Abstract
The actin-associated protein Pdlim7 is essential for heart and fin development in zebrafish; however, the expression and function of this PDZ-LIM family member in the mammal has remained unclear. Here, we show that Pdlim7 predominantly localizes to actin-rich structures in mice including the heart, vascular smooth muscle, and platelets. To test the requirement for Pdlim7 in mammalian development and function, we analyzed a mouse strain with global genetic inactivation of Pdlim7. We demonstrate that Pdlim7 loss-of-function leads to significant postnatal mortality. Inactivation of Pdlim7 does not disrupt cardiac development, but causes mild cardiac dysfunction in adult mice. Adult Pdlim7 -/- mice displayed increased mitral and tricuspid valve annulus to body weight ratios. These structural aberrations in Pdlim7 -/- mice were supported by three-dimensional reconstructions of adult cardiac valves, which revealed increased surface area to volume ratios for the mitral and tricuspid valve leaflets. Unexpectedly, we found that loss of Pdlim7 triggers systemic venous and arterial thrombosis, leading to significant mortality shortly after birth in Pdlim7 +/- (11/60) and Pdlim7 -/- (19/35) mice. In line with a prothrombotic phenotype, adult Pdlim7 -/- mice exhibit dramatically decreased tail bleed times compared to controls. These findings reveal a novel and unexpected function for Pdlim7 in maintaining proper hemostasis in neonatal and adult mice.
Introduction
The PDZ-LIM family of proteins has been shown to regulate diverse biological functions, including bone morphogenesis, cardiac and skeletal muscle development and maintenance, neuronal signaling, and tumor cell growth [1,2]. Ten members comprise this protein family: Pdlim1-5, Pdlim7, LDB3, LMO7, and LIMK1 and LIMK2, sharing similar domain structures including one PDZ domain and one or more LIM domains. PDZ and LIM domains act as modular protein-binding interfaces to facilitate dynamic interactions with the actin cytoskeleton (α-actinin and β-tropomyosin), nuclear factors (Tbx4 and Tbx5), and signaling molecules (protein kinase C, ret/ptc2, and β1-integrin) [1,2]. Binding with multiple cofactors allows PDZ-LIM proteins to take on a variety of biological roles in different contexts. Importantly, several PDZ-LIM proteins have been found to function in cardiac and skeletal muscle development and maintenance in zebrafish and mice [3-9]. For example, mice lacking either Pdlim3, Pdlim5, or LDB3 develop dilated cardiomyopathy, and the latter knockout mice die within 5 days of birth due to severe striated muscle defects [4,5,8]. Pdlim1 is the only PDZ-LIM protein described to function in platelets [10], and loss of the protein in mice results in arterial thrombosis [11].
We have previously demonstrated that Pdlim7, which contains one PDZ and three LIM domains, associates with cytoskeletal actin [12], and dynamically regulates both the subcellular localization and activity of the nuclear transcription factor Tbx5 [13]. Further, knockdown of pdlim7 in zebrafish results in loss of both cardiac valve tissue and pectoral fin outgrowth [3,14]. However, the functional importance of Pdlim7 in mammalian organogenesis has remained elusive. To gain insight into the biological significance of Pdlim7 in the mouse, we genetically inactivated the Pdlim7 gene in all tissues. In contrast to the zebrafish, the global loss of Pdlim7 does not disrupt mouse cardiac development, but causes mild cardiac dysfunction and valve structural defects in adults. Interestingly, loss of or reduced levels of Pdlim7 in homo- and heterozygous mutant mice, respectively also results in systemic, occlusive thrombosis leading to significant early lethality with survivors displaying decreased tail bleed times. These findings reveal an unexpected and previously unknown in vivo function for Pdlim7 in maintaining hemostasis.
Materials and Methods
Global deletion of the Pdlim7 gene in ES cells by retroviral insertion
Pdlim7 mutant mice were generated by Lexicon Genetics Inc. from 129Sv/Ev embryonic stem (ES) cells (OST445990) using a gene trap approach as previously described [15,16]. Pdlim7 was disrupted by insertion of the VICTR37 gene trap vector in ES cells: this allele is therefore named Pdlim7 Gt(VICTR37)445990Lex. For simplicity, we will refer to Pdlim7 +/Gt(VICTR37)445990Lex mice as Pdlim7 +/- and Pdlim7 Gt(VICTR37)445990Lex/ Gt(VICTR37)445990Lex as Pdlim7 -/-. Direct sequencing of products from 3’ rapid amplification of cDNA ends (RACE) determined the integration site to be in intron 2 of the Pdlim7 gene. Pdlim7 +/- hybrid mice were backcrossed 5 generations onto the C57BL/6 background. Pdlim7 mutant mice were genotyped using multiplex PCR analysis of tail genomic DNA using the following primers to detect mutant and WT Pdlim7: (WT Pdlim7 forward primer) 5’ACCAGCTTAGCCCTCACATTT3’: (WT Pdlim7 reverse primer) 5’TACGTGTGATGCTAACACTCAGGC3’: (viral LTR2 reverse primer) 5’ATAAACCCTCTTGCAGTTGCATC3’. All protocols involving animals in this work were approved by the Institutional Animal Care and Use Committee of Northwestern University and the Ann and Robert H. Lurie Children’s Hospital of Chicago Research Center.
Semi-quantitative RT-PCR expression analysis
Total RNA from adult uteri was prepared using the NucleoSpin RNAII kit including DNase treatment (Clontech, Mountain View, CA) as previously described [12]. RNA from the human and murine megakaryocyte cell lines, K562 and Y10, respectively, were a kind gift from Dr. John Crispino. Total RNA from washed mouse platelets was prepared as previously described by Rowley et al [17]. Oligonucleotides specific for the individual genes were: Human GAPDH: (FWD) 5’-CGTCATGGGTGTGAACCATGAGAA, (REV) 5’-GCC AGT AGA GGC AGG GAT GAT GTT; Mouse GAPDH: (FWD) 5’-TGTGATGGGTGTGAACCACGAGAA, (REV) 5’- ACCAGTGGATGCAGGGATGATGTT; Human Pdlim7: (FWD) 5’-AACAACGGCAAGACTCCCGTGTGT, (REV) 5’-TCTTGCACTTGGCACAGCTGGGTG; Mouse Pdlim7: (FWD) 5’-AACAACGGCAAGACTCCTGTATGC, (REV) 5’-TCTTGCATTTGGCACAGTTGGGTG.
Quantitative RT-PCR expression analysis
Quantitative RT-PCR was performed on adult uteri RNA (see above) using pre-validated Solaris gene specific primers Pdlim7 and GAPDH (Thermo Scientific) on the Step One Plus Real-Time PCR System (Applied Biosystems) using the Solaris qPCR ROX Master Mix (Thermo Fisher) for detection of amplified DNA. Data was analyzed using the comparative ∆∆Ct method and normalized to GAPDH control. Normalized values were converted to relative values by using the corresponding WT uteri as the calibrator.
Western-blot analysis
Total protein from adult uteri and platelets (see below for platelet isolation protocol) was obtained by homogenization in RIPA buffer containing Halt Protease Inhibitor and HaltTM Phosphatase Inhibitor Cocktails (Thermo Scientific) according to manufacturer protocol. The homogenate was centrifuged at 16,000 x g for 20 minutes at 4°C and supernatant containing the protein removed. Protein concentrations were determined by a BCA assay (Pierce Biotechnology) for subsequent SDS-PAGE and immunoblot analysis using a BioRad ChemiDoc MP system with anti-Pdlim7 [13] and anti-GAPDH (Santa Cruz) antibodies followed by measurement of relative densitometry.
Beta-galactosidase staining
Pdlim7 mutant embryos from E9.5-18.5 were fixed with 4% paraformaldehyde and for sections, embedded in OCT medium and processed for cryosectioning on a Leica CM3050S cryostat (Leica Microsystems). Whole-mount embryos and sections were stained with 1 mg/ml X-gal overnight at 37°C as previously described [18,19]. Sections were briefly counter-stained with nuclear fast red to visualize overall morphology. Whole-mount embryos were imaged on a Leica MZ16 stereomicroscope fitted with a Leica DFC490 color camera using ImagePro MC (Media Cybernetics) software. For these and all subsequent colorimetric stains, sections were imaged on a Leica DMR upright microscope equipped with a QImaging Retiga 4000R camera using OpenLab (Improvision) software. All images were processed using Photoshop CS4 (Adobe Systems, Inc).
Immunofluorescence
Indirect immunofluorescence detection was performed as previously described [13,20]. In short, tissue cryosections or washed platelets (see below for isolation protocol) were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 20% goat serum, 5% 20X blocking solution (50mM NH4Cl, 25mM Lysine, and 25mM Glycine), and 0.2% BSA in PBS. Tissues or platelets were incubated with primary antibodies including, anti-Pdlim7 [13], anti-PECAM (BD Biosciences) and anti-MF20 (Developed by D.A. Fischman, Developmental Studies Hybridoma Bank, University of Iowa) antibodies and detected using Alexa 488- or 564-conjugated secondary antibodies (Invitrogen). Filamentous actin and nuclei were detected using Alexa Fluor 546 or 633 (Invitrogen) and DAPI (Roche), respectively. Fluorescence staining was visualized on a Zeiss LSM510 META confocal microscope.
Echocardiography
Transthoracic echocardiography was performed using the VisualSonics 770 Imaging System (Toronto, Canada) equipped with a 30-mHz transducer. Mice were anesthetized with 1-2% isoflurane inhalation with heart rate (360-500 beats per minute) and core temperature (36-37°C) continuously monitored. Annulus dimensions were obtained in the apical 4-chamber view during end diastole with mitral and tricuspid valves in the closed position. Pulsed wave doppler interrogation was performed on the mitral valve inflow in the apical 4-chamber view using a sample volume toggle to optimally assess flow velocities. All measurements were obtained using an angle of interrogation <30° [21]. The isovolumetric relaxation time (IVRT), isovolumetric contraction time (IVCT), and ejection time (ET) were also measured from the mitral inflow doppler signal [22-24]. The Myocardial Performance Index (MPI) or Tei index was calculated as follows: (IVRT + IVCT) / ET. Higher values in the MPI indicate worsening cardiac function. Cardiac chamber dimensions, wall thickness, and left ventricular cardiac function (ejection fraction and fractional shortening) were measured from images obtained using two-dimensional M-mode echocardiography in the parasternal long and short axis views [25]. All measurements were obtained in triplicate and averaged.
Histological Analysis
Specimens were fixed in 4% paraformaldehyde, dehydrated, embedded in paraplast, and sectioned on a Leica RM2265 microtome (Leica Microsystems). For the embryonic and perinatal studies, sections were stained with 1% Alcian blue pH2.5/nuclear fast red and Hematoxylin & Eosin, respectively.
For the three-dimensional morphometric analysis, 3-month old hearts were first heparinized, relaxed, and perfused prior to fixation [25] to obtain better visualization of valve tissue. Specifically, 50 µL of 1,000 U/mL heparin was injected directly into the right ventricle and allowed to circulate to prevent clotting of the blood. The abdominal aorta and descending vena cava were then cut for fluid overflow. Next, 500 µL of relaxin solution was injected into each ventricle to relax the heart and keep the atrioventricular valves open, which was followed by perfusion with a total of 5 mL of PBS. The heart was then removed from the chest cavity and a 22G teflon catheter inserted into the aorta (past the aortic and mitral valves) followed by perfusion with 5mL PBS. Hearts were serial sectioned and stained with either 1% Alcian Blue pH2.5/nuclear fast red or Masson’s Trichrome staining.
Three-dimensional morphometric analysis
Adult heart serial sections were imported into Amira v5.4.0 (Visage Imaging) at a scaled unit pixel size of 1x1x1.285 to derive the comparative valve metrics. Sections were manually aligned, followed by a least squares computed alignment to minimize the number of non-overlapping pixels between two sequential images. The AV valve tissue was manually segmented with the aid of a threshold mask on a gray-scale image series by referencing the color images for each heart section. For the surface area and volume computation, a triangulated surface was computed from the voxel segmentation using the constrained smoothing algorithm option. The surface area and volume of the AV valves were computed using Amira’s Surface Area module.
Blood collection and counts
Blood was drawn by cardiac puncture from adult mice anesthetized with 2-3% isoflurane inhalation into either heparin or 3.2% sodium citrate at a ratio of 1:9. Blood cell counts were performed on a Beckman Coulter AcTdiff2 Analyzer. To obtain washed platelets, whole blood was centrifuged at 86 x g for 8 minutes to collect platelet-rich plasma (PRP). To increase platelet yield, the lower phase was washed 3 times with HEPES/Tyrode’s buffer and the obtained PRP were combined. The PRP was then spun in the presence of prostaglandin I2 (0.1 µg/mL) at 718 x g for 6 minutes. The platelet pellet was washed in HEPES/Tyrodes buffer pH7.4 (10mM HEPES, 12mM NaHCO3, 138mM NaCl, 5.5mM glucose, and 2.9mM KCl) [26] and either re-pelleted for Western blot or resuspended in HEPES/Tyrodes buffer pH7.4 containing 2 U/mL apyrase and 10µm indomethacin at a concentration of 50,000 plts/µL and allowed to spread on glass coverslips for 5 minutes at 37°C followed by indirect fluorescence immunostaining as described above. Platelet-poor plasma was obtained by centrifugation of whole blood at 2000 x g for 10 minutes. The liver chemistries, prothrombin, and partial thromboplastin times were performed by Antech Diagnostics (Oak Brook, IL).
Tail bleeding time assay
The tail bleeding time assay was carried out with the operator blinded to the genotype of the mice. Weaning age mice were sedated with Acepromazine (2mg/kg) and placed on a raised platform with tails protruding over the edge. Tails were positioned 5mm above filter paper and a 2-3mm cut was made at the distal tip of the tail with a scalpel. The bleeding time was defined as the time needed for the cessation of bleeding (no blood drops for 1 minute) [27].
Statistics
All values are expressed as mean SD. Comparisons between two groups were evaluated with the unpaired t test and between three groups with one-way ANOVA. The Kruskal-Wallis one-way analysis of variance was used to evaluate body weights, bleeding time, and the following blood cell and chemistry counts: monocyte, granulocyte, aspartate aminotransferase (AST), and total bilirubin, which were non-normally distributed. P<0.05 was considered statistically significant.
Results
Pdlim7 is dynamically expressed in actin-rich structures during murine development
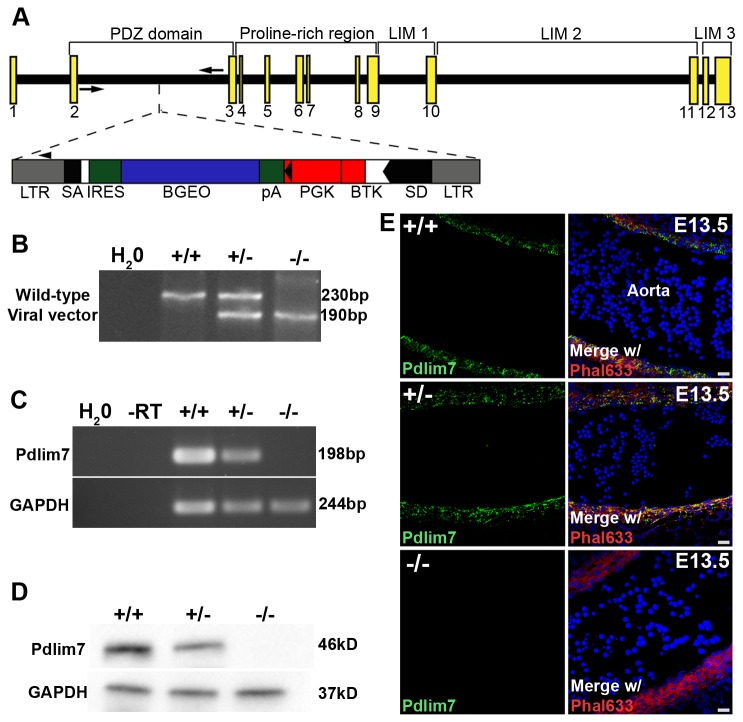
In order to study the functional role of Pdlim7 in the mammal, we chose a retrovirus insertion into intron two of the mouse Pdlim7 gene (Figure 1A-B), as this disruption is expected to globally abolish protein expression. Loss of Pdlim7 mRNA expression was verified in adult Pdlim7 mutant uteri using semi-quantitative RT-PCR (Figure 1C). The reduction in mRNA was further verified by quantitative RT-PCR, revealing a decrease in Pdlim7 transcript levels in the respective uteri of Pdlim7 +/- and Pdlim7 -/- mice of 61.27 15.25% and 99.37 0.06%, respectively (n≥3). Additionally, Western-blots with anti-Pdlim7 antiserum [13] recognized Pdlim7 protein in the smooth muscle-rich uterus of wild-type (WT) adult mice; however, protein expression in the respective uteri of Pdlim7 +/- and Pdlim7 -/- mice was only 29.36 14.92% and -1.18 2.64% of protein levels in WT mice (Figure 1D; n=5). In agreement with the Western-blot, immunohistochemistry with anti-Pdlim7 antiserum revealed Pdlim7 protein in the aorta of WT embryos while protein expression in the respective aorta of Pdlim7 -/- littermates could not be detected (Figure 1E). Thus, we conclude that the Pdlim7 allele generated is a null allele.
Figure 1. Generation of Pdlim7 mutant mice.
Schematic representation of the retroviral integration site within intron two of the Pdlim7 gene (A). Exons are depicted by yellow boxes with WT and viral-specific genotyping primers represented by arrows and an arrowhead, respectively. Genotyping reveals a 230bp PCR fragment representing the WT allele and a 190bp product for the viral integration (B). Semi-quantitative RT-PCR demonstrates absence of Pdlim7 gene transcripts in adult uteri of Pdlim7-/- mice using GAPDH as control (C). Western blot shows that Pdlim7-/- and Pdlim7+/- mice express only -1.18 ± 2.64% and 29.36 ± 14.92% Pdlim7 protein compared to WT controls, respectively (D). Sagittal section through the aorta demonstrates Pdlim7 antibodies (green) localize to filamentous actin (red) of smooth muscle in WT, but not Pdlim7-/- embryos (E), further verifying loss of Pdlim7 protein in null mice (control DAPI nuclei, blue). LTR = long terminal repeat; SA and SD = splice acceptor and donor sites, respectively; IRES = internal ribosomal entry site; BGEO = lacZ gene; pA = polyadenylation signal; PGK = phosphoglycerate kinase gene; BTK = Bruton’s tyrosine kinase gene.
Taking advantage of the gene trap containing a lacZ reporter in frame with Pdlim7 exon coding sequences (Figure 1A), we used β-galactosidase staining to establish a previously unknown expression profile of Pdlim7 in the mouse. In accordance with our studies in the zebrafish [14] and chicken [12,20], murine Pdlim7 was dynamically expressed throughout embryogenesis, particularly in the developing somites, forelimbs and hindlimbs, and heart (Figure 2A-C). We detected robust expression of Pdlim7 at E9.5 through E10.5 in a rostral to caudal gradient in the somites (Figure 2A-B) and at E11.5 in cells migrating from the myotome (Figure 2C). At E18.5, Pdlim7 was also detected throughout the embryonic smooth muscle including the stomach, bladder, duodenum, lungs, esophagus, and aorta (Figure 2F and Table S1 in Methods S1). In the developing heart, we detected Pdlim7 transcripts in the atria, trabeculated regions of the ventricles, and the interventricular and atrial septa (Figure 2D-E and Table S1 in Methods S1).
Figure 2. Pdlim7 is dynamically expressed in actin-rich structures throughout murine development.
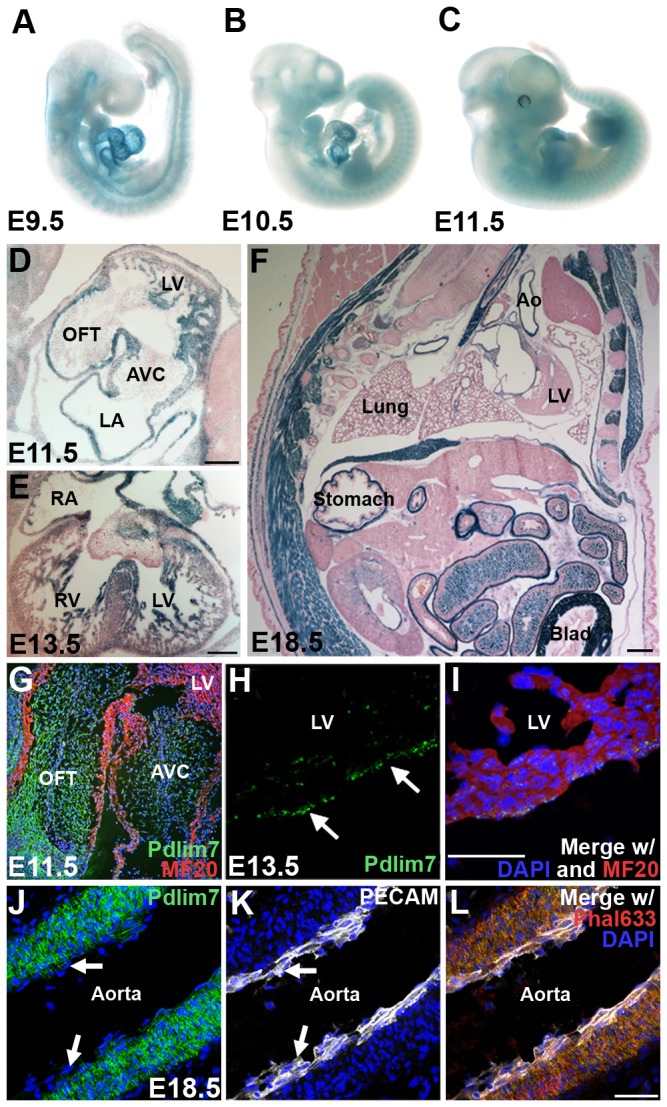
β-galactosidase staining of whole mount embryos identifies Pdlim7 early in the developing heart, forelimbs, hindlimbs, and somites (A-C). In E11.5 and E13.5 heart sections, β-galactosidase staining is localized to the trabeculated regions of the ventricles, atrial walls, and interventricular and atrial septa (D-E); of note: the β-galactosidase reaction time chosen does not reveal the lower levels of Pdlim7 expression in the cardiac cushion or valve cells. Sagittal sections of E18.5 embryos show Pdlim7 expression throughout the smooth muscle, including the lung alveoli, stomach, aorta, and bladder (F). Immunostaining of E11.5 hearts demonstrates Pdlim7 (green) localization to the outflow tract and atrioventricular cushions with MF20 positive myocardial cells (red) (G). Pdlim7 (green) proteins are also detected in the epicardium of E13.5 embryos (arrows, H-I) with MF20 positive myocardial cells (red) and DAPI nuclei (blue). In the aorta, Pdlim7 proteins (green) localize to filamentous actin (red) of smooth muscle, but not PECAM-positive endothelium (white) (arrows, J-L), control DAPI nuclei (blue). Scale bar = 200μm (D-E) and 50 μm (F, I, L). Ao = Aorta; AVC = atrioventricular cushion; Blad = bladder; LA = left atrium; LV = left ventricle; OFT = outflow tract; RA = right atrium; RV = right ventricle.
To obtain more detailed information on Pdlim7 protein distribution, we utilized immunohistochemistry with our anti-Pdlim7 antiserum [13], which is cross-reactive in mouse tissue (Figure 1E). We noted from NCBI searches that Pdlim7, like other PDZ-LIM proteins, is expressed in multiple splice variants (data not shown). Our Pdlim7 antibodies, however, were designed to exclusively detect the full-length protein composed of a PDZ domain, proline-rich region, and three LIM domains and, thus, was not expected in all cases to produce an overlapping pattern with the β-galactosidase staining. For example, we found Pdlim7 transcripts but no antibody reaction in the left atrium and trabeculated regions of the left ventricle of E11.5 embryos (Figure 2D and G), suggesting that these domains express shorter splice forms of the Pdlim7 gene. Consistent with protein distribution in the chicken [20], our immunostaining detected Pdlim7 proteins in the developing atrioventricular (AV) and outflow tract (OFT) cushions of the heart as well as the epicardium (Figure 2G-I). This more sensitive technique detected expression that was not readily apparent from β-galactosidase staining. We also confirmed Pdlim7’s selectivity to smooth muscle by performing immunostaining with anti-Pdlim7 and anti-platelet endothelial cell adhesion molecule (PECAM)-1 antibodies along with an actin stain (Phalloidin). We found Pdlim7 co-localized with filamentous (F)-actin in aortic smooth muscle, but not in the endothelial layer (arrows, Figure 2J-L). Overall, these expression data revealed that Pdlim7 predominantly localizes to actin-rich structures throughout mammalian development.
Non-Mendelian ratios are observed in Pdlim7 mutant mice
From Pdlim7 heterozygous crosses, we noticed that the percentage of Pdlim7 +/+ (23.2), Pdlim7+/- (39.2), and Pdlim7 -/- (12.8) were not obtained in the expected 25:50:25 Mendelian ratios at weaning (Table 1; n=94; Chi square test p=0.0006). Close observation revealed that non-surviving Pdlim7+/- (11/60) and Pdlim7 -/- (19/35) pups were born alive, but died within 48 hours of birth while the survivors lived over 1 year of age. Notably, the percent postnatal lethality strongly correlates with Pdlim7 -/- and Pdlim7 +/- mice expressing only -1.18 2.64% and 29.36 14.92% of protein levels in WT mice, respectively (Figure 1D).
Table 1. Mendelian ratios are not obtained at weaning from Pdlim7 heterozygous crosses.
| Age | N-value | Pdlim7+/+ (%) | Pdlim7+/- (%) | Pdlim7-/- (%) |
|---|---|---|---|---|
| Birth | 125 | 24.0 | 48.0 | 28.0 |
| P21 | 94 | 23.2 | 39.2 | 12.8 |
Gross assessment of Pdlim7 mutant mice revealed that at birth, Pdlim7 -/- pups (n=3) had comparable body weights to WT littermates (n=7; 1.34 0.11 g vs. 1.27 0.12 g, p=0.3608); however, at 3-months of age, Pdlim7 -/- mice (n=8; 22.4 0.5 g) exhibited a reduction in body weight compared to WT littermates (n=13; 26.3 2.3 g) with the Pdlim7 +/- mice (n=13; 25.1 1.8 g, p=0.0003) displaying an intermediate phenotype.
Pdlim7-deficient mice maintain normal cardiac development while adults exhibit mild cardiac dysfunction and aberrant cardiac valve shape
Knockdown of pdlim7 in zebrafish causes improper AV boundary formation, leading to loss of the cardiac chamber valve [14]. Therefore, we hypothesized that a primary cardiac anomaly, especially related to the AV valves, may contribute to the lethality in Pdlim7 mutant mice. In mice, valve development begins at the AV junction where specialized myocardial cells secrete growth factors of the TGF gene family (TGFβ1-3, BMP2 and 4) that induce endocardial epithelial-to-mesenchymal transition (EMT) and promote endocardial cushion formation [28-33]. The endocardial cushions undergo complex remodeling whereby the cushion mesenchyme differentiates into fibroblastic interstitial cells characteristic of mature valves [34-38].
To our surprise, cardiac valve development in Pdlim7 -/- mice appeared normal. A multi-stage histological analysis of Pdlim7-/- embryonic hearts with a focus on the endocardial cushions and developing valves did not reveal significant morphological abnormalities compared to WT controls (Figure S1; n>3; Materials S1). To assess at the cellular level whether endocardial EMT was abnormal in Pdlim7-deficient mice, we utilized an ex vivo AV cushion explant assay [39]. We were unable to detect significant differences in the transformation of epithelial-to-mesenchymal cells and subsequent invasion into the collagen gel between Pdlim7 -/- embryos and WT controls (Figure S2A-D; n=3; Materials S1). In addition to our histological and cellular assessment of EMT progression, we performed molecular analyses. Utilizing whole-mount in situ hybridization, we found the spatiotemporal expression of Bmp2, a key regulator of endocardial EMT [40,41], to be relatively normal in Pdlim7 -/- embryos during critical stages (E9.5 and E10.5) of cushion formation (Figure S2E-H; n>3; Materials S1). A gene pathway PCR array selected for genes involved in EMT further verified that genes known to be important for endocardial cushion development, (e.g. Erbb3, Notch1, Snai1, Tgfb2, Twist1, and Versican (Vcan)), including several downstream of Bmp2, remained unchanged following loss of Pdlim7 in mice (Figure S2I; <2-fold differential expression; Materials S1). Taken together, these findings indicate that in mice, Pdlim7 does not appear to be essential for early cardiac AV valve formation.
Several PDZ-LIM protein loss-of-function studies in mice indicated later onset cardiac defects [5,8], especially compared to similar analyses done in zebrafish [6]. Thus, we sought to determine whether Pdlim7 loss-of-function leads to later onset cardiac defects. Doppler transthoracic echocardiography on 3-month old Pdlim7 -/- (n=9) and WT mice (n=8), however, did not reveal significant differences in left ventricular output during the contracted phase (systole) of the cardiac cycle as measured by ejection fraction (EF) and fractional shortening (FS) (Figure 3C, Table S2 in Methods S1; p>0.05). We did find that adult Pdlim7-/- mice displayed an elevated myocardial performance index or Tei index (Figure 3A-B and D, Table S3 in Methods S1; 0.66 ± 0.09 vs. 0.53 ± 0.06, p=0.0036), which assesses cardiac function during both the relaxed and contracted phases of the cardiac cycle [24,42,43], indicating mild cardiac dysfunction. Additionally, we found that adult Pdlim7-/- mice (n=9) had increased mitral (0.12 ± 0.01 mm/g) and tricuspid (0.09 ± 0.01 mm/g) annulus diameter to body weight ratios in comparison to WT (Figure 3E-G; n=8, 0.09 ± 0.01 and 0.07 ± 0.01 mm/g, respectively p=0.000018 and 0.0096). Left ventricular wall thickness and inner chamber dimensions during the cardiac cycle remained unchanged (Table S2 in Methods S1; n≥8, p>0.05), suggesting that the increased annulus was not a result of an enlarged chamber, but rather a primary malformation related to valve size.
Figure 3. Adult Pdlim7-/- mice exhibit mild cardiac dysfunction with increased valve annulus dimensions.
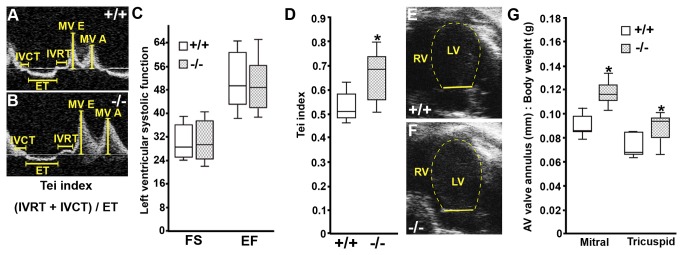
Representative Doppler profile in the apical-4 chamber view of mitral valve inflow velocities (A-B) and mitral valve annulus measurement (E-F) from WT (n=8) and Pdlim7-/- (n=9) mice. Box-and-whisker plots reveal normal fractional shortening (FS) and ejection fraction (EF) in Pdlim7-/- mice (C), but an increased Tei index (D) and increased mitral and tricuspid annulus dimensions to body weight ratios (G) as compared to controls, *p<0.01. A = late filling; AV = atrioventricular; E = early filling; ET = ejection time; IVCT = isovolumetric contraction time; IVRT = isovolumetric relaxation time; LV = left ventricle; MV = mitral valve; RV = right ventricle.
Following echocardiography, the 3-month old hearts were subjected to histological analysis in multiple orientations. Masson’s trichrome and Alcian blue staining revealed normal distribution of collagens and glycosaminoglycans in AV valve sections, respectively, but an elongation of the leaflets in Pdlim7 -/- hearts (arrows, Figure 4A-F). In order to reliably measure valve shape and size, we utilized AMIRA software to assemble a complete series of adjacent histological sections in both coronal and transverse orientations into 3-dimensional (3D) valve models. Using surface area to volume (SA:Vol) ratios as a measure of shape and size [44], we quantified the difference between Pdlim7 -/- and WT valve leaflets. We determined that Pdlim7 -/- mice have increased SA:Vol ratios for both the mitral (n=5; 1.015 0.157) and tricuspid (n=4; 0.917 0.111) valves in comparison to WT (Figure 4G-I and Video S1; n=6, 0.809 0.043, p=0.0127 and n=5, 0.782 0.038, p=0.0363, respectively). The increased SA:Vol ratio translates into longer, thinner valves, suggesting an abnormal remodeling of the AV valves in the absence of Pdlim7.
Figure 4. Atrioventricular valves of adult Pdlim7-/- mice display abnormal morphology.
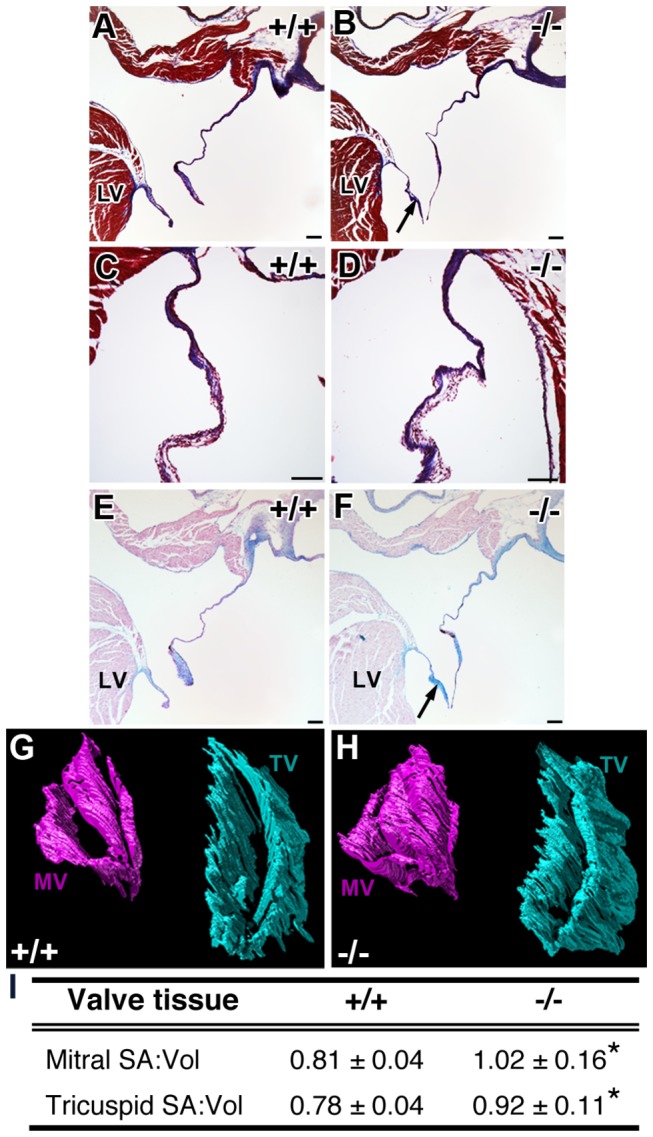
Histological sections from 3-month old WT and Pdlim7-/- hearts stained with Masson’s Trichrome (A-D) and Alcian blue (E-F) exhibit normal distribution of collagens (blue, A-D) and glycosaminoglycans (blue, E-F), but elongated mitral valves in Pdlim7-/- mice (arrows, B, F; n=5) compared to WT controls (A, E; n=3). Scale bar = 100 μm. Representative 3D reconstructions of serial sections of mitral (pink) and tricuspid (turquoise) valves from a WT (G) and Pdlim7-/- (H) mouse in the same anterodorsolateral perspective. Table providing quantification of increased surface area to volume ratios of atrioventricular valves in Pdlim7-/- (mitral, n=5; tricuspid, n=4) compared to WT (mitral, n=6; tricuspid, n=5) controls (I). *p<0.04. LV = left ventricle; MV = mitral valve; SA = surface area; TV = tricuspid valve; Vol = volume.
Occlusive vascular thrombosis leads to early mortality in Pdlim7 mutant mice
Unexpectedly, while examining adult Pdlim7 -/- hearts prior to perfusion, we noticed, in some cases, blood clots attached to the AV valves (Figure S3; Materials S1). These findings prompted an examination of a clotting issue that may explain the early mortality in Pdlim7 mutant mice. Gross morphological assessment of the non-surviving Pdlim7+/- (n=3) and Pdlim7-/- (n=9) pups revealed extensive vascular thrombi associated with dilatation of the atria and coronary vasculature as well as lung congestion (Figure 5B-C’). Histological analysis confirmed the presence of pre-mortem occlusive venous and arterial thrombi in several organ systems of non-surviving Pdlim7+/- (n=7) and Pdlim7-/- (n=6) pups, with each pup exhibiting multiple thrombi in more than one organ (Figure 5 and Table 2). For example, note the mural thrombus in the right ventricle of a Pdlim7 -/- heart (Figure 5E-E’) and the organized thrombus in the umbilical vessel with evidence of recanalization (Figure 5G-G’). We also found early organizing thrombi in the arterioles of lungs in almost all of the non-surviving Pdlim7-/- pups (5/6; Figure 5I-I’) with additional clots in the hepatic vein (4/6) and coronary vessels (4/6) (Table 2). Of note, we did not observe organizing thrombi in Pdlim7 -/- mice prior to birth (data not shown), indicating that the clots were forming after birth. These findings indicate that a deficiency in Pdlim7 results in systemic, pre-mortem vascular thrombosis leading to significant mortality.
Figure 5. Pathological analysis of non-surviving Pdlim7 mutant pups reveals pre-mortem thrombi.
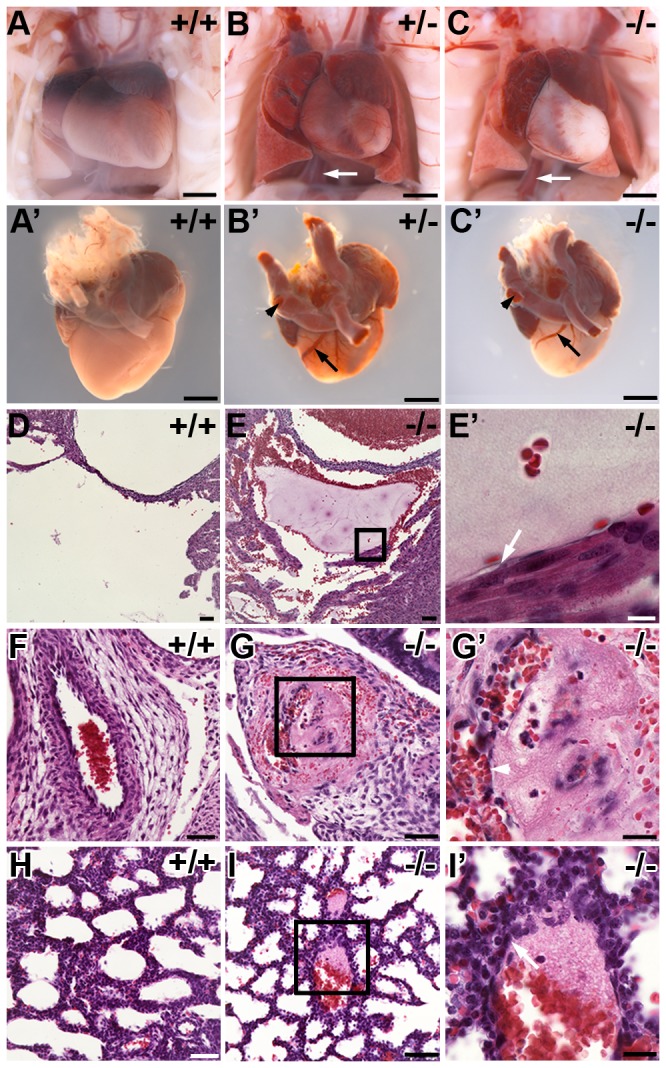
Pdlim7 +/- (n=3) and Pdlim7 -/- (n=11) perinatal lethal pups exhibit extensive blood clots in the heart, arteries (arrowhead), and veins (arrows, B-C’) associated with atrial dilation and lung congestion compared to WT (n=7) controls (A-A’). Example of pre-mortem blood clots in the right ventricle (E-E’), umbilical vessel (G-G’), and lung alveoli (I-I’) of a Pdlim7-/- perinatal lethal pup compared to WT control (D, F, H). Boxes depict location of high magnification image in E’, G’, and I’. Arrows point to attachment of the thrombus to the vessel wall (E’, I’) and arrowhead points to recanalization of the clot (G’). Scale bar = 1mm (A-C’), 50 µm (D-I), 20 µm (G’, I’), and 10µm (E’).
Table 2. Location and prevalence of pre-mortem thrombi in Pdlim7 mutant perinatal lethal pups.
| Cardiovascular System | Pdlim7+/- | Pdlim7-/- |
|---|---|---|
| Aorta | 2/7 | 2/6 |
| Coronary vessels | 4/7 | 4/6 |
| IVC / SVC | 1/7 | 2/6 |
| Atria | 3/7 | 3/6 |
| Ventricles | 1/7 | 2/6 |
| AV valves | 3/7 | 2/6 |
| Pulmonary System | ||
| Lung alveoli | 7/7 | 5/6 |
| Pulmonary artery | 0/7 | 1/6 |
AV = atrioventricular; IVC = inferior vena cava; SVC = superior vena cava.
Pdlim7 mutant survivors exhibit hemostatic dysfunction
Based on the neonatal findings and blood clots on the AV valves of Pdlim7-/- adult mice, we sought to further characterize a role for Pdlim7 in regulating adult hemostasis. To better understand the etiology of the hemostatic dysfunction, we performed a range of blood tests to assess any changes following loss of Pdlim7. Prothrombotic phenotypes in mice have almost exclusively been linked to deficiencies in plasma proteins [45-47], thus we also tested whether the plasma proteins related to the extrinsic (prothrombin time, PT) and intrinsic (partial thromboplastin time, PTT) coagulation pathways were affected. Both the PT and PTT assays revealed no significant difference between Pdlim7 -/- (PT and PTT n=6) and WT mice (Table 3; PT n=8 and PTT n=4; p>0.05), indicating that the coagulation pathways function normally in the absence of Pdlim7. The liver synthesizes clotting factors, coagulation inhibitors, and fibrinolytic proteins [48], but its central role in hemostasis is also emphasized by liver diseases that are accompanied by hemostatic changes, leading to either bleeding or thrombosis [48,49]. We conducted a liver chemistry panel, but did not find significant differences between Pdlim7 -/- (n=6) and WT (n=6; p>0.05) mice for any of the parameters tested, such as total bilirubin, albumin, aspartate aminotransferase (AST), or alanine aminotransferase (ALT) (Table 3). In addition to the liver chemistries, we performed a complete blood count to assess whether other components and features of the blood were abnormal in Pdlim7 mutant mice. We found slightly increased lymphocytes and monocytes in Pdlim7 -/- (n=5; 4.00 ± 1.50 and 0.32 ± 0.11 x103/µL, respectively) in comparison to WT (Table 3; n=5; 1.86 ± 0.63, p=0.0189 and 0.16 ± 0.05 x103/µL, p=0.0472, respectively). However, all other parameters tested including red blood cells, hemoglobin, and platelets were similar between Pdlim7 -/- (n=5) and WT (Table 3; n=5; p>0.05).
Table 3. Blood chemistry panel and complete blood cell counts in adult Pdlim7 -/- mice in comparison to WT controls.
| Parameter | Units | Pdlim7+/+ | Pdlim7-/- | P-value |
|---|---|---|---|---|
| Prothrombin time (PT) | sec | 8.78 ± 0.58 | 8.62 ± 0.69 | 0.6416 |
| Partial thromboplastin time (PTT) | sec | 34.40 ± 0.67 | 34.52 ± 4.77 | 0.9632 |
| Total protein | g/dL | 4.78 ± 0.12 | 4.77 ± 0.18 | 0.8501 |
| Albumin | g/dL | 2.43 ± 0.44 | 2.68 ± 0.16 | 0.2213 |
| Aspartate aminotransferase (AST) | IU/L | 108.8 ± 40.39 | 96.83 ± 40.81 | 0.2298 |
| Alanine aminotransferase (ALT) | IU/L | 41.67 ± 10.56 | 50.67 ± 26.57 | 0.8031 |
| Alkaline phosphatase | IU/L | 77.50 ± 6.41 | 86.83 ± 16.87 | 0.2339 |
| Gamma-glutamyl transpeptidase (GGT) | IU/L | 1.00 ± 0 | 1.00 ± 0 | NA |
| Total Bilirubin | mg/dL | 0.12 ± 0.04 | 0.1 ± 0 | 0.6310 |
| White blood cells | x 103/µL | 3.00 ± 0.87 | 4.44 ± 1.58 | 0.1118 |
| Red blood cells | x 106/µL | 7.59 ± 0.82 | 7.94 ± 0.52 | 0.4361 |
| Hemoglobin | g/dL | 12.32 ± 0.78 | 11.92 ± 0.81 | 0.4493 |
| Mean corpuscular hemoglobin | pg | 15.24 ± 0.30 | 15.00 ± 0.12 | 0.1411 |
| Red blood cell distribution width | % | 35.52 ± 3.79 | 35.32 ± 6.75 | 0.9553 |
| Platelets | x 103/µL | 550 ± 44.34 | 531 ± 76.01 | 0.6353 |
| Lymphocytes | x 103/µL | 1.86 ± 0.63 | 4.00 ± 1.50 | 0.0189* |
| Monocytes | x 103/µL | 0.16 ± 0.05 | 0.32 ± 0.11 | 0.0472* |
| Granulocytes | x 103/µL | 0.18 ± 0.05 | 0.24 ± 0.09 | 0.3272 |
To further evaluate the effect of Pdlim7 deficiency on hemostasis, we determined tail-bleeding times in normal and mutant mice (Figure 6A). The mean time to bleeding cessation was dramatically shorter in Pdlim7-/- mice (n=9; 5.33 ± 11.7 s) in comparison to WT (n=16, 84.9 ± 45.9 s) with Pdlim7 +/- mice exhibiting an intermediate phenotype (n=10, 38.3 ± 44.10 s, p=0.0007), suggesting a platelet dysfunction in Pdlim7-deficient mice. Next, we assessed Pdlim7’s expression in hematopoietic cells and detected Pdlim7 transcripts in murine and human bone marrow and megakaryocyte cell lines, the latter of which are the precursors to platelets (Figure 6B). In contrast to a previous report by Gupta et al. [11] that found no Pdlim7 (Enigma) mRNA expression in mouse platelets, using semi-quantitative RT-PCR, we could demonstrate Pdlim7 transcripts in WT murine platelets (Figure 6B). Additionally, we also demonstrate by Western-blot of platelet lysates that the RNA is translated into Pdlim7 proteins (Figure 6C). These findings are further supported by immunocytochemistry, revealing Pdlim7 co-localization with F-actin in resting and spreading platelets (Figure 6D-F). In context, these data reveal that despite approximately half of Pdlim7-/- mice escaping premature death, the survivors also exhibit abnormal blood coagulation. The decreased bleeding time, but normal function of plasma coagulation factors, suggests that the prothrombotic phenotype in Pdlim7 -/- mice is likely either platelet- or vascular smooth muscle-derived. These findings identify a novel and unexpected role for Pdlim7 in regulating mammalian hemostatic function.
Figure 6. Pdlim7 is expressed in platelets and adult Pdlim7 deficient mice display hemostatic dysfunction.
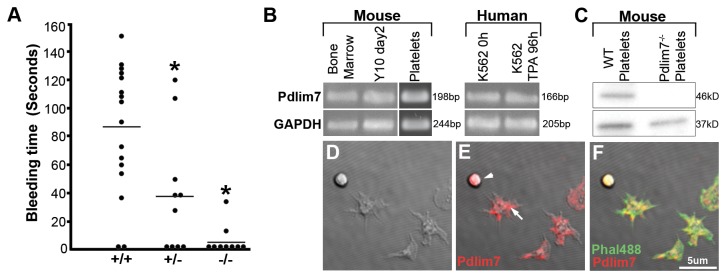
Upon tail clip, blood from 3-week old Pdlim7-/- (n=9) mice clotted immediately compared to WT (n=16) controls with Pdlim7+/- mice (n=10) exhibiting an intermediate phenotype (A). Horizontal line denotes mean bleed time for each genotype *p<0.01. Semi-quantitative RT-PCR identifies Pdlim7 gene transcripts in WT mouse bone marrow, megakaryocyte cell line Y10 (undifferentiated, day2), and platelets as well as human bone marrow cell line K562 (undifferentiated, 0h; differentiated into proplatelets using TPA for 96h) using GAPDH as a control (B). Western blot analysis of platelets from WT mice demonstrates the presence of Pdlim7 proteins and the absence of Pdlim7 proteins in Pdlim7-/- mice using GAPDH as a loading control (C). Confocal images of WT platelets spread on glass and immunostained with anti-Pdlim7 antibodies (red) and counterstained for F-actin (green) (D-F). Notably, Pdlim7 is distributed in resting (arrowhead) and spreading platelets (arrow) (E). Scale bar = 5µm.
Discussion
In the current study, we provide the first characterization of a global Pdlim7 loss-of-function mouse. As Pdlim7 is widely distributed during embryogenesis, with robust expression in the heart, we hypothesized that a significant number of Pdlim7 mutants die prematurely from a heart defect. However, we found that loss of Pdlim7 in mice does not cause detectable abnormalities in heart formation during embryogenesis. We demonstrate that Pdlim7 deficiency is associated with mild cardiac dysfunction and gross defects in hemostasis in both neonatal and adult mice, revealing a new and unexpected function for this protein.
As a gene trap approach was used to generate the Pdlim7 mutant mice, we cannot rule out the possibility that insertion of the Bgeo cassette into the genome has an effect on neighboring genes, leading to potential complications of interpretation of phenotypes. The fact that the intermediate phenotypes in body weight, bleeding time, and postnatal lethality correlate with the reduction of Pdlim7 transcript and protein levels in heterozygous mice, indicate that the demonstrated problems are the result of Pdlim7 loss-of-function. Considering the phenotypes in Pdlim7 +/- mice, it appears that the significantly reduced Pdlim7 transcript and protein levels in the heterozygous mice leads to haploinsufficiency.
Pdlim7 is dispensable for heart valve development in mice, but is important for adult heart function
The apparent lack of molecular and cellular consequences of Pdlim7 inactivation on valve development in mice are in stark contrast to the dramatic problems in valve formation in the two-chambered zebrafish heart following knockdown of pdlim7 [14]. Recent live imaging studies of zebrafish cardiac development demonstrated that the AV valve develops through direct invagination of AV endocardial cells without a complete EMT [50,51] as in higher vertebrates and mammals [28,31,52]. Thus, one possible explanation is that certain Pdlim7-dependent steps related to valve development in zebrafish are not recapitulated in the four-chambered mouse heart.
In mice, endocardial cells are considered to be the main contributors to mature AV valves [19]; therefore, we focused our attention on endocardially-derived valve defects in Pdlim7-deficient mice. Our investigation demonstrated that Pdlim7 is not required for AV cushion formation. However, we did find morphological aberrations in mature valves. Recent work by Wessels et al. revealed that in late fetal development, a subset of the AV valve leaflets largely consist of epicardial-derived cells (EPDCs) [53], shifting the long held paradigm that endocardial cells are the main contributors of valves. In both chicken [20] and mouse, we could detect Pdlim7-expressing cells in the epicardium, and it is therefore possible that Pdlim7 has a yet unknown function in EPDCs that supports valve remodeling. In light of these recent findings, future studies that would include EPDC lineage tracing to determine differences in the epicardial contribution to the AV valves following loss of Pdlim7 would be of great interest. However, given that Pdlim7-deficient adult mice also exhibit hemostatic dysfunction, we cannot rule out the possibility that the AV valve abnormalities are secondary to blood-related defects in these mice.
Multi-species functional analyses of PDZ-LIM proteins are not well-documented; however, loss-of-function studies using the PDZ-LIM protein Ldb3 revealed less severe phenotypes and later onset in mice as compared to zebrafish and Drosophila [5,6,9]. In context with our Pdlim7 loss-of-function studies in zebrafish [3,14] and the mouse data presented here, an understanding is emerging that PDZ-LIM proteins are dispensable during mammalian organogenesis, but are rather important for organ function and homeostasis. The difference in presentation of the Pdlim7 -/- phenotype between zebrafish and mouse may also lie in the fact that several PDZ-LIM proteins are expressed in overlapping domains in the mammalian heart, including Pdlim3 in the developing endocardial cushions, which would allow for other family members to compensate to some degree for the loss of Pdlim7 in our mutant model [4,5,8,54]. The possibility of functional redundancies of PDZ-LIM protein family members in the mouse complicate the analysis of single protein knockouts, and warrants further studies with PDZ-LIM protein double-mutant mice to uncover the full range of biological functions in the heart.
Loss of Pdlim7 in mice causes unique hypercoagulopathy
The systemic, pre-mortem thrombi observed in the non-surviving Pdlim7 mutant mice and decreased tail bleed time in adult Pdlim7-/- mice provides the first indication that Pdlim7 may function in regulating hemostasis. A predisposition to thrombosis has been reported to include three major factors: damage to the endothelium, abnormal blood flow, and/or aberrant hemostatic properties of the blood. We detected Pdlim7 proteins co-localized with F-actin in the vascular smooth muscle, but not the endothelium, suggesting that the hypercoagulopathy in Pdlim7 mutant mice does not arise from primary defects in the endothelial layer. However, other PDZ-LIM proteins have been reported to function in the maintenance of muscle contractility [4,5,8,55], thus it is plausible that Pdlim7 could also function in smooth muscle contraction, and thereby influence blood flow. We report significant mortality of Pdlim7 mutant mice shortly after birth. One possible explanation for this is that the stress endured by pups during labor and delivery causes an abnormal constriction of the blood vessels resulting in reduced blood flow, and triggering clot formation in the predisposed Pdlim7 mutant mice. In a similar manner, smooth muscle contraction in the small muscular arteries of the tail could also explain the significantly decreased tail bleed time observed in adult Pdlim7-/- mice. In the context of functional redundancy, it is worthwhile noting that, to our knowledge, Pdlim7 and Pdlim3 are the only PDZ-LIM family members with documented expression in mammalian smooth muscle [56], suggesting a lower likelihood of compensatory function in this muscle type. Future studies with Pdlim7-/- bone marrow transplants into WT marrow-ablated mice will clarify whether there is a primary vessel issue or if a blood-specific defect has to be considered.
Platelets are also a major player in coagulation. Upon vessel injury, they become activated and rapidly reorganize their actin cytoskeleton to adhere to the site of endothelial damage, triggering the formation of a fibrin-rich plug to prevent further blood loss [57,58]. The decreased bleeding time, but normal liver function and plasma coagulation factors in Pdlim7 -/- survivors suggest that the etiology of the hypercoagulopathy may also be platelet-derived. Previous studies have shown that the PDZ-LIM protein Pdlim1 associates with actin stress fibers during shape change and spreading of human platelets [10]. A recent report by Gupta et al. revealed that lack of functional Pdlim1 in mice leads to marked hyperactivity of the collagen-glycoprotein VI (GPVI) signaling pathway in platelets, resulting in a prothrombotic phenotype [11]. While Gupta et al. found no other Pdlim genes expressed in murine platelets, we find Pdlim7-specific transcripts in developing and mature platelets, and demonstrate by Western-blot and immunocytochemistry that Pdlim7 proteins are dynamically distributed in platelets. Moreover, normal platelet counts in Pdlim7 mutant mice indicate that platelet formation and survival are likely not affected, which supports the notion that the origin of the thrombi in Pdlim7 mutant mice could be caused by abnormal platelet function. In light of Pdlim1’s role in platelet signaling, the analysis of platelet function in Pdlim7 mutant mice is of great interest, and these investigations are currently ongoing in our laboratory.
The global inactivation of the Pdlim7 gene in the mouse presented an unexpected, but unique hypercoagulopathy phenotype. While Pdlim7 mutations in humans have not been reported, these new findings suggest that future studies investigating the clinical relevance of Pdlim7 may improve our understanding of the molecular basis of cardiac function and hemostasis in humans.
Supporting Information
Pdlim7-deficient embryos do not reveal significant morphological defects during atrioventricular valve formation. Embryos were sectioned in sagittal and transverse orientations and stained with Alcian blue/nuclear fast red. At E11.5, the developing AV cushions of Pdlim7-/- (B; n=5) appeared relatively similar to WT controls (A; n=3). At E13.5 and E15.5, the differentiating AV valves of Pdlim7-/- embryos (D; n=5 and F; n=5) were also similar to WT controls (n=4, C and n=3, E). Scale bar = 100µm (A-B) and 200µm (C-F). A = atrium; LV = left ventricle; MV = mitral valve; OFT = outflow tract; TV = tricuspid valve.
(TIF)
Pdlim7-/- embryos undergo endocardial EMT and exhibit a normal Bmp2 expression pattern at the AV canal. Atrioventricular cushion explants from WT (A-B; n=3) and Pdlim7 -/- (C-D; n=3) embryos cultured for 48 hours on collagen gels. Mesenchymal cells visualized at the gel surface (0µm, A, C) and at a depth of 40µm into the gel (B, D, arrows) did not show differences in distribution. Whole-mount in situ hybridization of Bmp2 at E9.5 and E10.5 in WT (E, G; n=5 and 4, respectively) and Pdlim7 -/- embryos (F, H; n=3 and 4, respectively) demonstrates similar expression patterns in Pdlim7 mutant hearts. qRT-PCR array analysis of AV canals of E10.5 Pdlim7 -/- embryos reveals normal expression of several genes important for endocardial EMT compared to WT controls (I). A = atrium, LV = left ventricle, Myo = myocardium.
(TIF)
Adult Pdlim7-/- mice exhibit blood clots attached to the atrioventricular valves. In some cases, prior to perfusion, blood clots were observed attached to the mitral valve in 3-month old Pdlim7 -/- adult mice (asterisk and arrowhead, B-B’), but not in WT littermates (A). Scale bar = 200 µm (A-B), 20 µm (B’).
(TIF)
Three-dimensional reconstructions of adult WT and Pdlim7-/- atrioventricular valves. Serial sections of adult hearts stained with either Masson’s trichrome or Alcian blue to distinguish valve tissue were aligned to extract the 3D voxels used to compute a 3D reconstruction of the cardiac valves.
(MOV)
Description of the materials and methods for the supporting information.
(PDF)
Acknowledgments
We thank Mona Cornwell and Elise V. Allender and Drs. Yukiko Sugi, Robert Hinton, and Shekhar Mayanil for expert technical assistance. We are grateful to Drs. Roger Markwald, Yukiko Sugi, Robert Hinton, John Crispino, Hau Kwaan, and Gilbert White II for inspiring discussions. In particular, we would like to thank Drs. Robert Dettman and Michael Klüppel for constructive criticism and reading of the manuscript, and all members of the Simon Laboratory for helpful discussions.
Funding Statement
This work was supported by the National Heart, Lung, and Blood Institute (Grant F31HL090031), the Children’s Research Fund (to J. Krcmery), the Lurie Children’s Hospital of Chicago Research Center Pilot Grant and The Family of Michael P. Polsky (to H.G. Simon). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Krcmery J, Camarata T, Kulisz A, Simon HG (2010) Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development. Bioessays 32: 100-108. doi: 10.1002/bies.200900148. PubMed: 20091751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. te Velthuis AJ, Bagowski CP (2007) PDZ and LIM domain-encoding genes: molecular interactions and their role in development. ScientificWorldJournal 7: 1470-1492. doi: 10.1100/tsw.2007.232. PubMed: 17767364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Camarata T, Snyder D, Schwend T, Klosowiak J, Holtrup B et al. (2010b) Pdlim7 is required for maintenance of the mesenchymal/epidermal Fgf signaling feedback loop during zebrafish pectoral fin development. BMC Dev Biol 10: 104. doi: 10.1186/1471-213X-10-104. PubMed: 20950450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pashmforoush M, Pomiès P, Peterson KL, Kubalak S, Ross J Jr. et al. (2001) Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat Med 7: 591-597. doi: 10.1038/87920. PubMed: 11329061. [DOI] [PubMed] [Google Scholar]
- 5. Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, et al. (2001) Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J Cell Biol 155: 605-612 [DOI] [PMC free article] [PubMed]
- 6. van der Meer DL, Marques IJ, Leito JT, Besser J, Bakkers J et al. (2006) Zebrafish cypher is important for somite formation and heart development. Dev Biol 299: 356-372. doi: 10.1016/j.ydbio.2006.07.032. PubMed: 16982050. [DOI] [PubMed] [Google Scholar]
- 7. Ott EB, van den Akker NM, Sakalis PA, Gittenberger-de Groot AC, Te Velthuis AJ et al. (2008) The lim domain only protein 7 is important in zebrafish heart development. Dev Dyn 237: 3940-3952. doi: 10.1002/dvdy.21807. PubMed: 19035355. [DOI] [PubMed] [Google Scholar]
- 8. Cheng H, Kimura K, Peter AK, Cui L, Ouyang K et al. (2010) Loss of enigma homolog protein results in dilated cardiomyopathy. Circ Res 107: 348-356. doi: 10.1161/CIRCRESAHA.110.218735. PubMed: 20538684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jani K, Schöck F (2007) Zasp is required for the assembly of functional integrin adhesion sites. J Cell Biol 179: 1583-1597. doi: 10.1083/jcb.200707045. PubMed: 18166658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bauer K, Kratzer M, Otte M, de Quintana KL, Hagmann J et al. (2000) Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 96: 4236-4245. PubMed: 11110697. [PubMed] [Google Scholar]
- 11. Gupta S, Braun A, Morowski M, Premsler T, Bender M et al. (2012) CLP36 is a negative regulator of glycoprotein VI signaling in platelets. Circ Res 111: 1410-1420. doi: 10.1161/CIRCRESAHA.112.264754. PubMed: 22955732. [DOI] [PubMed] [Google Scholar]
- 12. Krause A, Zacharias W, Camarata T, Linkhart B, Law E et al. (2004) Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol 273: 106-120. doi: 10.1016/j.ydbio.2004.05.024. PubMed: 15302601. [DOI] [PubMed] [Google Scholar]
- 13. Camarata T, Bimber B, Kulisz A, Chew TL, Yeung J et al. (2006) LMP4 regulates Tbx5 protein subcellular localization and activity. J Cell Biol 174: 339-348. doi: 10.1083/jcb.200511109. PubMed: 16880269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Camarata T, Krcmery J, Snyder D, Park S, Topczewski J et al. (2010a) Pdlim7 (LMP4) regulation of Tbx5 specifies zebrafish heart atrio-ventricular boundary and valve formation. Dev Biol 337: 233-245. doi: 10.1016/j.ydbio.2009.10.039. PubMed: 19895804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C et al. (1998) Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392: 608-611. doi: 10.1038/33423. PubMed: 9560157. [DOI] [PubMed] [Google Scholar]
- 16. Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG et al. (1997) Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci U S A 94: 3789-3794. doi: 10.1073/pnas.94.8.3789. PubMed: 9108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN et al. (2011) Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood 118: e101-e111. doi: 10.1182/blood-2011-03-339705. PubMed: 21596849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Franco D, de Boer PA, de Gier-de Vries C, Lamers WH, Moorman AF (2001) Methods on in situ hybridization, immunohistochemistry and beta-galactosidase reporter gene detection. Eur J Morphol 39: 169-191. doi: 10.1076/ejom.39.3.0169. PubMed: 11910536. [DOI] [PubMed] [Google Scholar]
- 19. de Lange FJ, Moorman AF, Anderson RH, Männer J, Soufan AT et al. (2004) Lineage and morphogenetic analysis of the cardiac valves. Circ Res 95: 645-654. doi: 10.1161/01.RES.0000141429.13560.cb. PubMed: 15297379. [DOI] [PubMed] [Google Scholar]
- 20. Bimber B, Dettman RW, Simon HG (2007) Differential regulation of Tbx5 protein expression and sub-cellular localization during heart development. Dev Biol 302: 230-242. doi: 10.1016/j.ydbio.2006.09.023. PubMed: 17045582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hinton RB Jr., Alfieri CM, Witt SA, Glascock BJ, Khoury PR et al. (2008) Mouse heart valve structure and function: echocardiographic and morphometric analyses from the fetus through the aged adult. Am J Physiol Heart Circ Physiol 294: H2480-H2488. doi: 10.1152/ajpheart.91431.2007. PubMed: 18390820. [DOI] [PubMed] [Google Scholar]
- 22. Tei C (1995) New non-invasive index for combined systolic and diastolic ventricular function. J Cardiol 26: 135-136. PubMed: 7674144. [PubMed] [Google Scholar]
- 23. Adamo CM, Dai DF, Percival JM, Minami E, Willis MS et al. (2010) Sildenafil reverses cardiac dysfunction in the mdx mouse model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 107: 19079-19083. doi: 10.1073/pnas.1013077107. PubMed: 20956307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Broberg CS, Pantely GA, Barber BJ, Mack GK, Lee K et al. (2003) Validation of the myocardial performance index by echocardiography in mice: a noninvasive measure of left ventricular function. J Am Soc Echocardiogr 16: 814-823. doi: 10.1067/S0894-7317(03)00399-7. PubMed: 12878990. [DOI] [PubMed] [Google Scholar]
- 25. Jujo K, Hamada H, Iwakura A, Thorne T, Sekiguchi H et al. (2010) CXCR4 blockade augments bone marrow progenitor cell recruitment to the neovasculature and reduces mortality after myocardial infarction. Proc Natl Acad Sci U S A 107: 11008-11013. doi: 10.1073/pnas.0914248107. PubMed: 20534467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chrzanowska-Wodnicka M, Smyth SS, Schoenwaelder SM, Fischer TH, White GC 2nd (2005) Rap1b is required for normal platelet function and hemostasis in mice. J Clin Invest 115: 680-687. doi: 10.1172/JCI200522973. PubMed: 15696195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davidson DC, Hirschman MP, Spinelli SL, Morrell CN, Schifitto G et al. (2011) Antiplatelet activity of valproic acid contributes to decreased soluble CD40 ligand production in HIV type 1-infected individuals. J Immunol 186: 584-591. doi: 10.4049/jimmunol.1001911. PubMed: 21115729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eisenberg LM, Markwald RR (1995) Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res 77: 1-6. doi: 10.1161/01.RES.77.1.1. PubMed: 7788867. [DOI] [PubMed] [Google Scholar]
- 29. Kinsella MG, Fitzharris TP (1980) Origin of cushion tissue in the developing chick heart: cinematographic recordings of in situ formation. Science 207: 1359-1360. doi: 10.1126/science.7355294. PubMed: 7355294. [DOI] [PubMed] [Google Scholar]
- 30. Krug EL, Runyan RB, Markwald RR (1985) Protein extracts from early embryonic hearts initiate cardiac endothelial cytodifferentiation. Dev Biol 112: 414-426. doi: 10.1016/0012-1606(85)90414-2. PubMed: 3935503. [DOI] [PubMed] [Google Scholar]
- 31. Markwald RR, Fitzharris TP, Manasek FJ (1977) Structural development of endocardial cushions. Am J Anat 148: 85-119. doi: 10.1002/aja.1001480108. PubMed: 842477. [DOI] [PubMed] [Google Scholar]
- 32. Person AD, Klewer SE, Runyan RB (2005) Cell biology of cardiac cushion development. Int Rev Cytol 243: 287-335. doi: 10.1016/S0074-7696(05)43005-3. PubMed: 15797462. [DOI] [PubMed] [Google Scholar]
- 33. Kruithof BP, Duim SN, Moerkamp AT, Goumans MJ (2012) TGFbeta and BMP signaling in cardiac cushion formation: lessons from mice and chicken. Differentiation 84: 89-102. doi: 10.1016/j.diff.2012.04.003. PubMed: 22656450. [DOI] [PubMed] [Google Scholar]
- 34. Aikawa E, Whittaker P, Farber M, Mendelson K, Padera RF et al. (2006) Human semilunar cardiac valve remodeling by activated cells from fetus to adult: implications for postnatal adaptation, pathology, and tissue engineering. Circulation 113: 1344-1352. doi: 10.1161/CIRCULATIONAHA.105.591768. PubMed: 16534030. [DOI] [PubMed] [Google Scholar]
- 35. Goldsmith EC, Hoffman A, Morales MO, Potts JD, Price RL et al. (2004) Organization of fibroblasts in the heart. Dev Dyn 230: 787-794. doi: 10.1002/dvdy.20095. PubMed: 15254913. [DOI] [PubMed] [Google Scholar]
- 36. Kruithof BP, Krawitz SA, Gaussin V (2007) Atrioventricular valve development during late embryonic and postnatal stages involves condensation and extracellular matrix remodeling. Dev Biol 302: 208-217. doi: 10.1016/j.ydbio.2006.09.024. PubMed: 17054936. [DOI] [PubMed] [Google Scholar]
- 37. Lincoln J, Alfieri CM, Yutzey KE (2004) Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn 230: 239-250. doi: 10.1002/dvdy.20051. PubMed: 15162503. [DOI] [PubMed] [Google Scholar]
- 38. Hinton RB Jr., Lincoln J, Deutsch GH, Osinska H, Manning PB et al. (2006) Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res 98: 1431-1438. doi: 10.1161/01.RES.0000224114.65109.4e. PubMed: 16645142. [DOI] [PubMed] [Google Scholar]
- 39. Runyan RB, Markwald RR (1983) Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol 95: 108-114. doi: 10.1016/0012-1606(83)90010-6. PubMed: 6825921. [DOI] [PubMed] [Google Scholar]
- 40. Ma L, Lu MF, Schwartz RJ, Martin JF (2005) Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132: 5601-5611. doi: 10.1242/dev.02156. PubMed: 16314491. [DOI] [PubMed] [Google Scholar]
- 41. Sugi Y, Yamamura H, Okagawa H, Markwald RR (2004) Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol 269: 505-518. doi: 10.1016/j.ydbio.2004.01.045. PubMed: 15110716. [DOI] [PubMed] [Google Scholar]
- 42. Tei C, Ling LH, Hodge DO, Bailey KR, Oh JK et al. (1995) New index of combined systolic and diastolic myocardial performance: a simple and reproducible measure of cardiac function--a study in normals and dilated cardiomyopathy. J Cardiol 26: 357-366. PubMed: 8558414. [PubMed] [Google Scholar]
- 43. Bruch C, Schmermund A, Marin D, Katz M, Bartel T et al. (2000) Tei-index in patients with mild-to-moderate congestive heart failure. Eur Heart J 21: 1888-1895. doi: 10.1053/euhj.2000.2246. PubMed: 11052862. [DOI] [PubMed] [Google Scholar]
- 44. Ward AD, Hamarneh G, Ashry R, Schweitzer ME (2007) 3D shape analysis of the supraspinatus muscle: a clinical study of the relationship between shape and pathology. Acad Radiol 14: 1229-1241. doi: 10.1016/j.acra.2007.06.014. PubMed: 17889340. [DOI] [PubMed] [Google Scholar]
- 45. Cui J, Eitzman DT, Westrick RJ, Christie PD, Xu ZJ et al. (2000) Spontaneous thrombosis in mice carrying the factor V Leiden mutation. Blood 96: 4222-4226. PubMed: 11110695. [PubMed] [Google Scholar]
- 46. Bugge TH, Flick MJ, Daugherty CC, Degen JL (1995) Plasminogen deficiency causes severe thrombosis but is compatible with development and reproduction. Genes Dev 9: 794-807. doi: 10.1101/gad.9.7.794. PubMed: 7705657. [DOI] [PubMed] [Google Scholar]
- 47. Lay AJ, Liang Z, Rosen ED, Castellino FJ (2005) Mice with a severe deficiency in protein C display prothrombotic and proinflammatory phenotypes and compromised maternal reproductive capabilities. J Clin Invest 115: 1552-1561. doi: 10.1172/JCI24030. PubMed: 15902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kujovich JL (2005) Hemostatic defects in end stage liver disease. Crit Care Clin 21: 563-587. doi: 10.1016/j.ccc.2005.03.002. PubMed: 15992673. [DOI] [PubMed] [Google Scholar]
- 49. Northup PG, Sundaram V, Fallon MB, Reddy KR, Balogun RA et al. (2008) Hypercoagulation and thrombophilia in liver disease. J Thromb Haemost 6: 2-9. PubMed: 17892532. [DOI] [PubMed] [Google Scholar]
- 50. Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY (2008) High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development 135: 1179-1187. doi: 10.1242/dev.010694. PubMed: 18272595. [DOI] [PubMed] [Google Scholar]
- 51. Staudt D, Stainier D (2012) Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu Rev Genet 46: 397-418. doi: 10.1146/annurev-genet-110711-155646. PubMed: 22974299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Armstrong EJ, Bischoff J (2004) Heart valve development: endothelial cell signaling and differentiation. Circ Res 95: 459-470. doi: 10.1161/01.RES.0000141146.95728.da. PubMed: 15345668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM et al. (2012) Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol 366: 111-124. doi: 10.1016/j.ydbio.2012.04.020. PubMed: 22546693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Passier R, Richardson JA, Olson EN (2000) Oracle, a novel PDZ-LIM domain protein expressed in heart and skeletal muscle. Mech Dev 92: 277-284. doi: 10.1016/S0925-4773(99)00330-5. PubMed: 10727866. [DOI] [PubMed] [Google Scholar]
- 55. Zheng M, Cheng H, Li X, Zhang J, Cui L et al. (2009) Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum Mol Genet 18: 701-713. PubMed: 19028670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pomiès P, Macalma T, Beckerle MC (1999) Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem 274: 29242-29250. doi: 10.1074/jbc.274.41.29242. PubMed: 10506181. [DOI] [PubMed] [Google Scholar]
- 57. Hartwig JH, Barkalow K, Azim A, Italiano J (1999) The elegant platelet: signals controlling actin assembly. Thromb Haemost 82: 392-398. PubMed: 10605729. [PubMed] [Google Scholar]
- 58. Bearer EL, Prakash JM, Li Z (2002) Actin dynamics in platelets. Int Rev Cytol 217: 137-182. doi: 10.1016/S0074-7696(02)17014-8. PubMed: 12019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pdlim7-deficient embryos do not reveal significant morphological defects during atrioventricular valve formation. Embryos were sectioned in sagittal and transverse orientations and stained with Alcian blue/nuclear fast red. At E11.5, the developing AV cushions of Pdlim7-/- (B; n=5) appeared relatively similar to WT controls (A; n=3). At E13.5 and E15.5, the differentiating AV valves of Pdlim7-/- embryos (D; n=5 and F; n=5) were also similar to WT controls (n=4, C and n=3, E). Scale bar = 100µm (A-B) and 200µm (C-F). A = atrium; LV = left ventricle; MV = mitral valve; OFT = outflow tract; TV = tricuspid valve.
(TIF)
Pdlim7-/- embryos undergo endocardial EMT and exhibit a normal Bmp2 expression pattern at the AV canal. Atrioventricular cushion explants from WT (A-B; n=3) and Pdlim7 -/- (C-D; n=3) embryos cultured for 48 hours on collagen gels. Mesenchymal cells visualized at the gel surface (0µm, A, C) and at a depth of 40µm into the gel (B, D, arrows) did not show differences in distribution. Whole-mount in situ hybridization of Bmp2 at E9.5 and E10.5 in WT (E, G; n=5 and 4, respectively) and Pdlim7 -/- embryos (F, H; n=3 and 4, respectively) demonstrates similar expression patterns in Pdlim7 mutant hearts. qRT-PCR array analysis of AV canals of E10.5 Pdlim7 -/- embryos reveals normal expression of several genes important for endocardial EMT compared to WT controls (I). A = atrium, LV = left ventricle, Myo = myocardium.
(TIF)
Adult Pdlim7-/- mice exhibit blood clots attached to the atrioventricular valves. In some cases, prior to perfusion, blood clots were observed attached to the mitral valve in 3-month old Pdlim7 -/- adult mice (asterisk and arrowhead, B-B’), but not in WT littermates (A). Scale bar = 200 µm (A-B), 20 µm (B’).
(TIF)
Three-dimensional reconstructions of adult WT and Pdlim7-/- atrioventricular valves. Serial sections of adult hearts stained with either Masson’s trichrome or Alcian blue to distinguish valve tissue were aligned to extract the 3D voxels used to compute a 3D reconstruction of the cardiac valves.
(MOV)
Description of the materials and methods for the supporting information.
(PDF)