulated as a result of ER stress secondary to proteasome insufficiency. However, Zhang et al. showed that ER stressors had no effect on Nrf1 activity [48]. Alternatively, Nrf1 expression may be regulated by a feedback mechanism given that the Nrf1 protein is degraded by the by the ubiquitin proteasome pathway. These possibilities will require further studies.
Our results show that the unfolded protein response (UPR) pathway is activated in Nrf1LKO livers. As proteasome dysfunction can lead to ER stress, this suggests that the UPR is induced by lowered proteasome function in Nrf1LKO livers. In accord with this, Nrf1 heterozygous mice are prone to liver ER stress induced by pharmacological inhibition of proteasome function. In addition, our data suggest that the attenuation of ER stress with PBA can prevent the development of steatosis. These findings draw a potential link between proteasome impairment and steatohepatitis, and we speculate that ER stress plays an important role in the pathogenesis of steatohepatitis that is characteristic of livers deficient in Nrf1. Studies have demonstrated that ER stress induces an inflammatory response via different UPR transducers including IRE1α and PERK pathways [49, 50]. Aside from inflammation, hepatic steatosis has also been attributed to ER stress. For instance, mice that are deficient in UPR signaling proteins ATF6α and IRE1α have been found to develop steatosis after treatment with compounds that induce ER stress [38]. It is thought that ER stress impedes folding of apoproteins in hepatocytes, which leads to disruption of VLDL secretion and lipid metabolism. In contrast to ATF6α and IRE1α knockouts however, ER stress and steatosis is already apparent in Nrf1LKO livers even without drug treatment. In this regard, it is interesting to note that a liver specific knockout of CBF-B, a transcription factor that is activated by ER stress, results in a phenotype similar to Nrf1LKO mice. CBF-B liver knockout mice are also characterized by hepatic cell death and steatohepatitis at 4 weeks of age that was preceded by the activation of the ER stress pathway [51]. While these findings suggest ER stress as an important cause of liver pathologies in Nrf1 knockout livers, Hirotsu et al. recently showed that Nrf1 regulates Lipin1 and PGC-1B. Both Lipin1 and PGC-1B are involved in controlling expression of fatty acid oxidation genes [52]. Hence, a defect in fatty acid oxidation also contributes to the fatty liver phenotype in Nrf1 liver knockouts.
In summary, our findings demonstrate that Nrf1 is important in maintaining proteasome homeostasis in the liver, and suggests that compromised proteasome function plays a role in the pathogenesis of steatohepatitis. The findings here also raise the possibility that Nrf1 may be a target for exploring therapeutic strategies for steatohepatitis as well as other liver diseases associated with abnormal proteasome function.
Materials and Methods
Reagents
Dulbecco’s modified Eagle’s medium (DMEM), Alpha minimum essential medium, fetal bovine serum (FBS), and Superscript III reverse transcriptase were purchased from Invitrogen (Carlsbad, CA). Sulfurophane and antibody for beta-actin (A1978) was from Sigma (St. Louis, MO). Antibody for phospho-Perk, Perk, phospho-eif2α, eif2α, GRP94, CHOP, ATF4, and Ubiquitin was from Cell Signaling (Beverly, MA). Biotinylated anti-rabbit IgG and 3,3′ diaminobenzadine (DAB) substrate kit, M.O.M. Immunodetection Kit were from Vector labs (Burlingame, CA). Proteasome substrate Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin was from Enzo Life Sciences (Farmingdale, NY). UltraSpec RNA Isolation Reagent was purchased from Biotecx (Houston, TX). 2X FastStart SYBR Green Master ROX for quantitative RT-PCR was purchased from Roche (Indianapolis, IN). RNeasy MinElute Cleanup Kit was from Qiagen (Valencia, CA). Bortezomib was purchased from Selleckchem (Houston, TX). 4-Phenylbutyric acid was purchased from Sigma-Aldrich. Hematoxylin and aqueous medium were from Vector laboratories (Burlingame, CA). Enhanced chemiluminescence substrate kit was from Pierce Biotechnology (Rockford, IL).
Mice
All animal experiments were approved by, and conducted in accordance with, the National Institutes of Health standards for the care and use of experimental animals and the University of California Irvine Institutional Animal Care and Use Committee. Nrf1 knockout mice have been described previously [29]. All efforts were made to minimize animals from suffering. Mice showing evidence of pain or distress after drug treatments by intraperitoneal injections were euthanized by CO2 overdose.
RNA Isolation and Quantitative Real-Time PCR
RNA was extracted using UltraSpec RNA reagent and then RNA was further purified using RNeasy MinElute Cleanup Kit. cDNA synthesis was generated with Superscript III first strand synthesis kit according to the manufacturer’s recommendation. Amplification of cDNA occurred in a Step One Plus PCR machine (ABI) using the FastStart SyBr Green reagent in duplicate 10-μl reactions. 18s rRNA was used as an endogenous control and relative expression was calculated with the equation 2(Ct target gene - Ct 18s). Fold change in expression was determined from the difference between the averaged expression levels relative to control.
Western Blotting
Protein lysates were prepared using Nonidet P-40 lysis buffer, and protein concentrations were determined using Bio-Rad protein assay reagent with BSA as protein standard. Samples were run on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were then blocked in 5% milk at room temperature for 1 h, and incubated with primary antibody over night at 4°C. Subsequently, blots were washed and peroxidase-conjugated secondary antibody added and incubated for 1 h. Blots were visualized using a chemiluminescent detection system.
Histology and Immunohistochemistry
Livers were fixed in 10% neutral buffered formalin and imbedded in paraffin. For immunohistochemical staining, paraffin sections were dewaxed, and rehydrated through a series of alcohols. Antigen retrieval was conducted by heating in 10 mM citrate buffer (pH 6.5). Sections were stained with primary antibody in blocking solution overnight at 4 °C, rinsed in Tris-buffered saline Tween 20 buffer, and then incubated for 1 h with biotinylated anti-rabbit IgG (1:300). Tissue Sections were visualized and examined using a Nikon microscope equipped with a CCD camera.
Triglyceride Assay
Liver tissues were homogenized in a 1:2 chloroform/methanol solution, followed by extraction with 1:1 chloroform/water mixture and centrifugation at 3000rpm for 15 minutes to separate the aqueous and organic phases. The organic phase was collected, air-dried, and then dissolved in isopropanol prior to triglyceride determination. Triglycerides were quantified using a Serum Triglyceride Determination Kit (Sigma Chemicals, St. Louis, MO) following manufacturer’s recommendations.
Oil Red O staining
Staining of neutral lipids was performed on 10μm thick liver frozen sections. Frozen sections were fixed in formalin and washed with running tap water for 10mins, followed by quick rinsing in 60% isopropanol. Fixed sections were then stained with Oil Red O working solutions for 15min and rinsed with 60% isopropanol. The nuclei were lightly stained with hematoxylin. The slides were air dried and mounted.
Proteasome activity assay
Freshly dissected livers were homogenized (10% wt/vol) in ice-cold buffer [10% glycerol, 25 mM Tris-HCL (pH 7.4), 10 mM MgCl2, 4 mM ATP, and 1 mM DTT] using a polytron and centrifuged at 12,000 × g for 15 min at 4 °C to remove debris. Protein concentrations were determined by the Bradford assay using BSA as the protein standard. Chymotrypin-like activity was determined by fluorometric assay using Suc-Leu-Leu-Val-Try-7-amino-4-methylcoumarin (Suc-LLVY-AMC as substrate. Reactions were initiated by adding 50 μM substrate followed by incubation at 37°C for 30 minutes. Release of fluorogenic AMC was monitored at 360 nm excitation and 460 nm emission using a Molecular Devices fluorometric plate reader. Fluorescence units were converted to AMC concentration by using standard curves generated from free AMC. Rates were expressed as μmol of AMC/sec/g liver tissue protein. For in-gel assays, equal amounts of liver lysates were prepared for the 1-month old control mice and the Nrf1LKO mice. These liver lysate samples were separated by native gel electrophoresis and assayed for 26S and 20S proteasome complexes by overlying gels with substrates. Liver extracts were electrophoresed on 4.5% nondenaturing polyacrylamide gel with 2.5% stacking gel and run at 90 V for 4 h at 4 °C. After electrophoresis, gels were incubated with proteasome substrate Suc-LLVY-amc for 30 min at 37 °C. The 20S core particle was detected by incubating the gel in the presence of 0.02% SDS to activate the 20S core. Active proteasome bands were then visualized by exposure to UV light at 360 nM, using Fujifilm Global (LAS-4000). Band intensity was quantified by using Gel & Graph Digitizing Software (Silk Scientific, Inc).
Statistical Analysis
The data is expressed as means ± SEM. For statistical comparison, the Student t test was used. P values of <0.05 were considered statistically significant.
Primers list
| Gene | Forward | Reverse |
|---|---|---|
| PsmA6 | AACGGAAAGCATTGGCTGTGT | TGCACAGCATGTCCACAGGAA |
| PsmA7 | CGGCCCTAATTGTGGGTTTTG | GTTTTTGCCACCTGACTGGAC |
| PsmB2 | CCTGGCTGGCTATGACGAGCA | CGGACACTGAAGGTGGGCAGA |
| PsmB4 | TCCGGCTCCAGGGCAGTTTTA | ACTCCGCCGTCGAACTTCACC |
| PsmB5 | AACCACCACCCTGGCCTTCAA | AGCCAACAACCGCTCCCAGAA |
| PsmB6 | GCAGGCTGGACCCTCAAGAA | CAAGCGGATCACCCCTCCACT |
| PsmB7 | TCTGGCTCCTTGGCAGCAATG | GGCCAAGCCTGGTCCCTTTCT |
| PsmC2 | GATGTTGGTGGCTGTAAGGAA | CCCAATAACTCGAATGAAGCA |
| PsmC5 | CGGCCTGGACAAGCAGATCAA | CTGGGGGTCCGTAGAGCAGGA |
| PsmD3 | CGGCTCTGTGCCTGGTCTCTG | TCCCAGCTGCTGCCTCCTCTT |
| PsmD4 | TCTCCTATTCTGGCTGGTGAA | CATGCTGCTTAGGTCTGGAAG |
| PsmD11 | AGGCAGACAGAAGCATTGAAA | GGTCCAAAATCCCATGAAACT |
| PsmD12 | GTGGCCGAACCTGGGGTACAG | TCTTGAAGCCGCCCTTCCTTG |
| PsmB8 | AATGCAGCCCACCGCATTCCT | AGGCGAGTGTGGTTGTGCCG |
| PsmB9 | AGAAGTCCACACCGGGACAACCAT | GTCGAACACGCGGTTCACCACT |
| PsmB10 | AACATGACGCTGGAGGCTGCG | CTGCAGCTTGGCACCCCCTG |
| Pomp | TTGGGTCGGAGCTGAAGGACA | GGGGAGCAAACAGACCCTGGA |
| Herp | ACTCCTCGCTGAGCAGATTT | CTCTGTCTGAACGGAAACCA |
| Atf4 | TCGACCAGGTTGCCCCCTTTA | CCAGGTAGGACTCCGGGCTCA |
| Bip | GGCCTGCTCCGAGTCTGCTTC | CCGTGCCCACATCCTCCTTCT |
| Chop | CCAGGGCCAACAGAGGTCACA | TCCGCTCGTTCTCCTGCTCCT |
| Grp94 | TGGGTCAAGCAGAAAGGAG | TCTCTGTTGCTTCCCGACTT |
| Gadd34 | CCCGAGATTCCTCTAAAAGC | CCAGACAGCAAGGAAATGG |
| Pdi | AACGGGAGAAGCCATTGTA | AGGTGTCATCCGTCAGCTCT |
| NQO1 | GCATTGGCCACACTCCACCAG | ATGGCCCACAGAGAGGCCAAA |
| GCLC | GCACGGCATCCTCCAGTTCCT | TCGGATGGTTGGGGTTTGTCC |
Acknowledgments
Financial support: This research was supported by NIH grant CA091907
Abbreviations
- ARE
antioxidant response element
- Btz
Bortezomib
- CNC
Cap and Collar
- ER
endoplasmic reticulum
- Nrf1
Nuclear factor erythroid-derived 2-related factor 1
- Nrf2
Nuclear factor erythroid-derived 2-related factor 2
- PBA
sodium 4-phenylbutyric acid
- SFN
sulforaphane
- PQC
protein quality control
- UPS
ubiquitin proteasome system
Footnotes
Conflict of Interest: The authors have no conflicts of interest to disclose
REFERENCES
- 1.Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 3.Silva GM, Netto LE, Simoes V, Santos LF, Gozzo FC, Demasi MA, Oliveira CL, Bicev RN, Klitzke CF, Sogayar MC, Demasi M. Redox control of 20S proteasome gating. Antioxid Redox Signal. 2012;16:1183–94. doi: 10.1089/ars.2011.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golab J, Bauer TM, Daniel V, Naujokat C. Role of the ubiquitin-proteasome pathway in the diagnosis of human diseases. Clin Chim Acta. 2004;340:27–40. doi: 10.1016/j.cccn.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 6.Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9:S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciechanover A, Brundin P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 2003;40:427–46. doi: 10.1016/s0896-6273(03)00606-8. [DOI] [PubMed] [Google Scholar]
- 8.McNaught KSP, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat Rev Neurosci. 2001;2:589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- 9.Banner BF, Savas L, Zivny J, Tortorelli K, Bonkovsky HL. Ubiquitin as a Marker of Cell Injury in Nonalcoholic Steatohepatitis. Am J Clin Pathol. 2000;114:860–866. doi: 10.1309/4ubb-bf78-f55v-50ka. [DOI] [PubMed] [Google Scholar]
- 10.Takagi M, Yamauchi M, Takada K, Ohkawa K. Serum Ubiquitin-Protein Conjugates in Normal Subjects and Patients With Alcoholic Liver Diseases: Immunoaffinity Isolation and Electrophoretic Mobility. Alcohol Clin Exp Res. 2002;26:1692–1696. doi: 10.1097/01.ALC.0000036924.31671.0E. [DOI] [PubMed] [Google Scholar]
- 11.Shirahashi H, Sakaida I, Terai S, Hironaka K, Kusano N, Okita K. Ubiquitin and hepatocellular carcinoma. Liver. 2002;22:413–418. doi: 10.1034/j.1600-0676.2002.01541.x. [DOI] [PubMed] [Google Scholar]
- 12.French S, Bardag-Gorce F. Ubiquitin-Proteasome Pathway in the Pathogenesis of Liver Disease. In: Dufour JF, Clavien PA, Trautwein C, Graf R, editors. Signaling Pathways in Liver Diseases. Springer; Berlin, Heidelberg: 2005. pp. 377–389. [Google Scholar]
- 13.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 14.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–71. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 15.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Mol Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 16.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci USA. 1993;90:11371–5. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: Binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26:512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 19.Kwong M, Kan YW, Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in gamma-gcs(l) and gss expression in mouse fibroblasts. J Biol Chem. 1999;274:37491–8. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- 20.Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–62. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci USA. 1996;93:14960–5. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myhrstad MC, Husberg C, Murphy P, Nordstrom O, Blomhoff R, Moskaug JO, Kolsto AB. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochim Biophys Acta. 2001;1517:212–9. doi: 10.1016/s0167-4781(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 23.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing W, Singgih A, Kapoor A, Alarcon CM, Baylink DJ, Mohan S. Nuclear factor-E2-related factor-1 mediates ascorbic acid induction of osterix expression via interaction with antioxidant-responsive element in bone cells. J Biol Chem. 2007;282:22052–61. doi: 10.1074/jbc.M702614200. [DOI] [PubMed] [Google Scholar]
- 25.Narayanan K, Ramachandran A, Peterson MC, Hao J, Kolsto AB, Friedman AD, George A. The CCAAT enhancer-binding protein (C/EBP)beta and Nrf1 interact to regulate dentin sialophosphoprotein (DSPP) gene expression during odontoblast differentiation. J Biol Chem. 2004;279:45423–32. doi: 10.1074/jbc.M405031200. [DOI] [PubMed] [Google Scholar]
- 26.Berg DT, Gupta A, Richardson MA, O’Brien LA, Calnek D, Grinnell BW. Negative regulation of inducible nitric-oxide synthase expression mediated through transforming growth factor-beta-dependent modulation of transcription factor TCF11. J Bio Chem. 2007;282:36837–44. doi: 10.1074/jbc.M706909200. [DOI] [PubMed] [Google Scholar]
- 27.Lee CS, Lee C, Hu T, Nguyen JM, Zhang J, Martin MV, Vawter MP, Huang EJ, Chan JY. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc Natl Acad Sci USA. 2011;108:8408–13. doi: 10.1073/pnas.1019209108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad of Sci USA. 2005;102:4120–5. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, Kruger E. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of Mammalian proteasomes. J Biol Chem. 2003;278:21517–25. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 31.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, Bailey C, Joseph M, Libermann TA, Treon SP, Munshi NC, Richardson PG, Hideshima T, Anderson KC. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA. 2002;99:14374–9. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243:170–92. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 33.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Bio. 2003;23:8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szokalska A, Makowski M, Nowis D, Wilczynski GM, Kujawa M, Wojcik C, Mlynarczuk-Bialy I, Salwa P, Bil J, Janowska S, Agostinis P, Verfaillie T, Bugajski M, Gietka J, Issat T, Glodkowska E, Mrowka P, Stoklosa T, Hamblin MR, Mroz P, Jakobisiak M, Golab J. Proteasome inhibition potentiates antitumor effects of photodynamic therapy in mice through induction of endoplasmic reticulum stress and unfolded protein response. Cancer Res. 2009;69:4235–43. doi: 10.1158/0008-5472.CAN-08-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nawrocki ST, Carew JS, Dunner K, Jr, Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 2005;65:11510–9. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- 37.Chauhan D, Singh A, Brahmandam M, Podar K, Hideshima T, Richardson P, Munshi N, Palladino MA, Anderson KC. Combination of proteasome inhibitors bortezomib and NPI-0052 trigger in vivo synergistic cytotoxicity in multiple myeloma. Blood. 2008;111:1654–64. doi: 10.1182/blood-2007-08-105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–75. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatology. 2011;54:495–508. doi: 10.1002/hep.24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–40. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang HY, Wek SA, McGrath BC, Lu D, Hai T, Harding HP, Wang X, Ron D, Cavener DR, Wek RC. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol Cell Biol. 2004;24:1365–77. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–77. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–71. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 44.Vilatoba M, Eckstein C, Bilbao G, Smyth CA, Jenkins S, Thompson JA, Eckhoff DE, Contreras JL. Sodium 4-phenylbutyrate protects against liver ischemia reperfusion injury by inhibition of endoplasmic reticulum-stress mediated apoptosis. Surgery. 2005;138:342–51. doi: 10.1016/j.surg.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Welch WJ, Brown CR. Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperones. 1996;1:109–15. doi: 10.1379/1466-1268(1996)001<0109:iomacc>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–87. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Lucocq JM, Hayes JD. The Nrf1 CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem J. 2009;418:293–310. doi: 10.1042/BJ20081575. [DOI] [PubMed] [Google Scholar]
- 49.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–6. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 50.Gentile CL, Frye MA, Pagliassotti MJ. Fatty acids and the endoplasmic reticulum in nonalcoholic fatty liver disease. Biofactors. 2011;37:8–16. doi: 10.1002/biof.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo R, Klumpp SA, Finegold MJ, Maity SN. Inactivation of CBF/NF-Y in postnatal liver causes hepatocellular degeneration, lipid deposition, and endoplasmic reticulum stress. Sci Rep. 2011;1:136. doi: 10.1038/srep00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirotsu Y, Hataya N, Katsuoka F, Yamamoto M. NF-E2-Related Factor 1 (Nrf1) Serves as a Novel Regulator of Hepatic Lipid Metabolism through Regulation of the Lipin1 and PGC-1beta Genes. Mol Cell Biol. 2012;32:2760–70. doi: 10.1128/MCB.06706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]