Abstract
We describe and characterize a stromal-cell independent culture system that efficiently supports pro-B cell to IgM+ B-cell development with near normal levels of IgH and Igκ diversity. Pro-B cells present in non-adherent bone marrow cells proliferate in the presence of IL-7 and subsequent to the removal of IL-7 and addition of BAFF, differentiate normally into IgM+ B cells. B-cell development in vitro closely follows the patterns of development in vivo with culture derived (CD) B cells demonstrating characteristic patterns of surface antigen expression and gene activation. IgM+ CD B cells respond to TLR stimulation by proliferation and differentiation into antibody-secreting cells. Self-reactive IgM+ B-cell development is blocked in 3H9 IgH knockin mice; however, cultures of 3H9 IgH knockin pro-B cells yields high frequencies of “forbidden”, autoreactive IgM+ B cells. Furthermore, serum IgG autoantibody exceeded that present in autoimmune, C4−/− animals following the reconstitution of RAG1−/− mice with IgM+ CD cells derived from BL/6 mice.
Keywords: B-cell, Development, Autoimmunity, Interleukin-7, BAFF
1. Introduction
The utility of in vitro culture methods to identify factors necessary for B-lymphopoiesis is well known (Ray et al., 1998; Cho et al., 1999; Hess et al., 2001). Most culture systems used to study lymphopoiesis and the development of hematopoietic progenitor cells (HPC) employ stromal cell lines, e.g., OP9 (Nakano et al., 1994) and S17 (Collins and Dorshkind, 1987), to provide necessary growth and differentiation factors (Billips et al., 1992). These stromal cells provide essential cytokines that support hematopoiesis (Baird et al., 1999) and express adhesion molecules that in vivo, define specialized bone marrow (BM) niches that promote B-cell development (Tokoyoda et al., 2004). The capacity of stromal cell culture systems to support the development of B-lineage cells from HPC has been well characterized (Rolink et al., 1991a; Nakano et al., 1994) and contributed to the discovery of IL-7 as a key cytokine for the stromal-dependent phase of mouse B-cell development (Cumano et al., 1990; Billips et al., 1992). However, stromal cell cultures are incapable of efficient de novo production of IgM+ B cells without the addition of exogenous cytokines (Flt-3L) or B-cell mitogens (LPS) (Ray et al., 1998; Cho et al., 1999).
In part, the presence of IL-7 in stromal cell cultures limits IgM+ B-cell development. B-cell precursors maintained in the presence of IL-7 continue to proliferate, retain their Igκ and Igλ gene loci in germ-line configuration and, therefore, do not express surface IgM (ten Boekel et al., 1995). Withdrawal of IL-7 from B-cell cultures decreases proliferation and is associated with increased expression of genes (RAG1/2) that are required for light chain (LC) gene rearrangements (Rolink et al., 1991a; Rolink et al., 1991b). IL-7-mediated regulation of B-cell maturation is suggested to occur either by direct control of Ig recombinase gene activity (Billips et al., 1995) or by controlling cell cycle (Li et al., 1996). B-cell receptor (BCR) transgenic (Tg) pre-B cells require IL-7 for proliferation but are able to bypass IL-7-mediated developmental blockade as LC rearrangement is no longer required to continue differentiation (Melamed et al., 1997; Tze et al., 2000). Several groups have demonstrated that B lymphopoiesis can occur in the absence of BM stromal cells if the proper cytokines are provided (Tze et al., 2000; Luo et al., 2009) Indeed, Claudio et al. reported that the removal of IL-7 and addition of BAFF to their B-cell culture system yields surface IgM+ B cells (Claudio et al., 2002).
The B-cell activating factor belonging to the TNF family (BAFF) is a cytokine that promotes B-cell survival (Schneider et al., 1999) and B-cell maturation (Batten et al., 2000; Rolink et al., 2002; Gorelik et al., 2004). BAFF−/− and BAFF-R−/− mice show an increase T1 B-cell compartment and have substantially reduced amounts of more mature B-cell subsets (Schiemann et al., 2001; Thompson et al., 2001). Conversely, BAFF Tg mice have elevated numbers of mature B cells and develop autoimmune-like manifestations (Mackay et al., 1999). BAFF signaling is mediated through three independently regulated receptors on B cells: B cell activating factor receptor (BAFF-R), transmembrane activator and CAML interactor (TACI) and B cell maturation antigen (BCMA) (reviewed in (Mackay et al., 2003)).
One clear limitation of stromal-independent cultures is that newly-formed B-cell populations have not been systematically characterized. We describe and detail a stromal-independent culture system that supports the survival, proliferation and differentiation of virtually all BM B-cell developmental stages. These culture-derived (CD) B-lineage cells are phenotypically and genotypically similar to their in vivo counterparts. Furthermore, we show that our culture system permits the development of autoreactive B cells which are normally purged during their development in the BM. CD cells were used to reconstitute peripheral lymphoid tissues of RAG−/− mice and restored both serum IgM and IgG to control levels. CD B cells maintained their bias toward autoreactive specificities even after transfer to RAG−/− hosts.
2. Materials and Methods
2.1 Mice
C57BL/6, RAG1 deficient (B6.129S7-Rag1tm1Mom/J) mice (both, The Jackson Laboratory, Bar Harbor ME), and congenic C4−/− (Fischer et al., 1996) mice were maintained in the Duke University vivarium. Congenic 3H9R HC-KI (Chen et al., 1995) mice were kindly provided by Dr. R. Eisenberg (University of Pennsylvania). RAG1 deficient mice were reconstituted with 2×107 CD B- and T cells by i.v. injection; these CD-RAG mice were held for 3–5 weeks before their use in experiments. Mice were housed in specific pathogen-free conditions at the Duke University Animal Care Facility and given sterile bedding, water and food; animals were entered into experiment protocols at 6–8 wk of age. All experiments were approved by the Duke University Animal Care and Use Committee.
2.2 Tissue preparation
Mice were sacrificed by cervical dislocation. BM was collected from long bones of the hind legs by flushing with 1 ml cold, Iscove’s modified Dulbecco’s Medium (IMDM) containing 10% defined fetal bovine serum (HyClone, Logan Utah), 5.5 × 10−5 M 2-ME, penicillin (10 Units/ml), and streptomycin (10µg/ml). BM was disaggregated and dispersed into single-cell suspensions by repeated pipetting. The preparation of cells from spleen, lymph node and thymus has been described (Chen et al., 2000a). Viable cells from dissociated tissues were enumerated in hemocytometers by Trypan Blue exclusion.
2.3 Flow cytometry
To identify, characterize, and isolate lymphocytes, mAb included: B220-PacificBlue (RA3-6B2), CD23-biotin (B3B4), CD93-APC (AA4.1), BP-1-PE (6C3), CD24-biotin (M1/69), CD24-FITC (M1/69), CD43-APC (S7) and APC-Alexa750-conjugated streptavidin were purchased from BD Pharmingen (San Diego, CA); and anti-mouse IgM-PEcy7 (eB121-15F9), anti-mouse IgD-FITC (11–26), CD21-PE (eBio8D9), CD5-PE (53-7.3), CD1d-PE (1B1), CD80-FITC (16-10A1), CD86-PE (GL1), CD4-PEcy7 (L3T4), CD8-PEcy5 (Ly-2), CD44-FITC (IM7), CD62L-PE (MEL-14), Gr-1-PEcy5 (Ly6G), CD11b-PEcy5 (M1/70) and TCRβ-APC (H57-597) were purchased from eBioscience (San Diego, CA). 106 cells were suspended in FACS Buffer and labeled with mAb described above. FACS buffer contained 1xPBS (pH7.2) with 3% FBS (Sigma) and 0.01% Sodium Azide. Propidium iodide (PI) was used to exclude dead cells from our samples. All FACS analysis was performed using a BD LSRII or Canto cytometer and presented with FlowJo software. Cell sorting was performed on a BD FACSVantage cytometer.
2.4 B-cell culture system
BM contents from single mice were dispersed in 5 ml of IMDM by repeated pipetting; cell suspensions from multiple mice were routinely pooled in 10 cm culture dishes. BM cells were incubated (15 min; 37°C) to permit cell attachment. Non-adherent cells were recovered by centrifugation (4°C; 400 x G; 5min), resuspended in 1 ml ACK buffer (1 min; 0°C) to remove erythrocytes, and immediately washed in 10 ml IMDM (Chen et al., 2000a). Washed live cells from BM were enumerated in Trypan Blue and transferred into T-75 culture flasks (7.5×105 cells/ml; 25 ml) for 4 d in IMDM supplemented with recombinant mouse IL-7 (10 ng/ml) (R&D Systems, Minneapolis, MN). Typically, non-adherent BM cells were twice cultured in IL-7 containing IMDM (4 days, each) followed by a single round of culture in IMDM containing BAFF (20–100 ng/ml; 3–4 days). Between sequential cultures, cells were washed and re-plated at the initial concentration.
2.5 ELISA
ELISA plates (BD Falcon) were coated (overnight, 4°C) with 2 µg/ml (50µl/well) of goat anti-mouse Ig(H+L) (Southern Biotechnology Associates, Birmingham, AL) in carbonate buffer (0.1M; pH9.5). Coated plates were washed with 1xPBS (pH7.4) containing 0.1% Tween-20 and 0.5% BSA (USB Corporation). Wells were incubated (2hrs; 25°C) with blocking buffer (PBS (pH7.4), 0.5% BSA). Serum samples were initially diluted 1:1000; followed by serial 3-fold dilutions. Purified mouse IgM (B1–8) and IgG (H33Lγ1) mAbs were used as a standard (30 µg/ml to 0.5 ng/ml) to determine serum Ab concentrations. HRP-conjugated goat anti-mouse IgM and goat anti-mouse IgG were used to detect bound antibody (Southern Biotechnology Associates, Birmingham, AL).
2.6 ELISpot
ELISpot plates (Millipore) were coated (overnight; 4°C) with 2 µg/ml (50 µl/well) of goat anti-mouse Ig(H+L) in carbonate buffer (0.1M; pH9.5). Coated plates were washed with PBS (pH7.4) containing 0.1% Tween-20 and 0.5% BSA. Wells were incubated (2hrs; 25°C) with blocking buffer (PBS (pH7.4), 0.5% BSA). B cells were incubated (3×104 cells/well; 200µl IMDM) with LPS (5µg/ml; 1–3 d). LPS-activated B cells were washed (3x; 5ml IMDM) and were incubated (0.5–1×103 cells/well) (37°C; 4hrs) in IMDM. Plates were washed with dH20 (2x; 200µl/well) and blocking buffer (1x; 200µl/well). Plated were incubated with blocking buffer (1–2 d; 4°C). Membranes were probed with goat-anti-mouse IgM-AP detection Ab (1hr; 25°C). SIGMA FAST BCIP/NBT reagent (Sigma) was used to develop spots (50µl/well; 20 min, 25°C); this reaction was stopped by flooding wells with dH20.
2.7 Immunofluorescence
NIH-3T3 cells (1–2×104 cells/ml; 10mls) were plated onto 10cm tissue culture plates (24hrs; 37°C) containing sterile glass coverslips. Coverslips were removed and immersed (10 min; −20°C) in methanol:acetone (1:1) for cell fixation. Slides containing C. luciliae (Scimedx Corporation, Denville, NJ) or coverslips containing NIH-3T3 cells were rehydrated (PBS (pH7.4); 30 min; 25°C). Samples were blocked (2 hr; 25°C) using PBS (pH7.2) containing rat anti-mouse CD16/CD32 (1%), purified rat IgG (5%), FBS (10%) and Tween-20 (0.1%). Samples were washed (1 min) in PBS (pH7.2) containing BSA (1%) and Tween-20 (0.1%). Samples were labeled with serum (1:20) or 2-ME treated hybridoma Ab (50µg/ml) (2hrs; 25°C) followed by extensive washing (2× 150mls; 10min each; 1× 150mls; overnight). Ab was detected using goat anti-mouse Igκ-FITC or goat anti-mouse Igλ-FITC Ab (2hrs; 25°C) followed by extensive washing (3× 150mls; 10min each). Coverslips were mounted to slides using Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL). Images were acquired using a Zeiss Axiovert 200M confocal immunofluorescent microscope.
2.8 Sequence analysis of V(D)J rearrangements
Genomic DNA was isolated from sorted B-cell subsets from cultures or BL/6 mice by phenol-chloroform extraction (Invitrogen). V(D)J rearrangements were amplified by semi-nested PCR using Pfu polymerase (Stratagene, La Jolla, CA) (Han et al., 1997) with 5` primers specific for VH1, Vκ4 or Vκ5 genes, and a reverse primer specific for JH2 or Jκ2 (Supplemental Table 1). This approach allows the amplification of VH- or Vκ- to JH1 or JH2 and Jκ1 or Jκ2, respectively. Amplified V(D)J products were gel purified and ligated into pCR2.1 plasmid (Invitrogen) and cloned by bacterial transformation (Jacob et al., 1991). Cloned V(D)J inserts were sequenced in an Applied Biosystems automated DNA sequencer and analyzed by IMGT/V-QUEST (http://imgt.cines.fr) and NCBI blast search (http://www.ncbi.nlm.nih.gov/BLAST) software. Light chain rearrangements of 3H9 CD hybridomas were amplified by PCR as previously described (Rohatgi et al., 2008) and sequenced as described above.
2.9 Quantitative real-time PCR
A. Genomic DNA was isolated from sorted B-cell subsets of cultures or BL/6 mice by phenol-chloroform extraction (Invitrogen). VDJ rearrangements were amplified from ∼20ng of genomic DNA by q-rtPCR method using SYBR Green PCR reagent (Applied Bioscience) with forward primers specific for VH1, VH2 or VH9 genes and a reverse primer specific for JH1 (Supplemental Table 1). The relative abundance of rearranged DNA was normalized to CD14 signal and was calculated by the comparative threshold cycle method (Ueda et al., 2007).
B. Expression of B-cell mRNA was measured by q-rtPCR. Briefly, total RNA was extracted from sorted cell populations (∼105 cells) using Trizol/chloroform extraction (Invitrogen). cDNA was prepared by standard methods (Ueda et al., 2005; Ueda et al., 2007) using oligo(dT)12–18 primers and Superscript III reverse transcriptase (both, Invitrogen, Carlsbad CA). PCR primers used in this study are listed (Supplemental Table 1) or previously described (Lazorchak et al., 2006). The relative expression levels of each message was normalized to Igβ message and were calculated by the comparative threshold cycle method (Ueda et al., 2007). All measurements were performed using a BioRad iCycler equipped with a MyiQ optical module (Bio-Rad).
3. Results
3.1 Efficient generation of IgM+IgD+ B cells in vitro
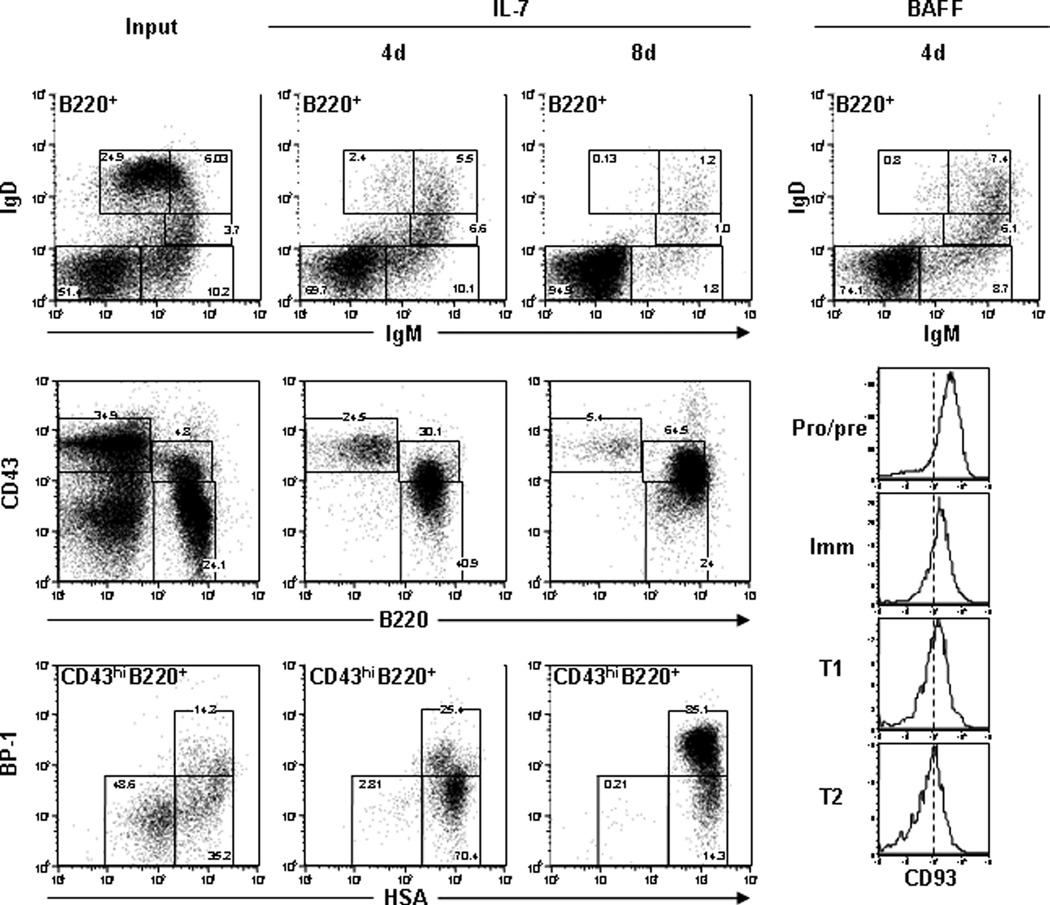
Non-adherent BM cells were collected and prepared for culture in tissue culture flasks (see 2.4). At the initiation of culture, 25–30% of BM cells were B220+; however, after 4 days with IL-7, ≥80% of 1° CD cells expressed B220 and cultures increased 2–3 fold in B220+ cell number (Table 1). To identify the B cells present in BM and recovered in 1° cultures, each cohort was labeled with mAb to B220, IgM, IgD, CD23 and CD21 to identify B-cell subsets (Fig. 1) (Ueda et al., 2007). The frequency of B220+ pro-/pre- (IgMnegIgDnegCD93hiCD23negCD21neg), immature (IgMloIgDnegCD93hiCD23negCD21neg), T1 (IgMhiIgDloCD93intCD23negCD21neg), T2 (IgMhiIgDhiCD93intCD23hiCD21int) and mature (IgMloIgDhiCD93negCD23hiCD21int) B cells was analyzed (Fig. 1). Each CD B-cell population expressed CD93, CD23 and CD21 at similar levels compared to BM B lymphocytes (data not shown). Pro-/pre-B cells were the most abundant population of 1° CD cells (∼80% of B220+ cells) (Fig. 1 and Table 1). Immature and T1 B cells were each present at 5–10% in the cultures; whereas, T2 and mature B cells were each present at ≤5% (Fig. 1 and Table 1). In addition to these characterizations, BM and 1° CD cells were analyzed using Hardy’s method (Hardy et al., 1991) to define stages of B-cell ontogeny. Fraction “B” (early pro-B) (B220loCD43hiBP-1loHSAint) and fraction “C-C`” (late pro- and preB-I) (B220loCD43hiBP-1hiHSAint) cells constituted ∼70% and ∼25%, respectively, of the B220loCD43hi CD cells (Fig. 1). Compared to BM, our cultures produced a substantial enrichment for pro- and preB-I compartments.
Table 1.
a. Generation of CD B cells (No.s)
| Input Cell No. (x106) |
B220+ (x106) |
Pro/pre (x106) |
Immature (x105) |
T1 (x105) |
T2 (x105) |
Mature (x105) |
|
|---|---|---|---|---|---|---|---|
| Input | 18.8 | 5.3 ± 0.5 |
3.9 ± 0.2 | 4.9 ± 1.4 | 2.4 ± 0.8 | 1.4 ± 0.4 | 3.1 ± 0.8 |
|
1° CD IL-7 |
18.8 | 15.5 ± 5.2 |
12.6 ± 4.2 |
14.1 ± 4.8 | 8.8 ± 3.3 | 3.3 ± 0.6 | 1.0 ± 0.3 |
|
2° CD IL-7 |
18.8 | 53.3 ± 12.3 |
51.8 ± 11.8 |
5.6 ± 1.9 | 2.5 ± 0.5 | 1.3 ± 0.4 | 0.3 ± 0.1 |
|
3° CD BAFF |
18.8 | 8.2 ± 1.3 |
5.5 ± 0.4 | 7.1 ± 1.7 | 6.3 ± 0.7 | 5.3 ± 0.5 | 0.6 ± 0.7 |
| b. Generation of CD B cells (%) | |||||||
|---|---|---|---|---|---|---|---|
| Input Cell No. (x106) |
B220+ (%) |
Pro/pre (%) |
Immature (%) |
T1 (%) |
T2 (%) |
Mature (%) |
|
| Input | 18.8 | 25.2 ± 4.2 |
59.2 ± 9.1 |
11.4 ± 3.0 | 3.8 ± 1.0 | 4.2 ± 2.9 | 16.2 ± 5.7 |
|
1° CD IL-7 |
18.8 | 84.8 ± 2.0 |
81.7 ± 1.8 |
9.2 ± 1.2 | 5.6 ± 0.6 | 2.1 ± 0.3 | 1.0 ± 0.3 |
|
2° CD IL-7 |
18.8 | 96.6 ± 1.0 |
97.2 ± 0.5 |
1.0 ± 0.1 | 0.5 ± 0.1 | 0.3 ± 0.1 | 0.3 ± 0.1 |
|
3° CD BAFF |
18.8 | 96.7 ± 0.9 |
67.8 ± 5.5 |
8.6 ± 0.9 | 7.7 ± 0.6 | 6.5 ± 0.5 | 0.6 ± 0.7 |
Figure 1. Efficient generation of IgM+IgD+ B cells in vitro.
Non-adherent BL/6 BM cells (left) were cultured with 10 ng/ml IL-7 for 4 d (1° CD) or 8 d (2° CD). Cells were labeled with mAbs to B220, IgM, IgD, CD23, CD21 and CD93 (top panels) to identify pro-/pre-B, immature B, T1 B, T2 B and mature B cells. Cells were also labeled with mAb to HSA, BP-1, CD43, B220 and IgM to identify early B-cell compartments using Hardy’s method (bottom panels). Additionally, 2° CD cells were washed to remove IL-7 and then cultured with 20ng/ml BAFF for 3–4 d (3° CD). Cells were labeled with mAb to B220, IgM, IgD, CD23, CD21 and CD93 (right panels). Histograms of CD93 expression on each population are shown (bottom right). Results are representative of multiple (n≥5) independent experiments.
CD B cells were re-cultured in new IL-7+ media (2° CD); then cultured cells were labeled with mAb to identify B-cell subsets (Fig. 1). The number of cells recovered from 2° CD cultures was 2–4 times greater than input values and 95% of cells expressed B220 (Table 1). The most abundant population (95%) of 2° CD cells were pro-/pre-B cells whereas IgM+ cells were minor populations (<5%) of cells (Fig. 1 and Table 1). 2° CD cells were analyzed using Hardy’s method, and we observed that Fraction “A” (prepro-B) (B220loCD43hiBP-1loHSAlo), “B” and “C-C`” cells comprised <1%, 10–15% and 80–85% of the cultures, respectively (Fig. 1).
B-cell progenitors occupy independent BM niches depending upon their relative maturity (Nagasawa, 2006; Pereira et al., 2009). While repeated IL-7+ cultures increased B220+ cell numbers (2- to 4-fold over each input value), this technique only expanded the pro-/pre-B cell compartments (Table 1). Earlier work showed that the removal of IL-7 and addition of BAFF resulted in the emergence of Ig+ B cells in vitro (Claudio et al., 2002), and we tested whether BAFF would enrich B220+IgM+IgD+ B cells in our cultures. 2° CD cells were cultured with BAFF for 3–4 days and then labeled with mAb to B220, IgM, IgD, CD23, CD21 and CD93 to identify B-cell subsets (Ueda et al., 2007). BAFF+ cultures (3° CD) yielded fewer numbers of cells as input values, but were enriched (>20%) for IgM+IgD+ B cells. 3° CD cells contained pro-/pre-B cells (65%), immature B cells (9%), T1 B cells (8%), T2 B cells (7%) and few (<1%) mature B cells (Table 1 and Fig. 1, right). CD93 was highly expressed on pro-/pre-B cell compartments and its expression decreased as maturation increased through the T2 B-cell compartment (Fig. 1, right). CD23 and CD21 expression increased at the transitional B-cell compartments similar to BM (Fig. 2). These results indicated that pro-/pre-B cells that expanded during the IL-7+ cultures retained their potential to differentiate. The sequential culture system of 1° (IL-7), 2° (IL-7) and 3° (BAFF) generated a large number of Ig+ B cells grown independently of the BM microenvironment.
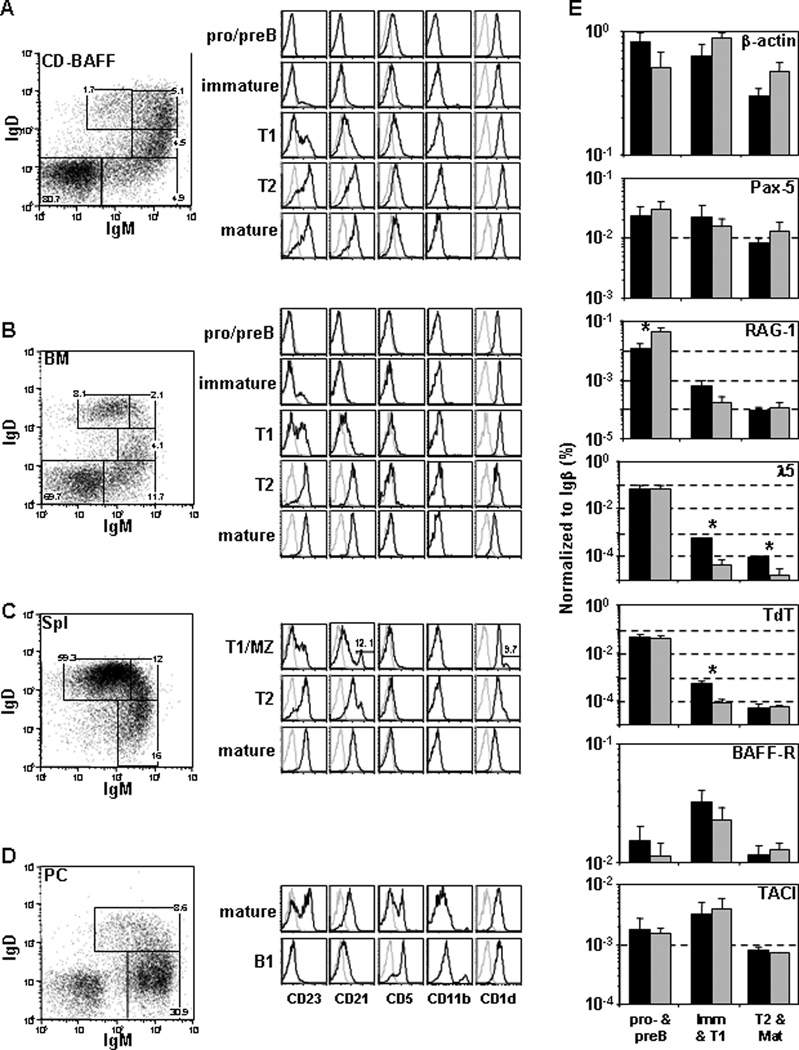
Figure 2. CD B cells have phenotypic and gene expression profile similar to BM B cells.
Non-adherent BL/6 BM cells were cultured using standard conditions (1°-IL-7, 2°-IL-7, 3°-BAFF) to generate (A) CD B cells. (B) BM, (C) spleen and (D) peritoneal cavity cells were harvested for comparison of surface antigen expression. Each group of cells were labeled with mAbs to B220, IgM, IgD and CD23, CD21, CD5, CD11b or CD1d. Flow diagrams were gated on live, single B220+ cells. Pro-/pre-, immature, T1, T2 and mature, MZ B and B1 B cells were identified as previously described. (E) CD B cells and ex vivo BM cells were labeled with mAbs to B220, IgM, IgD, CD23 and CD21 to identify pro-/pre-, immature/T1 and T2/mature B cells. RNA was isolated from FACS sorted ex vivo BM (black) and CD B-cell (gray) compartments. cDNA was created using oligo(dT)12–18 and Superscript III reverse transcriptase. q-rtPCR was performed on cDNA samples (n=2–3) from the B-cell compartments described above. Values for each reaction were normalized to Igβ expression. Significant differences (*; p < 0.05) between groups were determined by (two-tailed) Student’s t-test. Primers specific for β-actin, Igβ, BAFF-R, TACI, Pax5, λ5, TdT and RAG-1 are provided (Supplemental Table 2).
Since mature T cells are present at low frequencies within the BM (Supplemental Fig. 1A, top), we examined whether BM T cells survived in our cultures by FACS using mAbs for CD4, CD8, CD62L and CD44 (Kaech et al., 2002). In BM, we observed that ∼5% of B220neg cells expressed CD4 or CD8 and the majority (>50–60%) of these cells were CD62LhiCD44lo, a phenotype consistent with a mature, naïve T cells (Supplemental Fig. 1A). In 1° and 2° CD cultures, 5–20% of cells were B220neg (Fig. 1) and 25–50% of these CD cells expressed CD4 or CD8 (Supplemental Fig. 1A). Again, the majority (>80%) of these T cells exhibited a phenotype (CD62LhiCD44lo) consistent with naïve T cells. We determined the absolute number of cells with each T-cell phenotype that were recovered from 1° and 2° cultures (Supplemental Fig. 1B). Both CD4+CD62LhiCD44lo and CD8+CD62LhiCD44lo cells decreased ∼2-fold between 1° and 2° cultures. The number of CD4+CD62LloCD44hi and CD8+CD62LloCD44hi cells showed little or no change between 1° and 2° cultures. These data indicated that mature T cells survived through each IL-7+ phase of the culture system.
3.2 CD B cells have phenotypic and gene expression profile similar to BM B cells
We characterized CD B cells for surface antigens that are differentially expressed by distinct B-cell subsets (Carsetti et al., 2004; Lopes-Carvalho and Kearney, 2004; Pillai et al., 2005; Hardy, 2006) from BM, spleen and peritoneal cavity (PC). Cells from each tissue were labeled with mAb to B220, IgM, IgD and CD23, CD21, CD5, CD1d or CD11b. BM and CD B cells were characterized by their B220, IgM and IgD expression (Gorelik et al., 2004). Splenic B cells (B220+) were divided into mature follicular (mature) (IgMloIgDhiCD23hiCD21int), T2 (IgMhiIgDhiCD23hiCD21int), T1 (IgMhiIgDloCD23loCD21lo) and marginal zone (MZ) (IgMhiIgDloCD23loCD21hi) B cells (Lopes-Carvalho and Kearney, 2004). Resident PC cells were divided into B1 B cells (B220intIgMhiIgDloCD5hiCD23loCD21lo) and mature B cells as described (Martin and Kearney, 2001).
CD B cells most closely resembled BM B-cell compartments (Fig. 2). Early CD and BM B-cell fractions (pro-/pre- to T1) were indistinguishable for all markers tested (Fig. 2A–B). The T2 and mature B cells generated in vitro displayed increased expression of CD23 and CD21 when compared to their respective BM counterparts (Fig. 2A–B), consistent with previous reports that BAFF signaling induces increased CD23 and CD21 expression (Gorelik et al., 2004). CD B cells lack expression of CD5 and CD11b on each subset (Fig. 2A and 2D). CD1d is used for lipid-antigen presentation by B cells and is highly expressed on MZ B cells (Amano et al., 1998; Lee et al., 1998). Expression of CD1d increases during maturation from pro-/pre-B cells to T1 B cells, followed by decreased expression as B cells continue to mature (Fig. 2B). CD B cells express CD1d similar to BM B cells; however, T2 B cells sustain CD1d expression (Fig. 2A). Taken together, the surface phenotype of CD B cells suggests that we are generating B cells in vitro that are similar to BM-derived B cells.
Phenotypically normal cells may have gene expression rendering them dysfunctional. To directly compare gene expression, we sorted control and CD B cells (pro/pre, imm/T1 and T2/mature) to harvest mRNA for quantitative real-time PCR (q-rtPCR). We measured the expression of genes crucial for B-cell development, survival and V(D)J recombination of immunoglobulin (Ig) loci. RAG1/2 and TdT are critical enzymes in Ig gene rearrangement and diversification (Dudley et al., 2005). λ5 is an essential component of the pre-BCR, and necessary for survival beyond the pro-B cell stage (Shimizu et al., 2002). Pax-5 is a transcription factor required to induce and maintain B-cell identity (Cobaleda et al., 2007). The expression level and temporal regulation of each transcript was comparable between BM and CD B-cell fractions (Fig. 2E). We compared the expression of BAFF-R and TACI in each sorted B-cell compartment. The expression of each receptor gene was similar between BM and CD B cells for each B-cell population (Fig. 2E) demonstrating that CD B cells share the gene expression profiles of their BM counterparts.
3.3 CD B cells express diverse Igh and Igκ rearrangements
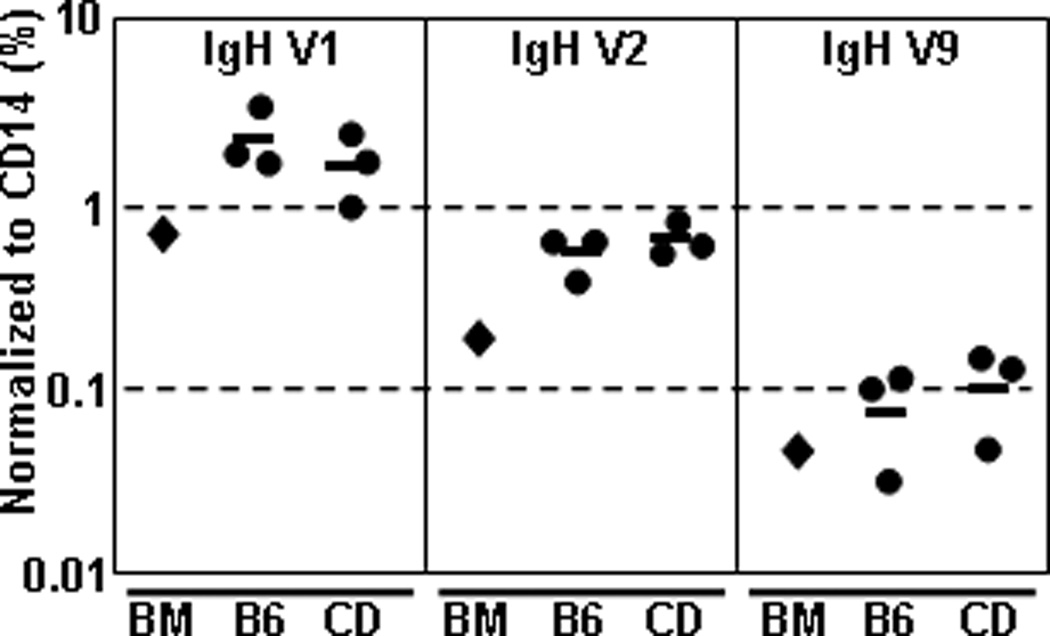
To ensure that CD B cells express a diverse set of heavy chain (HC) and LC genes, we amplified VH1-, VH2- and VH9- rearrangements to JH1 by PCR. A non-rearranging gene, CD14, was used to normalize the amount of input DNA (∼20ng). Genomic DNA from unsorted BL/6 BM served as a positive control for each PCR reaction; concurrently, sorted pro-/pre-B cells from BL/6 BM and CD B-cell samples were directly compared (Fig. 3). The VH1 family (69 members) is located at the 5` distal end of the Igh locus (IMGT/V-Quest). Both BM and CD samples showed equal VH1 usage (∼3% of CD14 signal) (Fig. 3). The VH2 family (9 members) is located at the 3` region of the Igh locus, proximal to the D cluster (IMGT/V-Quest). BM and CD samples showed similar (∼0.8% of CD14 signal) usage of VH2 gene segments (Fig. 3). Lastly, we examined VH9 (4 members), located in the center of the Igh locus (IMGT/V-Quest) and determined that BM and CD samples contained similar (∼0.1% of CD14 signal) amounts of these V-gene segments (Fig. 3). These results indicated that the stochastic nature of HC V-gene selection during rearrangement of the BCR is intact for B cells grown in our culture system.
Figure 3. CD B cells exhibit non-biased usage of several VH-family genes.
Non-adherent BL/6 BM cells were cultured (1°-IL-7, 2°-IL-7) to generate CD B cells. Cells were harvested and labeled with mAbs to B220, IgM and IgD. Pro-/pre-B cells were sorted from BM (B6) or 2°CD B cells (CD) and genomic DNA was extracted from 106 cells using proteinase K digestion and phenol-chloroform extraction. Whole BM (BM) gDNA sample was used as positive control for detection. ∼20 ng of gDNA was amplified by q-rtPCR using VH1-, VH2- or VH9-family specific 5` primer with an intronic JH-1 specific 3` primer. CD14-specific PCR reaction was used to normalize input quantities of gDNA. Values are represented as the percent of CD14 signal. Experiment was performed on sorted samples (n=3 each) of BL/6 BM and 2° CD pro-/pre-B cells.
3.4 CD B cells rapidly differentiate to AFC and upregulate co-stimulatory molecules on TLR stimulation
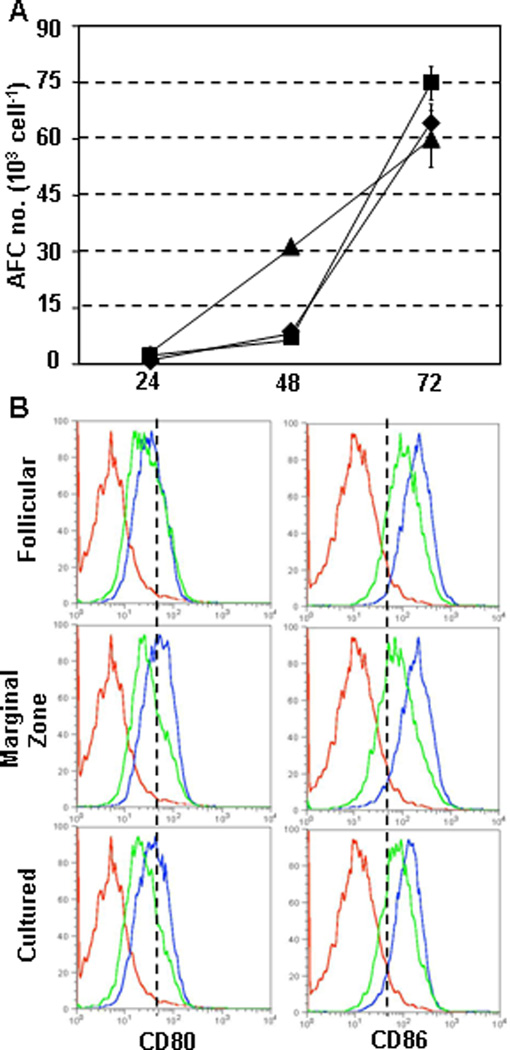
To determine whether CD B cells were functional, we compared the response of mature B cells, MZ B cells (Snapper et al., 1993; Oliver et al., 1997) and 3° CD B cells to Toll-like receptor (TLR) stimulation. Spleen cells were labeled with mAb to B220, IgM, IgD, CD23 and CD21 to sort mature and MZ B cells; while, CD B cells were sorted for B220+CD43negCD93lo cells. Sorted mature, MZ and CD B cells were stimulated with LPS for up to 3 days and were tested for IgM antibody-forming cell (AFC) differentiation. Mature- and MZ-derived AFC were low (∼1% of input cells) after 2 days of LPS stimulation, but increased (∼10% of input cells) by day 3 (Fig. 4A). CD B cells differentiated into IgM AFC by day 2 (∼3% of input cells) and reached levels similar to mature and MZ compartments by day3 (Fig. 4A). These results demonstrated that CD B cells were capable of rapid differentiation to AFC after LPS stimulation. Regulation of surface CD80 and CD86 expression after LPS stimulation is used to distinguish mature follicular (MF) and MZ B cells (Oliver et al., 1999). LPS-treated mature, MZ and CD B cells were analyzed for CD80 and CD86 expression. Each group increased CD80 and CD86 expression within 48hrs of LPS stimulation with MZ B cells expressing the highest levels (Fig. 4B). CD B cells expressed an intermediate level of each co-stimulation molecule compared to mature and MZ B cells (Fig. 4B). Each group reduced CD80/CD86 expression by the third day of LPS stimulation (Fig. 4B). These data demonstrate that CD B cells were able to differentiate in manners similar to splenic B-cell compartments in response to TLR ligation.
Figure 4. CD B cells rapidly differentiate to AFC and increase CD80/CD86 expression upon TLR stimulation.
CD43CD93lo CD B cells and mature splenic B-cell populations (MF and MZ) were sorted using FACS. Sorted cells were cultured with 5µg/ml LPS for 24hr, 48hr or 72hr. (A) Number of IgM AFC was determined by ELISPOT after LPS stimulation for mature (♦), MZ (▪) or CD (▲) B cells. ELISPOT plates coated with anti-mouse Ig(H+L) polyclonal Ab and assays were performed in triplicate for each condition tested. Values represent mean and standard deviation of triplicate samples. (B) Expression of CD80 and CD86 was measured on mature, MZ and CD43CD93lo CD B cells after LPS stimulation. Cells were labeled with mAbs to B220, CD80 and CD86 after LPS treatment for 48hr (blue) or 72hr (green) and compared to ex vivo splenocytes (red).
3.5 Elevated frequencies of autoreactive CD B cells
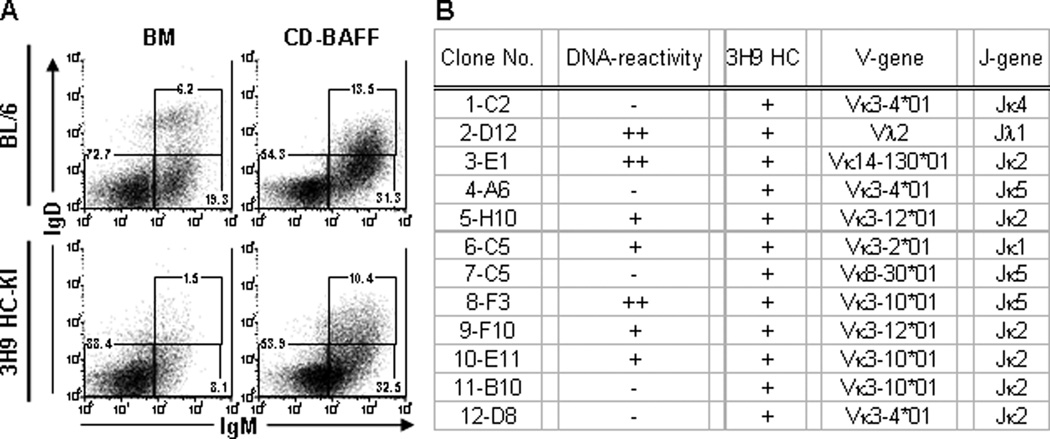
There are several co-receptors that modulate signaling through the BCR, such as CD19, CD22 and CD21 (Chen et al., 2000c; Nitschke, 2005). Mis-expression of these co-receptors can influence the intensity of down-stream signal transduction, sometimes resulting in autoimmunity (Tuscano et al., 2003). Since IgM+IgD+ CD B cells had elevated expression of CD23 and CD21 (Fig. 2A) and developed without the BM micro-environment associated with central tolerance (Sandel and Monroe, 1999; Sandel et al., 2001), we tested whether this culture system permitted the formation of autoreactive B cells. Previous work has demonstrated that the 3H9 HC encodes dsDNA and ssDNA binding with various LC (Shlomchik et al., 1990). The 3H9 IgH transgenic and “knock-in” (HC-KI) mice have been created to study the processes that promote self-tolerance (Erikson et al., 1991; Chen et al., 1995). We analyzed B-cell compartments of 3H9 HC-KI mice and confirmed that the frequency of BM immature and transitional B cells was substantially reduced (25–40% of BL/6 controls) (Fig. 5A, left). Furthermore, the number of immature/transitional B cells in BM of 3H9 HC-KI mice was reduced (20–30% of BL/6 controls) (Supplemental Fig. 2). After IL-7+ expansion of pro-/pre-B cells from 3H9 HC-KI and control BM, we tested whether 3H9 HC-KI immature and transitional B cells would develop in the presence of BAFF. Equal numbers of cells were recovered from BAFF+ cultures of 3H9 HC-KI and control cells (data not shown). The frequency of 3H9+ immature CD B cells was equal to control immature CD B cells; whereas, the frequency of 3H9+ transitional B cells approached (∼65%) that of control cultures (Fig. 5A, right). The frequencies of immature and transitional 3H9+ CD B cells were much greater (∼4-fold and ∼10-fold, respectively) than the 3H9 HC-KI BM compartments (Fig. 5A, bottom). These data suggested that autoreactive B cells lost during development in BM are recovered using this culture system.
Figure 5. Autoreactive transitional B cells are generated using BM culture system.
(A) Non-adherent BL/6 (top) and 3H9 HC-KI (bottom) BM cells (left) were cultured with 10 ng/ml IL-7 (4 d) followed by culture with 20ng/ml BAFF (4 d) (right). Cells were labeled with mAbs to B220, IgM, IgD, CD23, CD21 and CD93 to identify B-cell subsets. Representative flow diagrams were pre-gated on live, single B220+CD93+ cells. FACS plots are representative of multiple (n=3) independent experiments. (B) Summary of 3H9 CD hybridomas for labeling of C. luciliae (see Supplemental Fig. 2B for representative images), expression of 3H9 transcripts and identification of Vκ/Jκ and Vλ/Jλ usage.
To directly assess whether DNA-reactive B cells were generated in 3H9 HC-KI BM cultures, we activated 3H9 CD B cells (Fig. 5A, bottom right) with LPS and BAFF for fusion with NS0-bcl2 cells to create hybridomas (Yu et al., 2008). Cells were sub-cloned at 0.3 cells/well and each mAb was characterized. All of 3H9 CD hybridoma lines were IgM and 11 (91.7%) of these 3H9 CD hybridomas utilized a κLC. One hybridoma (8.3%) utilized a λLC (Fig. 5B). mRNA from each cloned hybridoma was isolated to generate cDNA and to confirm the presence of 3H9 Igh transcripts (Fig. 5B). C. luciliae are commonly used to detect antibodies that react with native DNA (ADA) (Gilkeson et al., 1995). Seven of the 3H9 CD hybridomas (6/11 κLC and 1/1 λLC) reacted with native DNA (Fig. 5B and Supplemental Fig. 2B). Many families of light chains (Vκ3, Vκ4, Vκ8, Vκ9, Vκ21, Vκ23 and Vλ1) combine with the 3H9 HC and impart DNA-reactivity (Shlomchik et al., 1990; Radic et al., 1993). We sequenced the LC loci of our 3H9 CD hybridomas and readily detected Vκ- and Vλ-family members that are “permissive” for DNA binding (Fig. 5B). These data demonstrate that autoreactive B cells which are lost during development in BM are recovered using this culture system.
3.6 CD cells reconstitute peripheral immune tissue of lymphocyte-deficient hosts
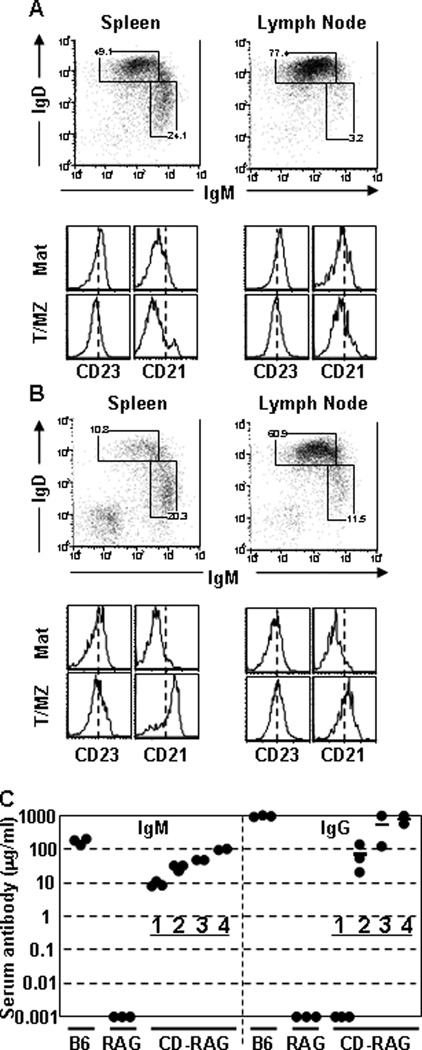
We tested whether CD cells could reconstitute lymphopenic recipients. BL/6 BM cells were cultured in IL-7+ media to generate 2° CD B cells which were then cultured with BAFF to generate IgM+ CD B cells for injection into B6.RAG1−/− recipients (CD-RAG mice) (section 2.1). Recipient mice were sacrificed 4wks post-transfer to analyze reconstitution of B-cell compartments in peripheral lymphoid tissue. Spleen and LN cells from control and CD-RAG mice were labeled with mAb to B220, IgM, IgD, CD23 and CD21 for comparison. BL/6 mice had a normal distribution of both splenic and LN B-cell populations as previously reported (Fig. 6A) (Allman and Pillai, 2008). The spleen of CD-RAG mice contained B cells (∼10% of splenocytes) that expressed elevated (∼2-fold) CD23 and CD21 and were enriched for IgMhiIgDlo cells (Fig. 6B), consistent with a MZ B cell phenotype. The LNs of CD-RAG animals contained B220+ cells that expressed levels of IgM, IgD, CD23 and CD21 similar to control LNs (Fig. 6B). CD B cells represented ∼30% of the lymph node cells recovered (data not shown). The recovery of B cells from spleen and LN demonstrate that CD B cells are able to traffic appropriately to peripheral lymphoid organs and survive for extended periods of time. Then, we tested whether the donor cells could reconstitute the serum Ab of recipient animals. Serum from CD-RAG mice was analyzed by IgM- and IgG-specific ELISA over the course of four weeks (Fig. 6C). CD B cells were able to restore serum IgM concentration progressively over the reconstitution period to near normal levels. Serum IgG was undetectable at day 7, but was comparable to normal controls by 4 wk post-reconstitution.
Figure 6. CD B cells reconstitute peripheral immune tissue of lymphocyte-deficient hosts.
Non-adherent BL/6 BM cells were cultured (1°-IL-7, 2°-IL-7, 3°-BAFF) to generate CD B cells for injection into B6.RAG−/− mice. At 4 wk post-transfer, spleen and LN cells from (A) BL/6 or (B) B6.RAG−/− mice reconstituted with 3° CD B cells were labeled with mAbs to B220, IgM, IgD, CD23 and CD21. Flow diagrams were pre-gated on live, single, B220+ cells and were representative of each mouse analyzed (n>3). Sera from control or CD-RAG mice were collected via retro-orbital eye bleeding at 7 d, 14 d, 21 d and 28 d post-transfer. (C) Concentrations of serum IgM and IgG were determined using anti-mouse IgM- or IgG-specific ELISAs including standard curves. Each group contained three mice (n=3) except for D21 & 28 time points (n=2).
We investigated whether reconstitution of lymphopoiesis occurred in CD-RAG mice after cell transfer. We observed very few (<1%) BM B220+ cells that co-expressed IgM+ (Supplemental Fig. 3A). The thymus did not contain CD4+CD8+ (DP), CD4+ (CD4-SP) or CD8+ (CD8-SP) T-cell progenitors, indicating that CD cells do not contain T-cell progenitors at the time of transfer to RAG-deficient mice (Supplemental Fig. 3B). However, mature T-cell populations were retained through our culture system (Supplemental Fig. 1) leading us to examine mature T-cell reconstitution of CD-RAG mice. We harvested the spleen and LNs of BL/6, RAG1−/− and CD-RAG mice to analyze mature T-cell compartments. The spleen and LNs of BL/6 mice contained both CD4+ and CD8+ cells that were predominately (>80%) CD62LhiCD44lo “naïve” phenotype and a small frequency (∼5–10%) of CD44hi “activated” or “memory” cells (Supplemental Fig. 3C–D, top). RAG-1−/− mice did not contain any CD4+ or CD8+ cells that we could detect (Supplemental Fig. 3C–D, middle). Mature T cells were present in the spleen and LNs of CD-RAG mice at ∼5% and ∼35% of the B220neg compartment, respectively, (Supplemental Fig. 3C–D, bottom). Both CD4+ and CD8+ cells in CD-RAG mice were predominately (∼70–80%) CD44hi, consistent with homeostatic driven proliferation (Kieper and Jameson, 1999). These data indicated that mature T cells which survived in vitro were able to partially reconstitute the peripheral lymphoid tissue of RAG1−/− mice.
3.7 Serum IgG autoantibody is present in CD-RAG mice
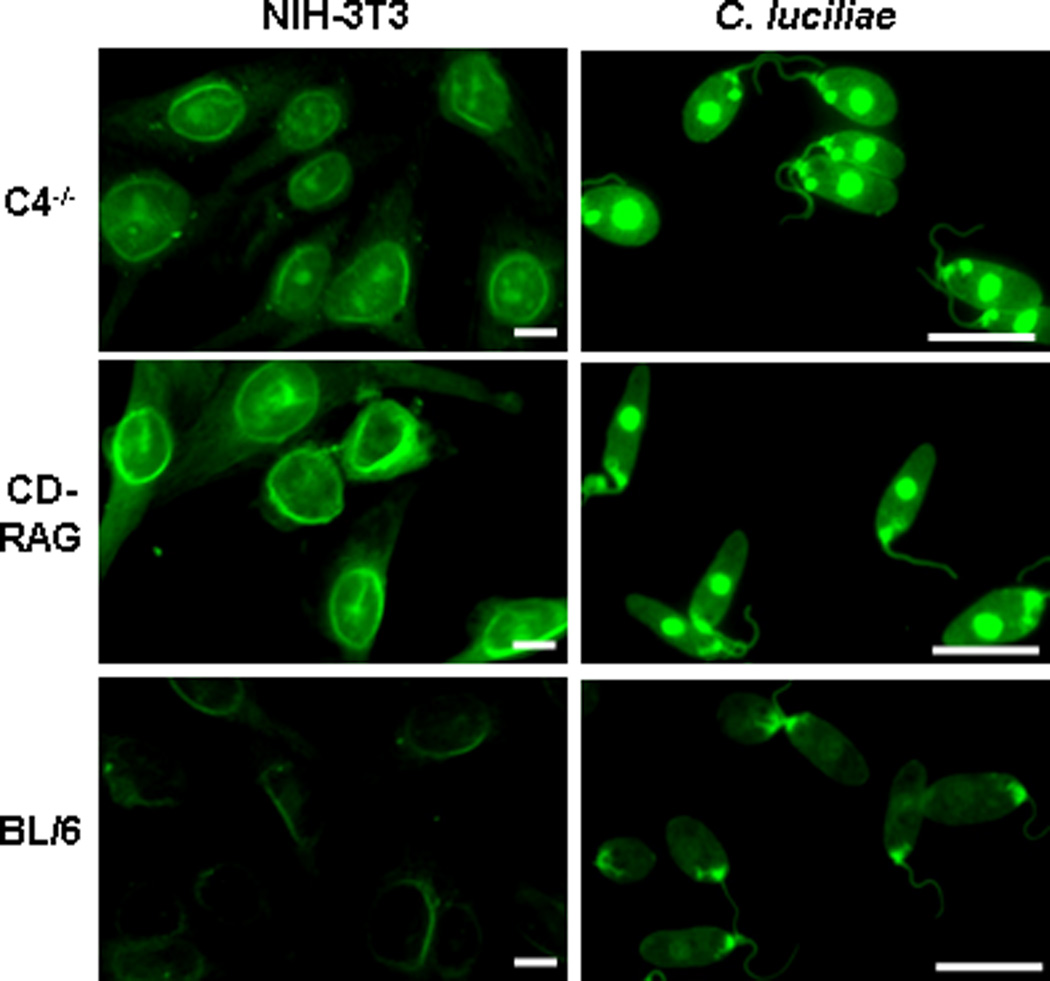
Autoreactive B cells can be eliminated from the mature B-cell repertoire by peripheral tolerance mechanisms (Hartley et al., 1991; Silveira et al., 2004). Therefore, we tested whether autoreactive B cells were retained in vivo after reconstitution of RAG−/− mice. We screened serum collected from CD-RAG animals for the presence of IgG autoantibody. We included NIH-3T3 cells as a source of mouse cellular antigens during these immunofluorescence studies. As a positive control, serum from C4−/− mice contained detectable levels of anti-nuclear antibodies (ANA) and ADA as previously reported (Fig. 7, top) (Chen et al., 2000b). Serum from BL/6 mice contained barely detectable levels of ANA and ADA (Fig. 7, bottom) and Ig-deficient RAG1−/− serum served as a negative control for labeling (data not shown). Conversely, the serum of CD-RAG mice contained ANA and ADA at levels similar to or higher than C4−/− mice (Fig. 7, middle). The recovery of serum IgG autoantibody suggested that RAG−/− hosts permitted autoreactive CD B cells survival long enough to contribute to serum Ab restoration.
Figure 7. Serum of CD-RAG mice is enriched for autoreactive IgG antibody.
Control serum was obtained from BL/6, B6.RAG−/− and C4−/− mice and from CD-RAG mice at 4wk post-transfer. Slides containing NIH-3T3 (left) and C. luciliae (right) were labeled with serum (1:20dil) from each mouse group. After overnight washing, Ab bound to cells was detected using rat anti-mouse IgG-FITC Ab. All images were acquired using a Zeiss Axiovert 200M confocal immunofluorescent microscope with an exposure time of 60ms (C. luciliae) or 100ms (NIH-3T3). Scale bar equals 10µm for all images.
4. Discussion
B-lymphocytes arise from committed lymphoid progenitors present in mouse BM (Kondo et al., 1997). The BM stromal compartment consists of heterogeneous cell populations that confound the investigation of B-cell development (Dorshkind, 1990) by providing distinct developmental niches for B-cell progenitors (Nagasawa, 2006; Pereira et al., 2009). Many investigators have discovered the utility of in vitro culture techniques to identify factors that are required for mouse and human B-lymphopoiesis (Fluckiger et al., 1998; Ray et al., 1998; Hess et al., 2001). Some B-cell culture systems use sequential conditions that require the addition or removal of factors to promote B-cell maturation (Claudio et al., 2002; Luo et al., 2009). We have described a stroma-independent culture system that supports the survival, proliferation and differentiation of virtually all BM B-cell developmental stages. While, IL-7 is a key cytokine required for the development and survival of T cells (reviewed in (Bradley et al., 2005)), de novo T-cell development, in vitro, requires stromal cells that express Notch ligands (Schmitt et al., 2004). Indeed, we did not observe any early T-cell compartments (DN-DP) that are present during thymic development (data not shown). Compared to stromal-dependant techniques, the clear advantages of our culture system is i) the ability to easily scale the size of cultures to suit our experimental requirements and ii) the ability to control the cytokine composition of our culture conditions.
The mature B-cell repertoire contains a diverse array of BCR specificities that promote antigen recognition and host survival (reviewed in (Market and Papavasiliou, 2003)). Several mechanisms, such as combinatorial association of variable- (V), diversity- (D) and joining- (J) gene segments, operate in developing B lymphocytes to promote BCR diversity (Chen and Alt, 1993; Benedict et al., 2000). Non-template (N-) nucleotide addition and imprecise joining of recombining VDJ gene segments further contribute to the diversity of the BCR repertoire (Benedict and Kearney, 1999). If few pro-/pre-B cells preferentially expanded during IL-7+ cultures, then IgM+ cells that arise during BAFF+ cultures would contain a restricted subset of IgH rearrangements. Therefore, we sequenced the complementarity determining region-3 (CDR3) regions of Igh and Igκ genes amplified from sorted pro-/pre- and im/T1 CD B cells. We recovered unique HC CDR3 sequences (∼100) and analyzed Igh rearrangements to both JH1 and JH2 gene segments in CD pro-/pre-B cells (subset shown in Table 2 and Supplemental Table 2). Over thirty distinct VH1 family members were recovered, representing ∼50% of VH1 gene segments available (Supplemental Table 2). These VH1 gene segments were associated with 11 different D gene segments (Table 2 and Supplemental Table 2) and contained both P- and N-nucleotide addition at V-D and D-J junctions, resulting in further diversification of the IgH repertoire (Table 2). This analysis was extended to the Vκ4 and Vκ5 LC loci in pre- and im/T1 CD B cells. Similar mechanisms of diversity, with the exception of N-nucleotide addition, were detected in the LC repertoire (Table 2 and Supplemental Table 2). Taken together, our sequencing data demonstrates that CD B-cell development supports typical, diverse V(D)J rearrangements that are observed in BM counterparts.
Table 2.
Sequence analysis of V(D)J CDR3 rearrangements in CD B cells
| Seq. # | VH-family | D-segment | 3` portion of V-segment | N or P nucleotides | D-region | N or P nucleotides | 5` portion of JH | J-region | f/nf* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | J558.10pg.100 | FL16.1 | TAT TGT GA | A CGG ACC C | TT ATT ACT ACG GTA GT | G | AC TGG | JH2 | nf |
| 2 | VH124 | SP2.2 | TAC TGT GCA AGA | TCG GGG | GAT TAC G | GC GTC AAC | TAC TTT GAC TAC TGG | JH2 | f |
| 3 | J558.67.166 | FL16.1 | TAC TGT GCA AGA | TA | C ATT ATT AC | T TCC GGA GTA GCG T | AC TAC TGG | JH2 | f |
| 4 | J558.75.177 | SP2.2 | TAC TGT GCA AGA | GAA GAG AGG C | AC TAT GAT TAC GAC | GGA GGA GGT | TAC TTT GAC TAC TGG | JH2 | f |
| 5 | J558.49.141 | SP2.9 | TAC TGT GCA AGA | AAA GGG | TGG TTA CTA C | GA A | AC TTT GAC TAC TGG | JH2 | f |
| 6 | J558.83.189 | FL16.1 | TTC TGT GCA AGA | T | TT ACT ACG GT | T | TAC TTT GAC TAC TGG | JH2 | f |
| 7 | J558.84.190 | FL16.1 | TTC TGT GCA AGA | GAG AGG GTA G | ACT ACG GTA GTA GCT | GGG GGT ACT | TTC GAT GTC TGG | JH1 | nf |
| 8 | J558.84.190 | SP2.10 | TTC TGT GCA AGA | TTT AGG ATC C | AC GAC | CCC C | AC TGG TAC TTC GAT GTC TGG | JH1 | f |
| 9 | J558.3.90 | SP2.2 | TAC TGT ACA AG | G T | TC TAC TAT GAT TAC | CCC TCT | TAC TGG TAC TTC GAT GTC TGG | JH1 | f |
| 10 | J558.55.149 | SP2.x | TAC TGT GCA AGA | GAA GC | A TAG TAA C | CC | C TAC TGG TAC TTC GAT GTC TGG | JH1 | nf |
| 11 | J558.51.167 | SP2.2 | TAC TGT GCA AGA | GAT TAC GAC | GGC GC | C TGG TAC TTC GAT GTC TGG | JH1 | f | |
| 12 | J558.78.182 | FL16.1 | TAC TGT GCA AGA | TGA GTG | TAC TAC GGT AGT AGC T | C | C TGG TAC TTC GAT GTC TGG | JH1 | f |
| Seq. # | Vic-family | 3` portion of V-segment | N or P nucleotides | 5` portion of Jκ | J-region> | f/nf * |
|---|---|---|---|---|---|---|
| 13 | km4 | TAC TGC CAT CAG CGG GGT TGT TAC CCA | TGG | CA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | nf |
| 14 | aj4 | TAC TGC CAG CAG TGG AGT AGT TCC CCG C | CA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | f | |
| 15 | kj4 | TAC TGC CAG CAG TGG AGT GGT TAC CCA TTC A | TG | CA CGT TCG GAG GGT GGA CCA AGC TGG | Jk2 | nf |
| 16 | 21–5 | TAC TGT CAG CAA AGT AAT GAG GAT CC | G | T ACA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | nf |
| 17 | 23–45 | TTC TGT CAA CAG AGT AAC AGC TGG CC | T ACA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | f | |
| 18 | 23–48 | TAC TGT CAA CAA AGT AAT AGC TGG CCA | A | A CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | nf |
| 19 | aa4 | TAC TGC CAG CAG TAT CAT AGT TAC CC | G | T GGA CGT TCG GTG GAG GCA CCA AGC TGG | Jk1 | nf |
| 20 | 23–39 | TAC TGT CAA AAT GGT CAC AGC TTT CC | T GGA CGT TCG GTG GAG GCA CCA AGC TGG | Jk1 | f | |
| 21 | 23–45 | TTC TGT CAA CAG AGT AAC AGC TGG CCT | G | T ACA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | nf |
| 22 | 23–45 | TTC TGT CAA CAG AGT AAC AGC TGG CCT | CA | T GGA CGT TCG GTG GAG GCA CCA AGC TGG | Jk1 | f |
| 23 | 23–48 | TAC TGT CAA CAA AGT AAT AGC TGG CCA | G | T ACA CGT TCG GAG GGG GGA CCA AGC TGG | Jk2 | nf |
| 24 | 23–48 | TAC TGT CAA CAA AGT AAT AGC TGG CCA | ACG | CGT TCG GTG GAG GCA CCA AGC TGG | Jk1 | f |
f = functional rearrangement nf = non-functional rearrangement
The molecular mechanism of Ig recombination permits the addition and/or removal of nucleotides which can generate frame-shift mutations, resulting in a truncated, non-functional immunoglobulin protein (Baumann et al., 1985). We determined the reading frame of our CDR3 sequences based on two criteria: a conserved cysteine residue of the framework-3 (FW3) region of V-gene segments and the JH to Cµ or JK to CK splice site present in each J-gene segment. The Igh and Igκ genes were analyzed for productive (P) and non-productive (nP) rearrangements and both P and nP rearrangements were observed (Table 2). Compared to previously described B-cell culture systems (Rolink et al., 1991a; Ray et al., 1998; Tze et al., 2000), we demonstrate that CD B cells utilize each element of V(D)J recombination that contributes to BCR diversity.
During development, self-reactive B cells are tolerized by apoptosis, anergy, or receptor editing (reviewed by(Goodnow, 1992)). These tolerizing processes have been examined in various transgenic mouse lines that express BCR for authentic (Nemazee and Burki, 1989; Erikson et al., 1991) or neo-self-antigens (Hartley et al., 1991). These experimental models defined immature and transitional-1 (T1) B cells as the targets of tolerizing apoptotic signals by developmental blockade (Hartley et al., 1993) and identified anergy (Adams et al., 1990) and receptor editing (Gay et al., 1993; Tiegs et al., 1993) by characterization of B-cell populations that escape apoptotic deletion. By introducing HEL antigen(s) to IL-7+ BM cultures, HEL-specific BCR Tg+ B cells were induced to undergo “de-differentiation” and “receptor editing”, defined as the loss of surface IgM, increased expression genes associated with earlier (pro- and pre-B) developmental stages and the initiation of endogenous LC rearrangement (Tze et al., 2000; Tze et al., 2003; Tze et al., 2005).
Clearly, factors that promote B-cell survival, such as Bcl-2 over-expression, alter B-cell selection and result in autoimmunity (Nisitani et al., 1993; Lang et al., 1997). BAFF Tg mice contain high levels of rheumatoid factors, circulating immune complexes, ADA and immunoglobulin deposition in the kidneys (Mackay et al., 1999). Our cultures support B-cell maturation with BAFF at 20ng/ml, which is ∼5–10 times greater than BL/6 serum BAFF concentration (data not shown). It remains unclear whether elevated BAFF-R signaling is required, in vitro, for self-reactive B-cell survival or that specific “death signals” are absent. Apoptotic thymocytes marked for clearance are rapidly removed by the mononuclear phagocyte system (Surh and Sprent, 1994). The possibility remains that autoreactive B-cell development would be abolished in the presence of phagocytes. However, autoreactive B cells survive in vivo and facilitate serum Ab reconstitution after transfer to RAG1−/− mice (Fig. 7). Consequently, we measured serum BAFF in B-lymphopenic animals, RAG1−/− and µMT mice, and determined the concentration to be ∼400–500ng/ml, roughly 100-fold higher than normal mice (data not shown). Would pre-treatment of RAG1−/− recipient animals with TACI-Ig, thereby lowering systemic BAFF availability, favor B-cell reconstitution of non-autoreactive B cells at the expense of autoreactive clones? These observations predict that B-cell autoimmunity could be modulated by controlling BAFF availability in the organism and, in fact, BAFF is a current target for therapeutic intervention in autoimmune disorders (Pelletier et al., 2003; Pranzatelli et al., 2008).
Recently, the mitigating effects of tolerizing processes in humans has been investigated by expressing IgH and IgL rearrangements from single immature, transitional, and mature B cells and determining the frequencies at which these Ab reacted with self-antigens (Wardemann et al., 2003; Wardemann et al., 2004). Our culture system allows for the systematic analysis of autoreactive B-cells removed from the primary repertoire by recovering cells reactive to common autoantigens and identifying their VH and Vκ/λ pairs. In humans, autoreactive Ab frequencies decline with increasing developmental maturity by virtue of apoptotic loss and receptor editing (Wardemann et al., 2003; Wardemann et al., 2004), even when cells were recovered from peripheral sites (Meffre et al., 2004; Tsuiji et al., 2006). Similarly, analysis of 3H9 Tg+ mouse BM cells revealed that IgM+ B cells did not co-label with an anti-idotypic 3H9 antibody, suggesting that 3H9 B cell-specificities were lost no later than the immature/transitional stage(s) (Gay et al., 1993). Furthermore, Ig heavy chain gene replacement occurs in some 3H9 HC-KI B cells as a mechanism of receptor editing (Chen et al., 1995). Since DNA-reactive 3H9+ B cells were readily recovered in our culture system, mechanisms that promote tolerance of developing lymphocytes can be investigated using this model by the manipulation of antigens, cell populations and/or survival factors.
Some bacterial pathogens avoid humoral immunity by mimicking host antigens, thereby exploiting a hole in the primary Ab repertoire achieved after tolerance is established (Bowes et al., 2002). The frequency of protective, broadly neutralizing Abs directed against conserved viral antigens is extraordinarily low (Burton et al., 2004; Ekiert et al., 2009). It has been suggested that HIV-1 limits humoral immunity to the conserved membrane-proximal external region (MPER) of gp41 by antigen mimicry (Haynes et al., 2005). Our culture system may provide a novel window into the “hidden” B-cell repertoire against pathogens that also cross-react with self-antigens. In conclusion, our B-cell culture system forms a suitable platform to provide substantial insight into areas of B-cell biology, such as development, selection and immunity.
Acknowledgments
These studies were supported by NIAID grants AI24335, AI56363 and AI81579 (G.K.) and the Bill and Melinda Gates Foundation (B.F.H. and G.K.). We would like to thank Masayuki Kuroaka PhD, Kwan-Ki Hwang PhD, Yoshihiro Ueda PhD and Dongmei Liao for support, both technical and theoretical, during these studies.
Abbreviations
- Ab
antibody
- APC
antigen presenting cell
- AFC
antibody forming cell
- BAFF
B-cell activating factor belonging to the TNF family
- BCR
B-cell receptor for antigen
- CD
culture-derived
- CD-RAG
RAG-deficient mice reconstituted with CD cells
- CSR
class-switch recombination
- CDR
complementarity determining region
- HPC
hematopoietic progenitor cell
- MF
mature follicular
- MZ
marginal zone
- MFI
mean fluorescence intensity
- MLN
mesenteric lymph node
- PC
peritoneal cells
- PI
propidium iodide
- P/S
penicillin/streptomyacin
- T1
transitional-1
- T2
transitional-2
- TLR
Toll-like receptor
References
- Adams E, Basten A, Goodnow CC. Intrinsic B-cell hyporesponsiveness accounts for self-tolerance in lysozyme/anti-lysozyme double-transgenic mice. Proc Natl Acad Sci U S A. 1990;87:5687–5691. doi: 10.1073/pnas.87.15.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Baumgarth N, Dick MD, Brossay L, Kronenberg M, Herzenberg LA, Strober S. CD1 expression defines subsets of follicular and marginal zone B cells in the spleen: beta 2-microglobulin-dependent and independent forms. J Immunol. 1998;161:1710–1717. [PubMed] [Google Scholar]
- Baird AM, Gerstein RM, Berg LJ. The role of cytokine receptor signaling in lymphocyte development. Curr Opin Immunol. 1999;11:157–166. doi: 10.1016/s0952-7915(99)80027-2. [DOI] [PubMed] [Google Scholar]
- Batten M, Groom J, Cachero TG, Qian F, Schneider P, Tschopp J, Browning JL, Mackay F. BAFF mediates survival of peripheral immature B lymphocytes. J Exp Med. 2000;192:1453–1466. doi: 10.1084/jem.192.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B, Potash MJ, Kohler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. Embo J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- Benedict CL, Kearney JF. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- Billips LG, Nunez CA, Bertrand FE, 3rd, Stankovic AK, Gartland GL, Burrows PD, Cooper MD. Immunoglobulin recombinase gene activity is modulated reciprocally by interleukin 7 and CD19 in B cell progenitors. J Exp Med. 1995;182:973–982. doi: 10.1084/jem.182.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billips LG, Petitte D, Dorshkind K, Narayanan R, Chiu CP, Landreth KS. Differential roles of stromal cells, interleukin-7, and kit-ligand in the regulation of B lymphopoiesis. Blood. 1992;79:1185–1192. [PubMed] [Google Scholar]
- Bowes T, Wagner ER, Boffey J, Nicholl D, Cochrane L, Benboubetra M, Conner J, Furukawa K, Furukawa K, Willison HJ. Tolerance to self gangliosides is the major factor restricting the antibody response to lipopolysaccharide core oligosaccharides in Campylobacter jejuni strains associated with Guillain-Barre syndrome. Infect Immun. 2002;70:5008–5018. doi: 10.1128/IAI.70.9.5008-5018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley LM, Haynes L, Swain SL. IL-7: maintaining T-cell memory and achieving homeostasis. Trends Immunol. 2005;26:172–176. doi: 10.1016/j.it.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Carsetti R, Rosado MM, Wardmann H. Peripheral development of B cells in mouse and man. Immunol Rev. 2004;197:179–191. doi: 10.1111/j.0105-2896.2004.0109.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–755. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Chen J, Alt FW. Gene rearrangement and B-cell development. Curr Opin Immunol. 1993;5:194–200. doi: 10.1016/0952-7915(93)90004-c. [DOI] [PubMed] [Google Scholar]
- Chen Z, Koralov SB, Gendelman M, Carroll MC, Kelsoe G. Humoral immune responses in Cr2−/− mice: enhanced affinity maturation but impaired antibody persistence. J Immunol. 2000a;164:4522–4532. doi: 10.4049/jimmunol.164.9.4522. [DOI] [PubMed] [Google Scholar]
- Chen Z, Koralov SB, Kelsoe G. Complement C4 inhibits systemic autoimmunity through a mechanism independent of complement receptors CR1 and CR2. J Exp Med. 2000b;192:1339–1352. doi: 10.1084/jem.192.9.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Koralov SB, Kelsoe G. Regulation of humoral immune responses by CD21/CD35. Immunol Rev. 2000c;176:194–204. doi: 10.1034/j.1600-065x.2000.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, Zuniga-Pflucker JC. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc Natl Acad Sci U S A. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Collins LS, Dorshkind K. A stromal cell line from myeloid long-term bone marrow cultures can support myelopoiesis and B lymphopoiesis. J Immunol. 1987;138:1082–1087. [PubMed] [Google Scholar]
- Cumano A, Dorshkind K, Gillis S, Paige CJ. The influence of S17 stromal cells and interleukin 7 on B cell development. Eur J Immunol. 1990;20:2183–2189. doi: 10.1002/eji.1830201006. [DOI] [PubMed] [Google Scholar]
- Dorshkind K. Regulation of hemopoiesis by bone marrow stromal cells and their products. Annu Rev Immunol. 1990;8:111–137. doi: 10.1146/annurev.iy.08.040190.000551. [DOI] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson J, Radic MZ, Camper SA, Hardy RR, Carmack C, Weigert M. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–334. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- Fischer MB, Ma M, Goerg S, Zhou X, Xia J, Finco O, Han S, Kelsoe G, Howard RG, Rothstein TL, Kremmer E, Rosen FS, Carroll MC. Regulation of the B cell response to T-dependent antigens by classical pathway complement. J Immunol. 1996;157:549–556. [PubMed] [Google Scholar]
- Fluckiger AC, Sanz E, Garcia-Lloret M, Su T, Hao QL, Kato R, Quan S, de la Hera A, Crooks GM, Witte ON, Rawlings DJ. In vitro reconstitution of human B-cell ontogeny: from CD34(+) multipotent progenitors to Ig-secreting cells. Blood. 1998;92:4509–4520. [PubMed] [Google Scholar]
- Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkeson GS, Pippen AM, Pisetsky DS. Induction of cross-reactive anti-dsDNA antibodies in preautoimmune NZB/NZW mice by immunization with bacterial DNA. J Clin Invest. 1995;95:1398–1402. doi: 10.1172/JCI117793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC. Transgenic mice and analysis of B-cell tolerance. Annu Rev Immunol. 1992;10:489–518. doi: 10.1146/annurev.iy.10.040192.002421. [DOI] [PubMed] [Google Scholar]
- Gorelik L, Cutler AH, Thill G, Miklasz SD, Shea DE, Ambrose C, Bixler SA, Su L, Scott ML, Kalled SL. Cutting edge: BAFF regulates CD21/35 and CD23 expression independent of its B cell survival function. J Immunol. 2004;172:762–766. doi: 10.4049/jimmunol.172.2.762. [DOI] [PubMed] [Google Scholar]
- Han S, Dillon SR, Zheng B, Shimoda M, Schlissel MS, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- Hardy RR. B-1 B cells: development, selection, natural autoantibody and leukemia. Curr Opin Immunol. 2006;18:547–555. doi: 10.1016/j.coi.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SB, Cooke MP, Fulcher DA, Harris AW, Cory S, Basten A, Goodnow CC. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72:325–335. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, Goodnow CC. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–769. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Hess J, Werner A, Wirth T, Melchers F, Jack HM, Winkler TH. Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc Natl Acad Sci U S A. 2001;98:1745–1750. doi: 10.1073/pnas.041492098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- Kieper WC, Jameson SC. Homeostatic expansion and phenotypic conversion of naive T cells in response to self peptide/MHC ligands. Proc Natl Acad Sci U S A. 1999;96:13306–13311. doi: 10.1073/pnas.96.23.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lang J, Arnold B, Hammerling G, Harris AW, Korsmeyer S, Russell D, Strasser A, Nemazee D. Enforced Bcl-2 expression inhibits antigen-mediated clonal elimination of peripheral B cells in an antigen dose-dependent manner and promotes receptor editing in autoreactive, immature B cells. J Exp Med. 1997;186:1513–1522. doi: 10.1084/jem.186.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin kappa locus in pre-B cells. Mol Cell Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Abeyratne A, Carson DA, Corr M. Induction of an antigen-specific, CD1-restricted cytotoxic T lymphocyte response In vivo. J Exp Med. 1998;187:433–438. doi: 10.1084/jem.187.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- Lopes-Carvalho T, Kearney JF. Development and selection of marginal zone B cells. Immunol Rev. 2004;197:192–205. doi: 10.1111/j.0105-2896.2004.0112.x. [DOI] [PubMed] [Google Scholar]
- Luo XM, Maarschalk E, O’Connell RM, Wang P, Yang L, Baltimore D. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood. 2009;113:1422–1431. doi: 10.1182/blood-2008-09-177139. [DOI] [PubMed] [Google Scholar]
- Mackay F, Schneider P, Rennert P, Browning J. BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol. 2003;21:231–264. doi: 10.1146/annurev.immunol.21.120601.141152. [DOI] [PubMed] [Google Scholar]
- Mackay F, Woodcock SA, Lawton P, Ambrose C, Baetscher M, Schneider P, Tschopp J, Browning JL. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market E, Papavasiliou FN. V(D)J recombination and the evolution of the adaptive immune system. PLoS Biol. 2003;1:E16. doi: 10.1371/journal.pbio.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Kearney JF. B1 cells: similarities and differences with other B cell subsets. Curr Opin Immunol. 2001;13:195–201. doi: 10.1016/s0952-7915(00)00204-1. [DOI] [PubMed] [Google Scholar]
- Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC. Surrogate Light Chain Expressing Human Peripheral B Cells Produce Self-reactive Antibodies. J Exp Med. 2004;199:145–150. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed D, Kench JA, Grabstein K, Rolink A, Nemazee D. A functional B cell receptor transgene allows efficient IL-7- independent maturation of B cell precursors. J Immunol. 1997;159:1233–1239. [PubMed] [Google Scholar]
- Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol. 2006;6:107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- Nisitani S, Tsubata T, Murakami M, Okamoto M, Honjo T. The bcl-2 gene product inhibits clonal deletion of self-reactive B lymphocytes in the periphery but not in the bone marrow. J Exp Med. 1993;178:1247–1254. doi: 10.1084/jem.178.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke L. The role of CD22 and other inhibitory co-receptors in B-cell activation. Curr Opin Immunol. 2005;17:290–297. doi: 10.1016/j.coi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Oliver AM, Martin F, Gartland GL, Carter RH, Kearney JF. Marginal zone B cells exhibit unique activation, proliferative and immunoglobulin secretory responses. Eur J Immunol. 1997;27:2366–2374. doi: 10.1002/eji.1830270935. [DOI] [PubMed] [Google Scholar]
- Oliver AM, Martin F, Kearney JF. IgMhighCD21high lymphocytes enriched in the splenic marginal zone generate effector cells more rapidly than the bulk of follicular B cells. J Immunol. 1999;162:7198–7207. [PubMed] [Google Scholar]
- Pelletier M, Thompson JS, Qian F, Bixler SA, Gong D, Cachero T, Gilbride K, Day E, Zafari M, Benjamin C, Gorelik L, Whitty A, Kalled SL, Ambrose C, Hsu YM. Comparison of soluble decoy IgG fusion proteins of BAFF-R and BCMA as antagonists for BAFF. J Biol Chem. 2003;278:33127–33133. doi: 10.1074/jbc.M305754200. [DOI] [PubMed] [Google Scholar]
- Pereira JP, An J, Xu Y, Huang Y, Cyster JG. Cannabinoid receptor 2 mediates the retention of immature B cells in bone marrow sinusoids. Nat Immunol. 2009;10:403–411. doi: 10.1038/ni.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- Pranzatelli MR, Tate ED, Hoefgen ER, Swan JA, Colliver JA. Therapeutic down-regulation of central and peripheral B-cell-activating factor (BAFF) production in pediatric opsoclonus-myoclonus syndrome. Cytokine. 2008;44:26–32. doi: 10.1016/j.cyto.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Radic MZ, Erikson J, Litwin S, Weigert M. B lymphocytes may escape tolerance by revising their antigen receptors. J Exp Med. 1993;177:1165–1173. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RJ, Stoddart A, Pennycook JL, Huner HO, Furlonger C, Wu GE, Paige CJ. Stromal cell-independent maturation of IL-7-responsive pro-B cells. J Immunol. 1998;160:5886–5897. [PubMed] [Google Scholar]
- Rohatgi S, Ganju P, Sehgal D. Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. J Immunol Methods. 2008;339:205–219. doi: 10.1016/j.jim.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO J. 1991a;10:327–336. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A, Streb M, Melchers F. The kappa/lambda ratio in surface immunoglobulin molecules on B lymphocytes differentiating from DHJH-rearranged murine pre-B cell clones in vitro. Eur J Immunol. 1991b;21:2895–2898. doi: 10.1002/eji.1830211137. [DOI] [PubMed] [Google Scholar]
- Rolink AG, Tschopp J, Schneider P, Melchers F. BAFF is a survival and maturation factor for mouse B cells. Eur J Immunol. 2002;32:2004–2010. doi: 10.1002/1521-4141(200207)32:7<2004::AID-IMMU2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Sandel PC, Gendelman M, Kelsoe G, Monroe JG. Definition of a novel cellular constituent of the bone marrow that regulates the response of immature B cells to B cell antigen receptor engagement. J Immunol. 2001;166:5935–5944. doi: 10.4049/jimmunol.166.10.5935. [DOI] [PubMed] [Google Scholar]
- Sandel PC, Monroe JG. Negative selection of immature B cells by receptor editing or deletion is determined by site of antigen encounter. Immunity. 1999;10:289–299. doi: 10.1016/s1074-7613(00)80029-1. [DOI] [PubMed] [Google Scholar]
- Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M, Frew E, Scott ML. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zuniga-Pflucker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer JL, Holler N, Ambrose C, Lawton P, Bixler S, Acha-Orbea H, Valmori D, Romero P, Werner-Favre C, Zubler RH, Browning JL, Tschopp J. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL. VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol. 2002;168:6286–6293. doi: 10.4049/jimmunol.168.12.6286. [DOI] [PubMed] [Google Scholar]
- Shlomchik M, Mascelli M, Shan H, Radic MZ, Pisetsky D, Marshak-Rothstein A, Weigert M. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J Exp Med. 1990;171:265–292. doi: 10.1084/jem.171.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira PA, Dombrowsky J, Johnson E, Chapman HD, Nemazee D, Serreze DV. B cell selection defects underlie the development of diabetogenic APCs in nonobese diabetic mice. J Immunol. 2004;172:5086–5094. doi: 10.4049/jimmunol.172.8.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper CM, Yamada H, Smoot D, Sneed R, Lees A, Mond JJ. Comparative in vitro analysis of proliferation, Ig secretion, and Ig class switching by murine marginal zone and follicular B cells. J Immunol. 1993;150:2737–2745. [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- ten Boekel E, Melchers F, Rolink A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–1019. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- Thompson JS, Bixler SA, Qian F, Vora K, Scott ML, Cachero TG, Hession C, Schneider P, Sizing ID, Mullen C, Strauch K, Zafari M, Benjamin CD, Tschopp J, Browning JL, Ambrose C. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–2111. doi: 10.1126/science.1061965. [DOI] [PubMed] [Google Scholar]
- Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokoyoda K, Egawa T, Sugiyama T, Choi BI, Nagasawa T. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 2004;20:707–718. doi: 10.1016/j.immuni.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Tsuiji M, Yurasov S, Velinzon K, Thomas S, Nussenzweig MC, Wardemann H. A checkpoint for autoreactivity in human IgM+ memory B cell development. J Exp Med. 2006;203:393–400. doi: 10.1084/jem.20052033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuscano JM, Harris GS, Tedder TF. B lymphocytes contribute to autoimmune disease pathogenesis: current trends and clinical implications. Autoimmun Rev. 2003;2:101–108. doi: 10.1016/s1568-9972(02)00148-9. [DOI] [PubMed] [Google Scholar]
- Tze LE, Baness EA, Hippen KL, Behrens TW. Ig light chain receptor editing in anergic B cells. J Immunol. 2000;165:6796–6802. doi: 10.4049/jimmunol.165.12.6796. [DOI] [PubMed] [Google Scholar]
- Tze LE, Hippen KL, Behrens TW. Late immature B cells (IgMhighIgDneg) undergo a light chain receptor editing response to soluble self-antigen. J Immunol. 2003;171:678–682. doi: 10.4049/jimmunol.171.2.678. [DOI] [PubMed] [Google Scholar]
- Tze LE, Schram BR, Lam KP, Hogquist KA, Hippen KL, Liu J, Shinton SA, Otipoby KL, Rodine PR, Vegoe AL, Kraus M, Hardy RR, Schlissel MS, Rajewsky K, Behrens TW. Basal immunoglobulin signaling actively maintains developmental stage in immature B cells. PLoS Biol. 2005;3:e82. doi: 10.1371/journal.pbio.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Kondo M, Kelsoe G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J Exp Med. 2005;201:1771–1780. doi: 10.1084/jem.20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, Liao D, Yang K, Patel A, Kelsoe G. T-independent activation-induced cytidine deaminase expression, class-switch recombination, and antibody production by immature/transitional 1 B cells. J Immunol. 2007;178:3593–3601. doi: 10.4049/jimmunol.178.6.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Hammersen J, Nussenzweig MC. Human autoantibody silencing by immunoglobulin light chains. J Exp Med. 2004;200:191–199. doi: 10.1084/jem.20040818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, Crowe JE., Jr An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods. 2008;336:142–151. doi: 10.1016/j.jim.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]