Abstract
Interleukin-23 receptor (IL23R) can interact with IL-23 and, thus, is involved in the T-helper 17 (Th17) cell-mediated inflammatory process as well as tumorigenesis. Recently, a functional single nucleotide polymorphism (SNP) rs10889677 has been identified in the 3’-untranslated region of IL-23R. It has been showed that the rs10889677AC SNP could increase the binding affinity of microRNA let-7f and downregulate IL-23R expression. Several case-control studies have examined the association between this SNP and genetic susceptibility of multiple solid tumors. However, the conclusions are conflicting. Therefore, we conducted this meta-analysis to systematically study the role of this functional IL-23R SNP in development of multiple solid tumors. There are a total of 5 studies are eligible (6731 cases and 7296 healthy controls). Either fixed-effect model or random-effect model was used to calculate pooled odds ratios (ORs) and the 95% confidence interval (95% CI). Significant association between this functional rs10889677 genetic variant and risk of multiple solid tumors were observed (CC genotype vs. AA genotype: OR = 0.59, 95% CI = 0.53-0.66, P < 0.001). These findings demonstrated that the IL-23R rs10889677 genetic variant might play an important part during malignant transformation of multiple solid tumors.
Introduction
Interleukin-23 receptor (IL23R) can interact with IL-23 and, thus, is involved in the T-helper 17 (Th17) cell-mediated inflammatory process as well as tumorigenesis [1,2]. Th17 cells belongs to pro-inflammatory CD4+ effecter T-cells and can mediate tissue inflammation by secreting high levels of the pro-inflammatory cytokine IL-17 in response to stimulation [3]. IL-23R is involved in multiple important biological processes, including Th17 cell-mediated immune response, tumor-promoting pro-inflammatory processes and the failure of the adaptive immune surveillance [1,2]. IL-23R could lessen immunosurveillance by CD8+ T cells and accelerate tumor proliferation as well [4]. For regulatory T cells (Tregs), the IL-23R signaling pathway might also promote the immunosuppressive function of Tregs facilitating evasion of the immune system by cancer cells [5-7]. Interestingly, both Th17 cells and Tregs can enhance proliferation of cancer cells [8]. These findings suggest that IL-23R might play an important part during carcinogenesis and progression.
Several single nucleotide polymorphisms (SNPs) have been identified in IL-23R locating on chromosome 1p31.3. Notably, there is an rs10889677AC SNP in the 3’-untranslated region (3’-UTR) of IL-23R, which could increase the binding affinity of microRNA let-7f and, thus, decreased IL-23R transcription ex vivo and in vivo. Biological studies indicated that rs10889677CC carriers had less Tregs and a faster T-cell proliferation than individuals carrying the rs10889677AA homozygous genotype [9]. Several case-controls studies have examined the association between rs10889677 SNP and risk of multiple solid tumors, such as breast cancer, lung cancer, ovarian cancer, gastric cancer, nasopharyngeal carcinoma, and oral cancer [9-13]. However, the results are inconsistent and inconclusive. Due to the importance of IL-23R during tumorigenesis, we systematically investigated the role of functional IL-23R rs10889677 genetic variant on multiple solid tumors through a meta-analysis.
Materials and Methods
Literature search and data extraction
The electronic literature searches were done with search terms of “Interleukin-23 receptor”, “IL-23 receptor”, “IL23R”, “polymorphism”, “variant”, “SNP”, “rs10889677”, “cancer”, “tumor”, as well as their combinations using HuGE Navigator (version 2.0), PubMed (US National Library of Medicine, National Institutes of Health), EMBASE and Web of science [14-16]. Case-control studies of the IL-23R rs10889677 SNP published from October, 2009 to June, 2013 were identified without language restrictions. Additional studies have also identified by screening reference lists of important studies and reviews. Criteria for selecting an eligible study: (i) original studies; (ii) studies that investigated the association between IL-23R rs10889677 polymorphism and solid tumor risk; (iii) studies that reported crude odds ratio (OR) with 95% confidence interval (CI) values or sufficient data to calculate crude OR and 95% CI. Criteria for exclusion of studies were (i) overlapping data and (ii) case-only studies, family-based studies and review articles. The raw data and demographic information, including first author, published year, population, sample size, cancer types and genotypes were independently extracted.
Statistical analysis
Association between IL-23R rs10889677 SNP and solid tumor risk were re-calculated using crude ORs together with their corresponding 95% CIs. If the P value of the heterogeneity test was ≥ 0.05, we used the fixed effect model to calculate the combined OR (the Mantel-Haenszel method) [17]. The fixed effect model assumed the same homogeneity of effect size across all studies. If the P value of the heterogeneity test was <0.05, there is statistically significant between-study heterogeneity. Then, we would use a random effects mode (the DerSimonian and Laird method) to calculate the combined OR [18]. Funnels plots was utilized to test publication bias. Egger’s test was used to evaluate the funnel plot asymmetry [19], which can indicate potential existing publication bias. The statistical analyses were performed using Stata Statistical package (version 11.0; Stata Corp., College Station, Tex). All P values were two-sided. Statistical power in relation to the minor allele frequencies (MAFs) of the IL-23R rs10889677 SNP were calculated with Quanto 1.2.4 software [20].
Results
Literature search and data extraction
After searching HuGE Navigator, NCBI PubMed , EMBASE and Web of science using the keywords “Interleukin-23 receptor”, “IL-23 receptor”, “IL23R”, “polymorphism”, “variant”, “SNP”, “rs10889677”, “cancer”, or “tumor”, and found 5 studies (6731 cases and 7296 healthy controls), which fulfilled inclusion criteria [9-13]. There are two studies on breast cancer [9,12], one on lung cancer [9], one ovarian cancer [10], one on gastric cancer [11], one on nasopharyngeal carcinoma [9], and one on oral cancer [13]. A database of genotype frequency and other information extracted from each study, was created. Table 1 showed the essential information, including first author, year of publication, SNPs genotyped, sample size,population, cancer types and genotyping methods.
Table 1. Studies included in the meta-analyses of association between IL-23R rs10889677 SNP and multiple solid tumors.
| No | Studies | Cases | Controls | Populations | Cancer types | Genotyping method |
|---|---|---|---|---|---|---|
| 1 | Zhang et al. 2010 | 96 | 115 | Chinese | ovarian cancer | PCR-RFLP |
| 2 | Chen et al. 2011 | 962 | 787 | Chinese | gastric cancer | PCR-RFLP |
| 3 | Zheng et al. 2012 | 1010 | 1014 | Chinese | breast, lung and nasopharyngeal cancers | MALDI-TOF MS |
| 4 | Wang et al. 2012 | 491 | 502 | Chinese | breast cancer | SNaP shot |
| 5 | Chien et al. 2012 | 240 | 240 | Chinese | oral cancer | PCR-RFLP |
Abbreviations: SNP, single nucleotide polymorphism; MALDI-TOF MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry; PCR-RFLP: PCR-Restriction Fragment Length Polymorphism.
Quantitative data synthesis
The association between the functional IL-23R rs10889677 SNP and risk of multiple solid tumors was investigated in 6731 cases and 7296 controls. Compared with the rs10889677 AA genotype, a 0.91-fold decreased risk to develop multiple solid tumors was observed for individuals with AC genotype in a random effect model (95% CI = 0.85-0.97, P = 0.006) (Figure 1A). The rs10889677 CC genotype was also associated with significantly decreased risk of multiple solid tumors compared to those AA genotype carriers (OR = 0.59, 95% CI = 0.53-0.66, P < 0.001) (Figure 1B). Interestingly, when we combined data of AC and CC genotypes, an OR of 0.89 (95%CI = 0.84-0.95, P < 0.001) was observed (Figure 1C).
Figure 1. Meta-analyses for functional IL-23R rs10889677 polymorphism in multiple solid tumors.
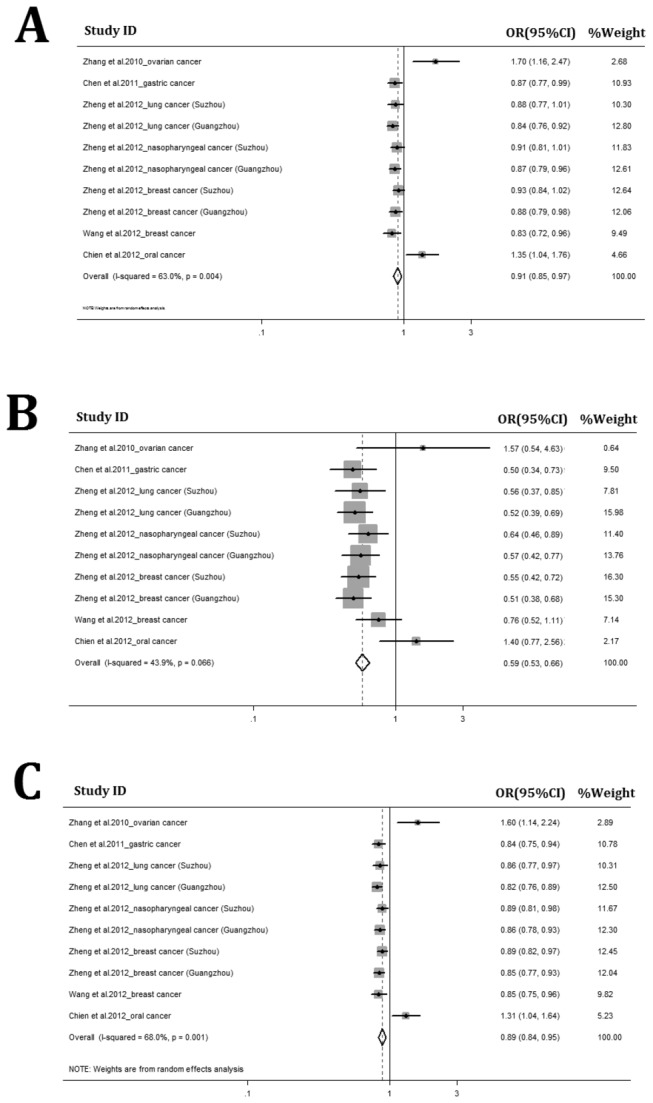
Forest plots for AC vs. AA comparison (A, CC vs. AA comparison (B), and AC + CC vs. AA comparison (C).
For the rs10889677 polymorphism with MAF equaling to 0.316 in healthy individuals, there is 94.6% or 99.9% statistical power to detect a 0.91-fold or 0.59-fold increased risk developing multiple solid tumors.
Bias diagnostics
To evaluate publication bias, genotypes of IL-23R rs10889677 polymorphism were plotted against the precision ones using a funnel plot. However, we did observe that there was publication bias on studies of IL-23R rs10889677 polymorphism (Egger’s test: P < 0.05 (Figure 2).
Figure 2. Funnel plots of the Egger’s test of IL-23R rs10889677 allele comparison for publication bias.
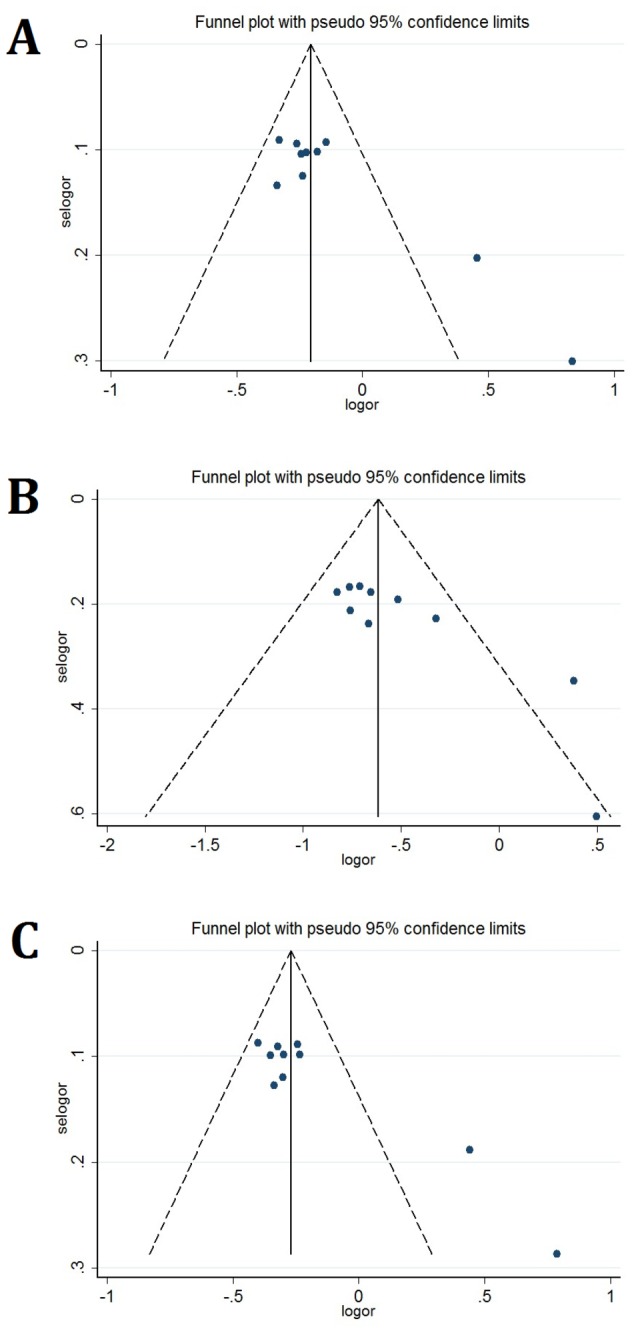
Funnel plots for AC vs. AA comparison (A, CC vs. AA comparison (B), and AC + CC vs. AA comparison (C).
Discussion
IL-23R is essential for initiating, maintaining and accelerating the IL-23/IL-17 inflammatory signal transduction pathway, which is important during malignant transformation [1,2]. A functional rs10889677AC SNP has been associated with genetic susceptibility of solid tumors, including breast cancer, lung cancer, ovarian cancer, gastric cancer, nasopharyngeal carcinoma, and oral cancer [9-13]. However, no conclusions on how the genetic variant influences cancer risk have been made. The current meta-analysis including five literatures (6731 cases and 7296 healthy controls) systematically examined association between the rs10889677AC SNP and multiple cancer risk. We found that rs10889677 SNP might be common genetic susceptible factor for multiple solid tumors.
Zheng et al demonstrated that IL-23R rs10889677 genetic variant is a regulatory genetic variant of IL-23R gene expression [9]. MiRNA let-7f can down-regulate IL-23R gene expression by binding to 3’UTR of the IL-23R mRNA and inhibit translation of IL23R protein. Interestingly, the rs10889677 A>C change locates in the binding sequence of let-7f, which could interfere let-7f-mediated repression of IL-23R expression. It has also been showed that there was higher expression level of IL-23R in peripheral blood mononuclear cells (PBMCs) from healthy individuals with rs10889677AA genotype than that among subjects with rs10889677AC or CC genotype. The IL-23R rs10889677 A allele carriers had more Tregs in vivo and lower T-cells proliferation rate in vitro compared to those with rs10889677 C allele [9]. All these findings might explain the observed decrease in cancer susceptibility in the meta-analysis of IL-23R rs10889677 SNP.
In all, we for the first time examined the involvement of functional IL-23R rs10889677 polymorphism during malignant transformation of multiple solid tumors through a systematic way. Our findings demonstrated that genetic variants that impact the efficacy of the immune system may modify cancer risk [21-23].
Supporting Information
PRISMA checklist.
(DOC)
Funding Statement
The authors have no support or funding to report.
References
- 1. Chen Z, Laurence A, O'Shea JJ (2007) Signal transduction pathways and transcriptional regulation in the control of Th17 differentiation. Semin Immunol 19: 400-408. doi: 10.1016/j.smim.2007.10.015. PubMed: 18166487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Volpe E, Servant N, Zollinger R, Bogiatzi SI, Hupé P et al. (2008) A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nat Immunol 9: 650-657. doi: 10.1038/nrm2446. PubMed: 18454150. [DOI] [PubMed] [Google Scholar]
- 3. Romagnani S (2008) Human Th17 cells. Arthritis Res Ther 10: 206. doi: 10.1186/ar2392. PubMed: 18466633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Langowski JL, Zhang X, Wu L, Mattson JD, Chen T et al. (2006) IL-23 promotes tumour incidence and growth. Nature 442: 461-465. doi: 10.1038/nature04808. PubMed: 16688182. [DOI] [PubMed] [Google Scholar]
- 5. Fontenot JD, Gavin MA, Rudensky AY (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4: 330-336. doi: 10.1038/ni904. PubMed: 12612578. [DOI] [PubMed] [Google Scholar]
- 6. Hori S, Nomura T, Sakaguchi S (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299: 1057-1061. doi: 10.1126/science.1079490. PubMed: 12522256. [DOI] [PubMed] [Google Scholar]
- 7. Kim JM, Rasmussen JP, Rudensky AY (2007) Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol 8: 191-197. doi: 10.1038/ni1428. PubMed: 17136045. [DOI] [PubMed] [Google Scholar]
- 8. Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH et al. (2009) Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc Natl Acad Sci U S A 106: 4793-4798. doi: 10.1073/pnas.0900408106. PubMed: 19273860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng J, Jiang L, Zhang L, Yang L, Deng J et al. (2012) Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis 33: 2409-2416. doi: 10.1093/carcin/bgs307. PubMed: 23042301. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Z, Zhou B, Zhang J, Chen Y, Lai T et al. (2010) Association of interleukin-23 receptor gene polymorphisms with risk of ovarian cancer. Cancer Genet Cytogenet 196: 146-152. doi: 10.1016/j.cancergencyto.2009.09.006. PubMed: 20082850. [DOI] [PubMed] [Google Scholar]
- 11. Chen B, Zeng Z, Xu L, Wu X, Yu J et al. (2011) IL23R +2199A/C polymorphism is associated with decreased risk of certain subtypes of gastric cancer in Chinese: a case-control study. Cancer Epidemiol 35: 165-169. doi: 10.1016/j.canep.2010.08.006. PubMed: 20863779. [DOI] [PubMed] [Google Scholar]
- 12. Wang L, Liu W, Jiang W, Lin J, Jiang Y et al. (2012) A miRNA binding site single-nucleotide polymorphism in the 3'-UTR region of the IL23R gene is associated with breast cancer. PLOS ONE 7: e49823. doi: 10.1371/journal.pone.0049823. PubMed: 23239971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chien MH, Hsin CH, Chou LS, Chung TT, Lin CH et al. (2012) Interleukin-23 receptor polymorphism as a risk factor for oral cancer susceptibility. Head Neck 34: 551-556. doi: 10.1002/hed.21779. PubMed: 21717520. [DOI] [PubMed] [Google Scholar]
- 14. Cai X, Yang M (2012) The functional MDM2 T309G genetic variant but not P53 Arg72Pro polymorphism is associated with risk of sarcomas: a meta-analysis. J Cancer Res Clin Oncol 138: 555-561. doi: 10.1007/s00432-011-1124-8. PubMed: 22205265. [DOI] [PubMed] [Google Scholar]
- 15. Yu D, Shi J, Sun T, Du X, Liu L et al. (2012) Pharmacogenetic role of ERCC1 genetic variants in treatment response of platinum-based chemotherapy among advanced non-small cell lung cancer patients. Tumour Biol 33: 877-884. doi: 10.1007/s13277-011-0314-y. PubMed: 22249976. [DOI] [PubMed] [Google Scholar]
- 16. Liu L, Wu G, Xue F, Li Y, Shi J et al. (2013) Functional CYP1A1 genetic variants, alone and in combination with smoking, contribute to development of head and neck cancers. Eur J Cancer 49: 2143-2151. doi: 10.1016/j.ejca.2013.01.028. PubMed: 23462525. [DOI] [PubMed] [Google Scholar]
- 17. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719-748. PubMed: 13655060. [PubMed] [Google Scholar]
- 18. Petitti DB (1994) Of babies and bathwater. Am J Epidemiol 140: 779-782. PubMed: 7977287. [DOI] [PubMed] [Google Scholar]
- 19. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629-634. doi: 10.1136/bmj.315.7109.629. PubMed: 9310563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu L, Wu G, Xue F, Li Y, Shi J et al. (2013) Functional CYP1A1 genetic variants, alone and in combination with smoking, contribute to development of head and neck cancers. Eur J Cancer 49: 2143-2151. doi: 10.1016/j.ejca.2013.01.028. PubMed: 23462525. [DOI] [PubMed] [Google Scholar]
- 21. Sun T, Zhou Y, Yang M, Hu Z, Tan W et al. (2008) Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res 68: 7025-7034. doi: 10.1158/0008-5472.CAN-08-0806. PubMed: 18757416. [DOI] [PubMed] [Google Scholar]
- 22. Yang M, Sun T, Wang L, Yu D, Zhang X et al. (2008) Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res 14: 3230-3236. doi: 10.1158/1078-0432.CCR-08-0177. PubMed: 18483392. [DOI] [PubMed] [Google Scholar]
- 23. Yang M, Sun T, Zhou Y, Wang L, Liu L et al. (2012) The functional cytotoxic T lymphocyte-associated Protein 4 49G-to-A genetic variant and risk of pancreatic cancer. Cancer 118: 4681-4686. doi: 10.1002/cncr.27455. PubMed: 22359319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(DOC)


