Abstract
This paper describes a simple, highly efficient and robust proteomic workflow for routine liquid-chromatography tandem mass spectrometry analysis of Laser Microdissection Pressure Catapulting (LMPC) isolates. Highly efficient protein recovery was achieved by optimization of a “one-pot” protein extraction and digestion allowing for reproducible proteomic analysis on as few as 500 LMPC isolated cells. The method was combined with label-free spectral count quantitation to characterize proteomic differences from 3,000–10,000 LMPC isolated cells. Significance analysis of spectral count data was accomplished using the edgeR tag-count R package combined with hierarchical cluster analysis. To illustrate the capability of this robust workflow, two examples are presented: 1) analysis of keratinocytes from human punch biopsies of normal skin and a chronic diabetic wound and 2) comparison of glomeruli from needle biopsies of patients with kidney disease. Differentially expressed proteins were validated by use of immunohistochemistry. These examples illustrate that tissue proteomics carried out on limited clinical material can obtain informative proteomic signatures for disease pathogenesis and demonstrate the suitability of this approach for biomarker discovery.
Keywords: Laser Capture Microdissection, Proteomics, Label-free, Biopsy, Mass Spectrometry
INTRODUCTION
Given the many factors that regulate protein transcription, it is critical to directly characterize cellular protein expression and identify proteomic signatures in order to determine the molecular and/or physiological dysfunctions that cause disease. The ability to obtain quantitative proteomic profiles from human clinical specimens is challenging because of tissue heterogeneity and the limited material available in a typical biopsy. Laser microdissection pressure catapulting (LMPC) is a technology that can extract homogeneous cell populations from complex tissues. Using frozen and formalin-fixed tissue sections, it is possible to retrieve and analyze RNA, DNA and proteins present in the LMPC captured cells [1–5]. However, in contrast to RNA and DNA, the ability to recover high quality protein and the time required for sample collection have severely constrained the routine use of LMPC-isolated samples for proteomic analysis of clinical biopsies. Increasingly more research groups are using laser capture as a method to examine the cellular proteome even for sample where fewer than 10,000 cells are captured [6, 7]. More routinely 10,000 to 100,000 cells are captured for mass spectrometry analysis [8–16]. Unfortunately, clinical needle biopsies typically provide less than 10,000 cells of a single (homogeneous) cell type.
In this report, we describe a proteomics workflow for routine quantitative liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of LMPC isolates from frozen biopsies. An important feature of this workflow is a “one-pot” protein extraction and digestion method. This sample preparation strategy uses commercially available reagents and delivers efficient protein recovery resulting in highly reproducible proteomics data appropriate for label-free spectral counting quantification. To demonstrate the capability of this approach with human tissues, we analyzed punch biopsies of normal skin and chronic wound keratinocytes from a diabetic patient and glomeruli from needle biopsies of patients with diabetic, lupus and genetic kidney diseases. Statistical analysis of protein abundances differences was possible by the use of the bioconductor package edgeR orginally developed for analysis of tag-count RNA-Seq data and recently applied to proteomics [17–20]. Validation of several differentially expressed proteins was performed by use of immunohistochemistry as well as by correlation with previously reported results. The data reported here for the proteomic characterizations of a chronic wound and diseased kidneys provide supporting evidence that the workflow is capable of routine biomarker candidate identification from frozen tissues.
EXPERIMENTAL
Tissue sectioning and fixation
Frozen tissue blocks were cut into 8 μm sections, mounted on a thermoplastic polyethylenenapthalate (PEN covered glass slide (Carl Zeiss MicroImaging GmbH, Munchen, Germany) and kept at −80°C until use. When needed, sections were briefly thawed and immediately fixed in ice cold 70% ethanol and rinsed in deionized H2O each for 1 min. After the water rinse, the wet sections were stained briefly (5–10 s) with hematoxylin and immediately rinsed in two changes of deionized H2O. The sections were then dehydrated in 70%, 95% and 100% ethanol for 2 min each. The dehydrated slide was allowed to air dry for immediate capture on a PALM technology microdissection system. All anonmymized samples used in this study were collected according to IRB approved protocols and stored in internal biobanks.
Laser capture and pressure catapulting (LMPC)
LMPC was performed using the laser microdissection system from PALM Technologies (Carl Zeiss MicroImaging GmbH, Munchen, Germany) containing a PALM MicroBeam and RoboStage for high-throughput sample collection and a PALM RoboMover (PALM Robo software, Version 2.2). Typical settings used for laser cutting were UV-Energy of 75–85 and UV-Focus of 52. Sections were cut and captured under a 10x ocular lens. Cut elements were catapulted into 25 μl of 0.5% Rapigest (Waters Corporation, Milford, MA) and resuspended in 50 mM NH3HCO3. Total number of cells captured was determined using the conversion factor of 50,000 μm2 = 200 cells for keratinocytes or by assuming an average cell diameter of 18 μm for kidney tissue. Upon completion of microdissection, the captured material was spun down into a 0.2 ml PCR tube and held at −80°C until protein retrieval.
Immunohistochemistry
Tissue specimens were sectioned (10 μm) and mounted on positively charged Superfrost slides (FisherScientific). Using the early wound and normal skin, immunohistochemical staining was performed following standard procedures, using the following antibodies all of which were IgG mouse monoclonal: anti-human keratin 6 from Cedarlane, (Burlinton NC), anti-human desmocollin-2 and S100 A9 from Abcam (Cambridge, MA), anti-human vimentin from BD Biosciences (San Jose CA) and anti-human keratin 14 from Covance (Emeryville Ca). In brief, the sections were fixed with ice-cold acetone for 1 min and then blocked with 10% normal goat serum to minimize non-specific binding before being incubated with the primary antibodies for 1 h at room temperature. The primary antibody was detected by using alexa-fluor tagged secondary antibodies from Molecular Probes. Dilutions used were per manufacturer recommendations. Stained sections were mounted in Vectamount (Vector labs, Burlingame, CA) containing Dapi and images were collected using a Zeiss Axiovert inverted microscope and Axiovision software. For the kidneys, Immunoperoxidase staining with a monoclonal antibody to fibulin −1 (Santa Cruz Biotechnology, Santa Cruz, CA) and a polyclonal antibody to podocin (Alpha Diagnostic Intl Inc., San Antonio, TX) was performed (dilution 1:100 and 1: 40 respectively) on formalin-fixed paraffin embedded tissue sections of the fibronectin glomerulopathy biopsies, and additional cases of diabetic nephritis and proliferative Class IV LN (different from the cases used for mass spectrometry). Fibronectin immunostaining was performed in the laboratory of the late Dr. Andrew Herzenberg, Toronto, Canada. The antibody used was a mouse monoclonal antibody directed to the N-terminus of fibronectin.
Protein Retrieval
Samples that had been stored at −80°C were thawed briefly and sonicated (in the original 0.2 ml tube) on the low power setting for a total instrument time of 60 s with a 3 s on 3 s off time interval using a Diagenode Bioruptor (model UCD-200, Sparta NJ.). The samples were placed in a boiling water bath for 5 min and cooled to room temperature. Trypsin was added in a ratio of 1:30 trypsin:protein assuming ~2 μg retrieved protein/10,000 isolated cells. After overnight incubation at 37°C with gentle shaking, formic acid was added to a final concentration of 30% and the suspension was incubated for 30 min at 37°C to precipitate the Rapigest. The samples were centrifuged 2x at 15,000 RPM at 4°C for 10 min to eliminate cellular debris and the precipitated Rapigest from the sample. The extracts were dried in a speedvac to dryness. Dried protein digests were re-suspended in 20 μl of a solution of 2% acetonitrile with 0.1% formic acid and sonicated for 1–2 min in a water bath sonicator at 4°C to ensure peptide solubilization. Final peptide concentrations were controlled by measurement of 280 nm absorbance on the tryptic digest using a Nanodrop ND-1000 spectrometer.
Mass Spectrometry
Peptides (1–2 μg) were separated by reversed-phase HPLC (Dionex Ultimate 3000 capillary/nano HPLC system, Dionex, Sunnyvale, CA) and mass analyzed with a Thermo Fisher LTQ Orbitrap XL, equipped with micro/nanospray ionization sources (Michrom Bioresources Inc., Auburn, CA). HPLC separations were carried out at a flow rate of 2 μL/min on a 0.2mm × 150 mm C18 column (5 μm, 300 Å, Michrom Bioresources Inc., Auburn, CA). In general, 1.5 – 2 μg of protein was loaded/sample if sample size was non-limiting; otherwise the entire sample was loaded for a single run with at least 1μg. The mobile phases consisted of HPLC grade water and acetonitrile (ACN) with formic acid added as ion-pairing reagent. A 10–32% gradient was run over 5 hours followed by a 32–90% gradient run over 40 minutes. The heated capillary temperature and electrospray voltage were set at 175°C and 2.0kV, respectively. Data were acquired in the top 5 data-dependent mode with dynamic exclusion and Orbitrap preview modes enabled. Dynamic exculsion settings are as follows: repeat count=2, repeat duration=30.00, exclusion list size=200, exclusion duration=350s and exclusion mass width of ±1.50 m/z. Protein identifications were obtained using the MassMatrix search engine searching the IPI Human (v3.65) FASTA database. Search parameters included: variable oxidation of methionine, two missed trypsin cleavages, precursor ion tolerance of 20 ppm and product ion tolerance of 0.8 Da. The false discovery rate (FDR) was estimated using the target decoy strategy by appending the target databases with reversed sequences. The protein identifications and respective spectral counts were parsed from each sample data file and then combined taking in consideration parsimony for protein groupings[21] using an in-house developed application written in python. The combined protein list was sorted by maximum protein score. Protein matches were retained above an FDR threshold of 1% based on max protein score for a row and one sample having at least two different unique peptide matches.
Statistical Analysis
Label free relative quantitation was accomplished by the spectral count approach [22, 23] in which the number of MS/MS spectra identified from the same protein is used to represent relative protein abundance. Spectral counts did not include modified, or semitryptic peptides and shared peptides from multiple protein isoforms contributed equally to each isoform. Significance analysis of relative protein abundance was determined by analysis of the spectral count data using the edgeR bioconductor package [19]. Peptide spectral counts were modeled as an overdispersed Poisson / negative binomial distribution in which an empirical Bayes procedure was used to moderate overdispersion across each protein [17–19]. An exact text for overdispersed data was then used to assess difference in protein abundance [18]. A Benjamini-Hochberg multi-test correction (α=0.05) was applied to final p-values to control for false discoveries [24].
RESULTS
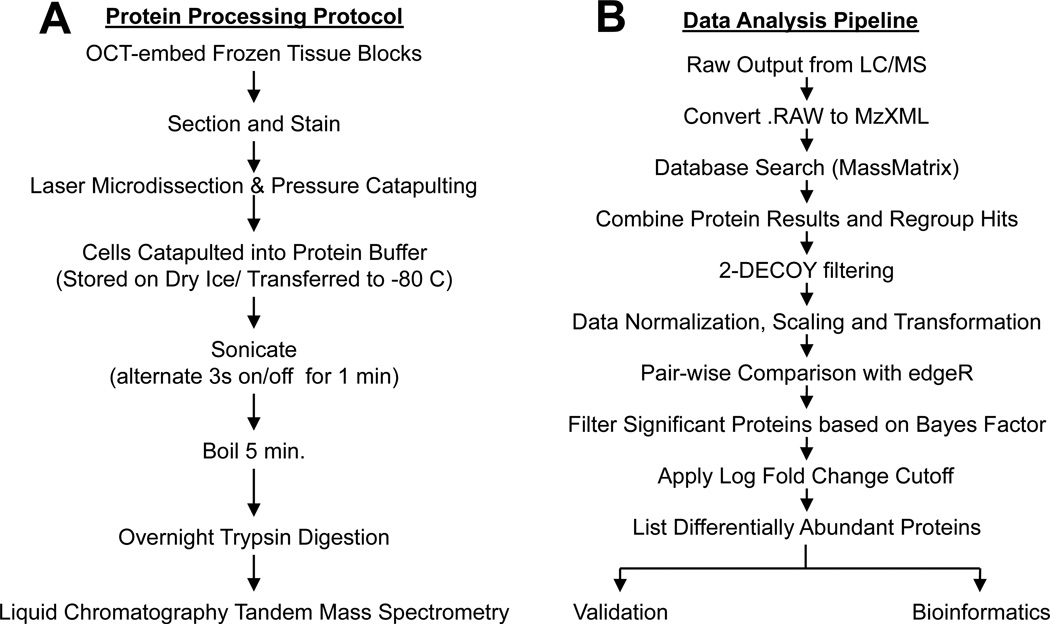
The protocol used to process the frozen biopsies is shown in Fig. 1A. The quantity and quality of peptides generated following trypsin digestion of proteins recovered from LMPC isolates was controlled for by monitoring peptide absorbance. A Nanodrop spectrometer (using 280 absorbance) was used to quantify peptides after tryptic digestion of capture tissue when > 5,000 cells were isolated. A typical Nanodrop spectrum of the trypsin digest recovered from a normal kidney LMPC glomerular isolate is provided in Supplemental Data 1. In general, tryptic digestion of proteins from 106 cells yields ~200–300 μg.[25] We estimate that the total available tryptic peptides from 10,000 LMPC isolated cells to be ~2–3 μg. Using this estimate, ~70–90% of the available tryptic digests was recovered from the LMPC isolates when samples were analysed for peptide concentration. Supplemental Data 1 shows examples of tryptic digestion recovery/efficiency from 4 individual keratinocyte LMPC isolations.
Fig. 1.
(A) Flowchart illustrating steps for sample preparation and protein recovery from LMPC isolates. (B) Flowchart describing spectral count data acquisition and analysis
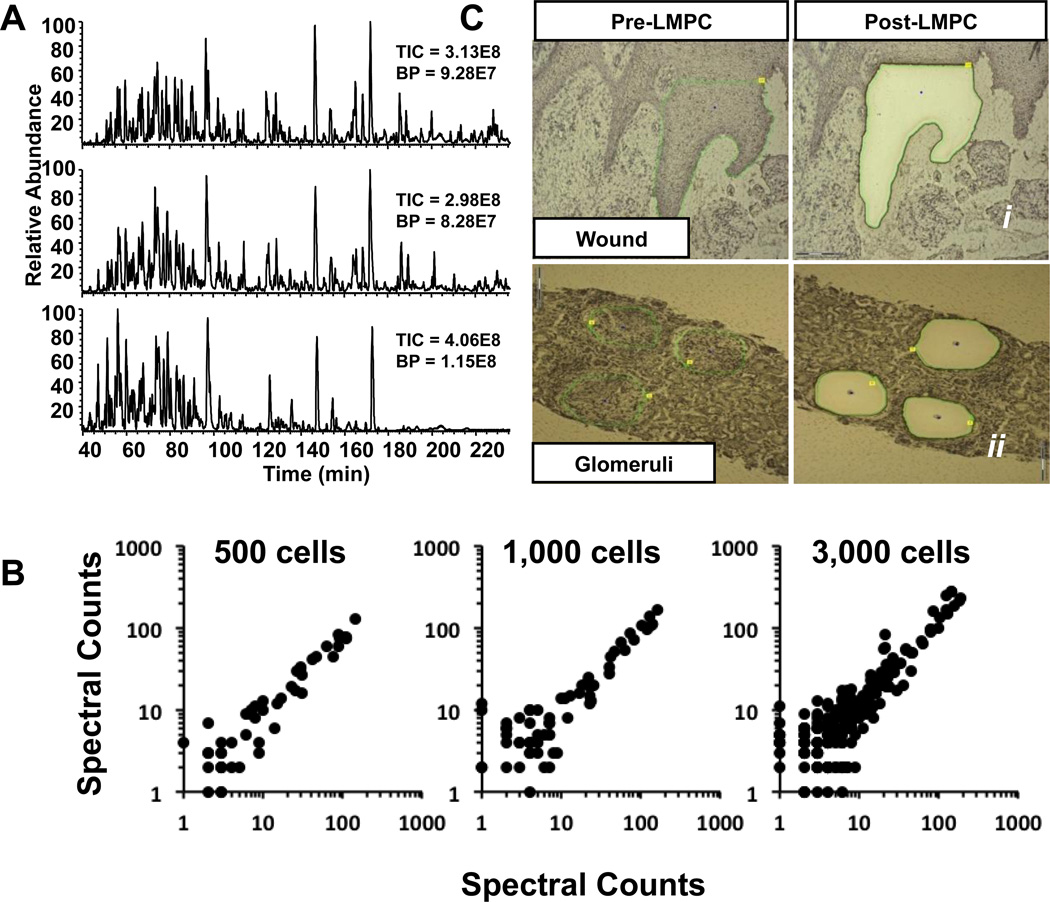
Examination of the LC-MS/MS base peak chromatograms of peptides from LMPC isolates demonstrates a high degree of sample complexity. LC-MS/MS base peak chromatograms from three separate LMPC keratinocyte captures (from the same late wound biopsy) indicate excellent reproducibility (Fig. 2A). Additionally, log-log scatter plots of spectral counts for 500, 1,000 and 3,000 LMPC cell isolates, each comparing spectral counts from 2 individual biological replicates provide further indication of the technique’s reproducibility (Fig. 2B). Spectral count data for the three replicates of 500, 1,000 and 3,000 cell captures is provided in Supplemental Data 2.
Fig. 2.
(A) Base peak chromatograms following LC-MS/MS analysis of LMPC captures of three biological replicates of 3,000 keratinocytes collected from a single chronic wound biopsy; (B) Log-log scatter plots comparing spectral count data from two biological replicates each from 500, 1,000 or 3,000 keratinocytes; (C) Photomicrographs of hematoxylin stained histological sections from frozen biopsies before and after sample capture using LMPC.
Fig. 2C i shows the chronic wound tissue section before and after LMPC collection of keratinocytes. Following LMPC sample processing and analysis, protein identifications and respective spectral counts were parsed from each sample data file and then combined taking in consideration parsimony for protein groupings using an in-house developed application. Spectral count data from normal skin and early and late wound keratinocyte LMPC isolates was analyzed using edgeR for pair-wise comparisons between identified proteins in normal vs early wound, normal vs. late wound and early wound vs late wound. The data analysis pipeline is shown in Fig. 1B.
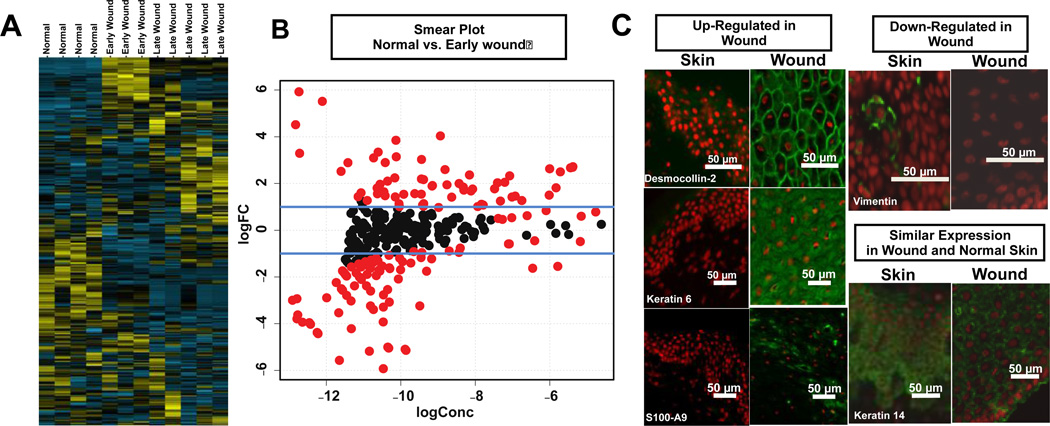
The statistical comparison of normal vs. early and late wound keratinocytes identified over 144 and 98 statistically significant differentially expressed proteins / protein groups. Comparison of the spectral count data of the early wound vs. late wound biopsies (collected one month apart) identified 59 statistically significant differentially expressed proteins / protein groups. The number of significant differentially expressed proteins represents a significant fraction of the proteins identified. This is not surprising as these cells types are differentiated between normal keratinocytes and those involved wound healing. The raw spectral counts and edgeR analysis of wound spectral counts comparing normal to early and late wound as well as the comparison of early to late wound keratinocytes are provided as a spreadsheet in Supplemental Data 3. The unsupervised hierarchical clustering of the spectra counts correctly grouped normal, early and late wound keratinocytes (Fig. 3A).
Fig. 3.
Analysis of keratinocytes from normal skin and chronic diabetic wound biopsies. (A) unsupervised hierarchical cluster analysis showing grouping of normal (4 biological replicates) vs. early (3 biological replicates) and late (5 biological replicates) from chronic wound with yellow indicating positive and blue indicating negative expression; (B) smear plots showing proteins (red) with BH corrected p-values > 0.05; (C) Immunohistochemistry shows expected staining of both differentially and non-differentially abundant proteins following spectral count analysis. Staining was performed on frozen sections from punch biopsies. Dapi (nuclei) = red, protein of interest = green.
Smear plots (log2 concentration vs. log2 fold change) of the normalized differential spectral count data from the keratinocytes of normal skin vs. the two chronic wounds show the distribution of significant protein expression changes as determined by the edgeR analysis (Fig. 3B). The differential expression of many of these identified proteins matches previous reports describing wound or chronic wound healing [26–30]. Immunohistochemistry was used to validate 3 over-abundant (desmocollin-2, S100-A9, Keratin 6), 1 under-abundant (vimentin) and one non-differential (keratin 14) protein in early wound biopsies (Fig. 3C).
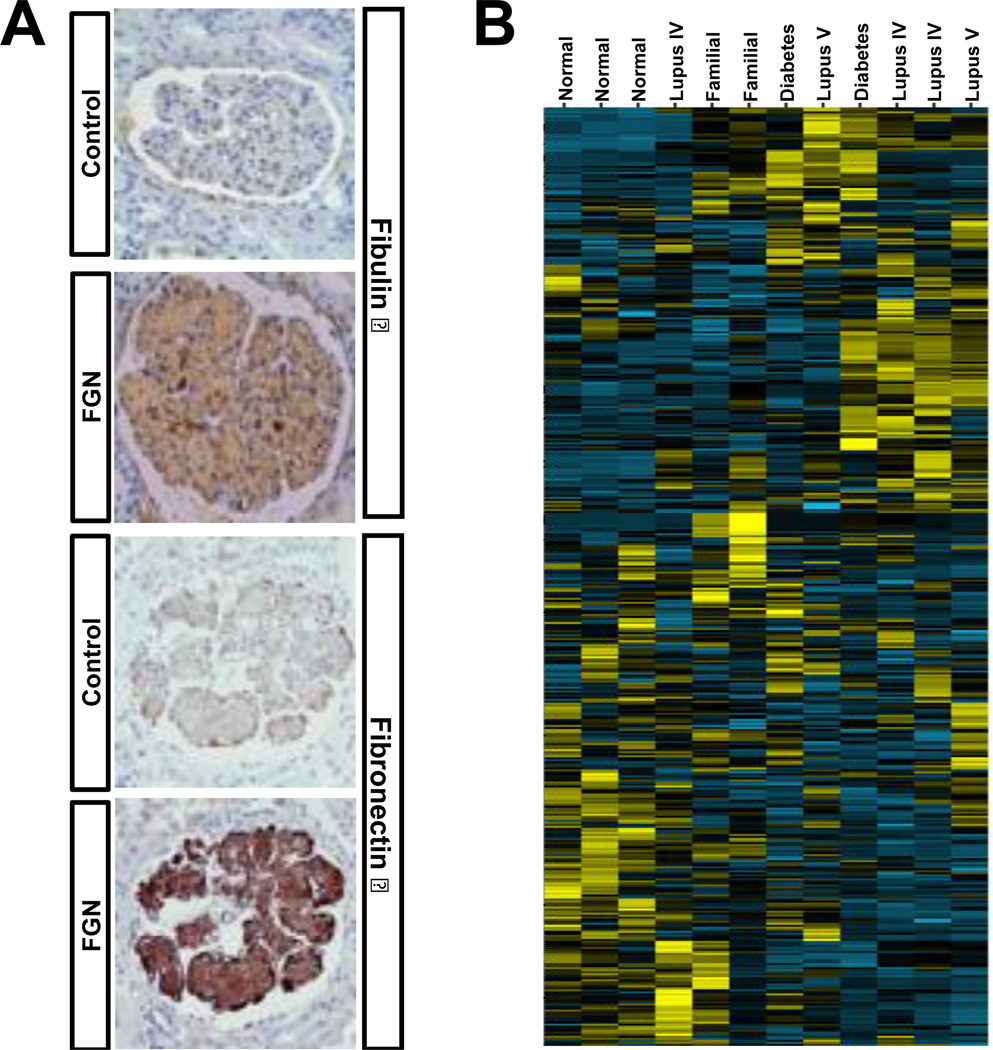
Glomeruli isolated with LMPC from needle biopsies of two patients known to have fibronectin glomerulopathy (FNG) were compared to glomeruli isolated from normal (n=3), diabetic (n=2), and lupus nephritis (class IV n=3 and class V n=2) needle biopsies. Fig. 2C ii shows pre- and post-LMPC collection of glomeruli. FNG is a rare familial kidney disease that eventually leads to kidney failure and is thought to be caused by abnormal glomerular deposition of fibronectin[31]. Although fibronectin deposits have been confirmed using immunohistochemistry in prior studies, fibronectin staining is often mild suggesting other proteins present in the glomerular deposits may be more informative. Proteomic analysis of the FNG glomeruli showed a significant increase in the presence of fibronectin isoforms 1, 3, 4, 5, 8. More interestingly, the spectral counts revealed several additional changes in protein abundance compared to normal glomeruli, including an increase in fibulin isoforms 1 and 5. Immunohistochemistry confirmed the increases in fibronectin 1 and fibulin 1 (Fig. 4A). Unsupervised cluster analysis of glomerular proteins indicated good correlation withn the normal and FNG biopsies (Fig. 4B). In lupus nephritis, a disease with a different pathogenesis than FNG, examination of spectral count data showed over abundance of IgG and complement factor-proteins that are well-known to be deposited in the glomeruli of patients during active glomerulonephritis. Similar to the wound/normal samples, a signifcant fraction of the proteins identified were found to be differentially expressed in FNG when compared to the normal kidney glomeruli. These results were also not surprising. The protein changes we are observing are due to protein accumulation in the glomeruli and the inflammatory response proteins associated with damage to the kidney. The complete list of spectral count data for glomerular proteins from all biopsies and the edgeR analysis of normal vs FNG are provided in Supplemental Data 4.
Fig. 4.
Analysis of glomerular proteins from normal and diseased diseased kidneys. (A) Immunohistochemistry showing expected over-abundance of fibulin 1 and fibronectin-1 in the FNG glomeruli; (B) Unsupervised hierarchical cluster analysis of 809 proteins from 3 normal and 9 diseased kidneys with yellow indicating positive and blue indicating negative expression showing grouping of normal and diseased kidneys.
DISCUSSION
These examples demonstrate the capability of this proteomic workflow to characterize samples with <10,000 cells obtained by LMPC from frozen biopsies. The ethanol used to fix the frozen biopsy sections on the LMPC PEN slides precipitates the proteins present in a frozen histological sample and avoids protein cross-linking. This method allows for efficient recovery of proteins from the ethanol fixed LMPC isolates collected from the frozen biopsies using the methods described here. Variations of the methods described here may also achieve successful protein digestion from a formalin fixed specimens, which is the more common method of biopsy preservation and also amenable to LMPC combined with proteomics [15, 32].
The ability to use LMPC to isolate homogeneous populations of cells from complex tissue samples such as the frozen clinical biopsies examined here affords one an opportunity to characterize and dissect the underlying physiological mechanisms that lead or contribute to a disease. The ability to gain insight into these underlying physiological mechanisms will provide for an enhanced capability to design novel and more effective treatment(s) for these disorders. The approach is robust, with all individual biopsies described here requiring only one LMPC capture and LC-MS/MS analysis. This was particularly critical for the analysis of the glomeruli given that the individual needle biopsies from the two FNG patients were each one-of-a-kind with no opportunity for a repeat biopsy or LMPC collection.
In order to function as an effective proteomics method for characterization of differential protein expression in small tissue samples such as the frozen needle biopsies (1 mm X 3 mm) examined in this study, it was presumed that the maximum number of LMPC homogenous collected cells should be 10,000 or less. This assumption requires both sensitive and highly reproducible methods. The base peak chromatograms of the three 3,000 cell biological replicates as well as the log-log scatter plots comparing spectral counts from two biological replicates clearly demonstrates the method reproducibility (Fig. 2A and B). Perhaps most remarkable is the ability of the 500 wound keratinocyte LMPC isolates to identify proteins / protein groups in a reproducible manner as demonstrated by the linearity of the log-log scatter plots. Additionally, the unsupervised cluster analysis of all of the biologic specimens described here further supports the capability of these methods to provide the reproducibility necessary for small sample analysis.
Pearson and Spearman rank correlation analysis were performed on the normal (N=4), early wound (N=3) and late wound (N=5) biopsies (Supplemental Data 5) to assess technical reproducibility. In addition principal component analysis (PCA) was performed on all three examples. The PCA analysis show tight clustering across the technical replicates for the wound example consistent with the hierarchical clustering results. In addition the Pearson and Spearman rank correlation analysis showed very good consistency within replicates (Normal r=0.9603–0.9822, Early Wound r=0.9881–0.9959, Late Wound r=0.9086–0.9825). These data are consistent with strong linearity observed when plotting protein spectral counts across replicates (see example in Supplemental Data 5). Not surprisingly the correlation between peptide spectral counts is not as strong due to fewer counts observed for each individual peptide. Protein IDs across samples was also very consistent. An example Venn diagram comparing three replicate normal biopsies is provided in Supplemental Data 5.
For the kidney data we also saw good reproducibility across biological replicate biopsies for normal individuals and the familial FNG patients. The diabetic and SLE patients showed much higher heterogeneity (although different from normal) and would therefore require a greater number of samples to better rank differentially expressed proteins between these groups. While this study set is smaller than would be needed to evaluate diabetic and SLE cases, the normal and FNG case demonstrate the applicability of this approach to clinic material. For future experiments where biological variability is high, the number of biological replicates would be increased and statistical analysis that includes mixture modeling or pairwise analysis would be employed to rank differentially expressed proteins.
In order to validate the capability of the method for biomarker discovery, immunohistochemistry studies were used to verify the differentially expressed proteins identified by analysis of spectral counts. The differential expression of 5 keratinocyte normal vs. early wound proteins selected from the edgeR analysis were confirmed using immunohistochemistry. As noted in the results, the methods employed here for protein isolation and data analysis were sensitive enough to identify the differential expression of desmocollin 2 in spite of the narrow difference in spectral counts between the normal and early wound isolates. Additionally, the previously reported FNG fibronectin 1 over-abundance was confirmed by both the spectral counts analysis and immunohistochemistry. Finally, the novel finding identifying increased accumulation of fibulin 1 in FNG was also confirmed using immunohistochemistry. These confirmations of the spectral count statistical analysis establish the ability of this technology to provide identification of biomarkers and function as a discovery tool for diseased tissue.
The ability of the spectral count data to identify which proteins are differentially expressed between disease and normal samples is also supported by standard clinical observations and/or previous reports. For instance, it is well established that both IgG and complement factors are two protein families involved in Lupus pathogenesis and are found deposited in the glomeruli of patients during active disease. Here, the spectral count data confirm the increased presence of complement proteins and immunoglobulins in Lupus glomeruli. The over-abundance of complement components in the FNG and diabetic patients, was unexpected, but clearly demonstrates the utility of this technique as a discovery tool. Furthermore, many of the proteins differentially expressed in the wound keratinocytes have been described in prior studies [26–30]. In an additional study, 65 genes identified as up or down-regulated using mRNA microarray analysis examining chronic wound biopsies are also found in the keratinocyte wound LMPC samples collected here [33]. The direction of protein expression determined by the edgeR analysis matched gene expression in 75% of these common identifications.
Confirmation of the edgeR spectral count analysis using immunohistochemstry substantiates the reliability of the large array of identified, differentially expressed proteins shown in the volcano plots. The ability to identify a large number of differentially expressed proteins provides opportunities to discover physiological mechanisms that are amenable to novel therapeutic intervention. For example, one of the proteins significantly down-regulated in both chronic early and late wounds is ATPase alpha 1 (ATP1A). This protein provides energy for the active transport of nutrients, and its down-regulation could impair the ability of keratinocytes to participate effectively in the healing process [34]. This report also describes the ability of glycosides such as ouabain or digoxin to up-regulate the expression of ATP1A, and use of ouabain for wound treatment was associated with improved wound healing in a rat wound model.
CONCLUSION
In conclusion, the robust qualities and sensitivity of the proteomic workflow described here will allow for routine proteomic analysis of frozen tissue biopsies. The efficiency of protein recovery from the LMPC isolates provides the capability for analysis of small samples (<10,000 cells). The general utility of this workflow will advance the capability to identify protein biomarkers in human tissues leading to greater understanding of disease molecular physiology for design, development and testing of novel therapeutic treatments. Additionally, the quantitative analysis of proteomic profiles may allow for better disease staging and provide clinicians with the ability to deliver more guided and appropriate treatment. The comparison of normal and FNG glomerular proteome profiles has already provided new insight into the underlying disease mechanism and may eventually “translate” into better targeted therapeutic intervention for treatment or improved individualized patient disease classification/stratification.
Supplementary Material
Highlights.
-
➢
highly efficient proteomic workflow for Laser Microdissection proteomics
-
➢
demonstrates proteomics analysis on as few as 500 captured cells
-
➢
easily combined with label-free spectral count quantitation
-
➢
tissue proteomics carried out on limited clinical biospecimens
ACKNOWLEDGEMENTS
The study was funded by The Ohio State University and the National Institutes of Health RR025755 (MF and SR), DK074661 (BHR), DK076566 (SR), GM 077185 (CKS) and GM 069589 (CKS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.P.S., C.K.S., S.R. and M.A.F. conceived the study; J.P.S. engineered the approach; S.B., C.K.S., and S.R. provided the wound biopsies and performed their IHC; A.S. and B.H.R., provided the kidney biopsies and performed their IHC; A.M.; C.T. and S.L. provided assistance with statistical analysis; J.P.S., A.S.M. B.H.R. and M.A.F wrote the paper.
REFERENCES
- 1.Anderson CE, McLaren KM, Rae F, Sanderson RJ, Cuschieri KS. Human papilloma virus in squamous carcinoma of the head and neck: a study of cases in south east Scotland. J Clin Pathol. 2007;60:439–441. doi: 10.1136/jcp.2005.033258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Espina V, Heiby M, Pierobon M, Liotta LA. Laser capture microdissection technology. Expert Rev Mol Diagn. 2007;7:647–657. doi: 10.1586/14737159.7.5.647. [DOI] [PubMed] [Google Scholar]
- 3.Gutstein HB, Morris JS. Laser capture sampling and analytical issues in proteomics. Expert Rev Proteomics. 2007;4:627–637. doi: 10.1586/14789450.4.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu BJ. Combining laser capture microdissection and proteomics: methodologies and clinical applications. Proteomics Clin Appl. 2010;4:116–123. doi: 10.1002/prca.200900138. [DOI] [PubMed] [Google Scholar]
- 5.Mustafa D, Kros JM, Luider T. Combining laser capture microdissection and proteomics techniques. Methods Mol Biol. 2008;428:159–178. doi: 10.1007/978-1-59745-117-8_9. [DOI] [PubMed] [Google Scholar]
- 6.Liu NQ, Braakman RB, Stingl C, Luider TM, Martens JW, Foekens JA, et al. Proteomics Pipeline for Biomarker Discovery of Laser Capture Microdissected Breast Cancer Tissue. J Mammary Gland Biol Neoplasia. 2012 doi: 10.1007/s10911-012-9252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braakman RB, Luider TM, Martens JW, Foekens JA, Umar A. Laser capture microdissection applications in breast cancer proteomics. Methods Mol Biol. 2011;755:143–154. doi: 10.1007/978-1-61779-163-5_11. [DOI] [PubMed] [Google Scholar]
- 8.Lawrie LC, Curran S. Laser capture microdissection and colorectal cancer proteomics. Methods Mol Biol. 2005;293:245–253. doi: 10.1385/1-59259-853-6:245. [DOI] [PubMed] [Google Scholar]
- 9.Cha S, Imielinski MB, Rejtar T, Richardson EA, Thakur D, Sgroi DC, et al. In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol Cell Proteomics. 9:2529–2544. doi: 10.1074/mcp.M110.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greengauz-Roberts O, Stoppler H, Nomura S, Yamaguchi H, Goldenring JR, Podolsky RH, et al. Saturation labeling with cysteine-reactive cyanine fluorescent dyes provides increased sensitivity for protein expression profiling of laser-microdissected clinical specimens. Proteomics. 2005;5:1746–1757. doi: 10.1002/pmic.200401068. [DOI] [PubMed] [Google Scholar]
- 11.Umar A, Luider TM, Foekens JA, Pasa-Tolic L. NanoLC-FT-ICR MS improves proteome coverage attainable for approximately 3000 laser-microdissected breast carcinoma cells. Proteomics. 2007;7:323–329. doi: 10.1002/pmic.200600293. [DOI] [PubMed] [Google Scholar]
- 12.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proc Natl Acad Sci U S A. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johann DJ, Rodriguez-Canales J, Mukherjee S, Prieto DA, Hanson JC, Emmert- Buck M, et al. Approaching solid tumor heterogeneity on a cellular basis by tissue proteomics using laser capture microdissection and biological mass spectrometry. J Proteome Res. 2009;8:2310–2318. doi: 10.1021/pr8009403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johann DJ, Mukherjee S, Prieto DA, Veenstra TD, Blonder J. Profiling solid tumor heterogeneity by LCM and biological MS of fresh-frozen tissue sections. Methods Mol Biol. 2011;755:95–106. doi: 10.1007/978-1-61779-163-5_8. [DOI] [PubMed] [Google Scholar]
- 15.Vrana JA, Gamez JD, Madden BJ, Theis JD, Bergen HR, 3rd, Dogan A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114:4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 16.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, et al. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14:1002–1014. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 17.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 18.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fei SS, Wilmarth PA, Hitzemann RJ, McWeeney SK, Belknap JK, David LL. Protein database and quantitative analysis considerations when integrating genetics and proteomics to compare mouse strains. J Proteome Res. 2011;10:2905–2912. doi: 10.1021/pr200133p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–3557. doi: 10.1021/pr070230d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 23.Colinge J, Chiappe D, Lagache S, Moniatte M, Bougueleret L. Differential proteomics via probabilistic peptide identification scores. Anal Chem. 2005;77:596–606. doi: 10.1021/ac0488513. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 25.Piedimonte G, Guetard D, Magnani M, Corsi D, Picerno I, Spataro P, et al. Oxidative protein damage and degradation in lymphocytes from patients infected with human immunodeficiency virus. J Infect Dis. 1997;176:655–664. doi: 10.1086/514087. [DOI] [PubMed] [Google Scholar]
- 26.Dover R, Watt FM. Measurement of the rate of epidermal terminal differentiation: expression of involucrin by S-phase keratinocytes in culture and in psoriatic plaques. J Invest Dermatol. 1987;89:349–352. doi: 10.1111/1523-1747.ep12471751. [DOI] [PubMed] [Google Scholar]
- 27.Mazzalupo S, Wong P, Martin P, Coulombe PA. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev Dyn. 2003;226:356–365. doi: 10.1002/dvdy.10245. [DOI] [PubMed] [Google Scholar]
- 28.Sharma A, Singh AK, Warren J, Thangapazham RL, Maheshwari RK. Differential regulation of angiogenic genes in diabetic wound healing. J Invest Dermatol. 2006;126:2323–2331. doi: 10.1038/sj.jid.5700428. [DOI] [PubMed] [Google Scholar]
- 29.Pollins AC, Friedman DB, Nanney LB. Proteomic investigation of human burn wounds by 2D-difference gel electrophoresis and mass spectrometry. J Surg Res. 2007;142:143–152. doi: 10.1016/j.jss.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moll I, Houdek P, Schafer S, Nuber U, Moll R. Diversity of desmosomal proteins in regenerating epidermis: immunohistochemical study using a human skin organ culture model. Arch Dermatol Res. 1999;291:437–446. doi: 10.1007/s004030050435. [DOI] [PubMed] [Google Scholar]
- 31.Castelletti F, Donadelli R, Banterla F, Hildebrandt F, Zipfel PF, Bresin E, et al. Mutations in FN1 cause glomerulopathy with fibronectin deposits. Proc Natl Acad Sci U S A. 2008;105:2538–2543. doi: 10.1073/pnas.0707730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alkhas A, Hood BL, Oliver K, Teng PN, Oliver J, Mitchell D, et al. Standardization of a Sample Preparation and Analytical Workflow for Proteomics of Archival Endometrial Cancer Tissue. Journal of proteome research. 2011 doi: 10.1021/pr2007736. [DOI] [PubMed] [Google Scholar]
- 33.Brem H, Stojadinovic O, Diegelmann RF, Entero H, Lee B, Pastar I, et al. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol Med. 2007;13:30–39. doi: 10.2119/2006-00054.Brem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Okdi N, Smaili S, Raju V, Shidyak A, Gupta S, Fedorova L, et al. Effects of cardiotonic steroids on dermal collagen synthesis and wound healing. J Appl Physiol. 2008;105:30–36. doi: 10.1152/japplphysiol.00119.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.