Abstract
The inherited bone marrow failure syndromes (IBMFS) are a set of clinically related yet heterogeneous disorders in which at least one hematopoietic cell lineage is significantly reduced. Many of the IBMFS have notably increased cancer risks as well as other physical findings. Highly penetrant germline mutations in key pathways, such as DNA repair, telomere biology, or ribosomal biogenesis are causative of Fanconi anemia (FA), dyskeratosis congenita (DC) and Diamond-Blackfan anemia (DBA), respectively.
Next-generation sequencing (NGS) generally refers to high-throughput, large-scale sequencing technologies and is being used more frequently to understand disease etiology. In the IBMFS, NGS has facilitated the discovery of germline mutations that cause thombocytopenia absent radii syndrome, a subset of DC and DBA, and other uncharacterized, but related, disorders. Panels of large numbers of genes are being used to molecularly characterize patients with IBMFS, such as FA and DBA. NGS is also accelerating the discovery of the genetic etiology of previously unclassified IBMFS. In this review, we will highlight recent studies that have employed NGS to ascertain the genetic etiology of IBMFS, namely FA, DC, DBA and TAR and discuss the translational utility of these findings.
INTRODUCTION
The inherited bone marrow failure syndromes (IBMFS) are a set of clinically related yet heterogeneous disorders in which at least one hematopoietic cell lineage is significantly reduced in number. Certain IBMFS, such as Fanconi anemia (FA), dyskeratosis congenita (DC), and Diamond-Blackfan anemia (DBA), are associated with increased risk of solid tumors and hematopoietic malignancies (1). The genetic etiology of the IBMFS includes germline mutations in several key biological processes (e.g., DNA repair, telomere biology, or ribosomal biogenesis) (1). Highly penetrant germline mutations have been identified that can explain approximately 95% of FA and 95% of Shwachman Diamond syndrome (SDS) (2). In contrast, the genetic cause is known in only about one-half of patients with Diamond-Blackfan anemia (DBA) and about 70% of patients with dyskeratosis congenita (DC) (3).
Prior to the advent of next generation sequencing (NGS) technology, identification of the genetic etiology of the IBMFS and other inherited disorders was conducted primarily through a combination of linkage studies and candidate gene sequencing. While successful, these approaches are limited because linkage studies require large families with multiple affected individuals; and candidate gene studies can consume significant resources by sequencing one gene at a time and success is predicated on the good fortune of picking the right gene in the right set of patients. The more expansive survey of the genome using NGS technologies has also led to rapid advances in understanding the structure of the human genome; and of mutations or single nucleotide polymorphisms (SNPs) associated with both rare and common diseases (4, 5).
NGS, also known as massively parallel sequencing or second generation sequencing refers to high-throughput, large-scale sequencing technologies (4, 6, 7). Different NGS approaches include whole genome, whole exome (e.g., targeting of all known exons in the reference data base), and/or sequencing of large panels of specific genes. In addition to sequence analysis and evaluation of single nucleotide substitutions, NGS also enables characterization of copy number variants (CNVs), insertions/deletions (indels), and structural rearrangements. However, the error rate for NGS is higher than SNP genotyping and most variants require validation by another sequencing method, such as traditional Sanger sequencing. NGS of RNA transcripts (e.g., RNA-Seq) allows for the quantification of transcript levels and the RNA sequence information that form the basis of what is now called the transcriptome (6). Chromosomal immunoprecipitation (ChIP) followed by DNA sequencing (ChIP-Seq) is an NGS technique that allows mapping of specific transcription factors and histone modifications to their genomic location (8). Other NGS methods include the identification and quantification of methylated DNA sites, histone-bound DNA, and protein-RNA interactions (7).
NGS has been applied to the discovery of IBMFS mutations. For example, whole exome sequencing (WES) has led to the discovery of new telomere biology genes associated with DC (e.g., mutations in the CTC1 and RTEL1genes) (9-11). The discovery of GATA1 mutations in DBA by WES has led to a novel connection between DBA and the distinct disorder of X-linked dyserythropoietic anemia and thrombocytopenia (12, 13). More recently, comparative genomic hybridization (CGH) identified RPL15 as a novel gene causing DBA (14). Complimentary genomic approaches, including CGH, exome sequencing, and targeted sequencing of the non-deleted allele, were required for the identification of mutations that explain a subset of the thromobocytopenia absent radii syndrome (TAR) (15, 16). Even in a complex syndrome such as FA, for which the genetic cause can usually be identified, new genomics approaches combining WES, CGH, and RNA-Seq are being used to develop a more efficient and cost effective approach to new patient characterization (17-22). This review will highlight recent advances in IBMFS genetics based on NGS and consider the role of genomics approaches in future studies of these disorders.
FANCONI ANEMIA (FA)
Clinical features and diagnosis of FA
FA is a chromosomal instability disorder caused by germline mutations resulting in defective DNA damage response. It is associated with a myriad of clinical features, including congenital anomalies, progressive bone marrow failure (BMF), and cancer predisposition (1, 23)(Table 1). Radial bone and thumb abnormalities, short stature, skin hyperpigmentation and/or café au lait macules are the most commonly reported features, although other organ systems may also be involved (24). Importantly, less than two-thirds of FA patients present with pathognomonic physical features; BMF or cancer can be the primary presenting sign in these individuals. The diagnostic test for FA is the detection of increased chromosomal breakage in cells cultured with a clastogen, such as diepoxybutane (DEB) or mitomycin C (MMC) (1).
TABLE 1. Clinical features of selected inherited bone marrow failure syndromes.
All disorders may present with or without family history or as a result of de novo mutations in the proband. Patients with these disorders may present with some, but not necessarily all of the features listed. Clinically silent carriers are possible.
| Disorder | Clinical features | Laboratory findings | Biologic al pathway |
Inheritance: known genes | Associated cancers |
|---|---|---|---|---|---|
| Fanconi Anemia | Radial ray anomalies, short stature, microcephaly, café au lait spots, structural renal anomalies, BMF |
Increased chromosome breakage in clastogenic assay, macrocytosis, elevated HbF |
DNA repair |
XLR: FANCB AR: FANCA, FANCC, FANCD1/ BRCA2, FANCD2, FANCE, FANCF, FANCG/ XRCC9, FANCI, FANCJ/BACH1/ BRIP1, FANCL, FANCM, FANCN/PALB2, FANCO/RAD51C FANCP/SLX4, FANCQ (ERCC4) |
Squamous cell cancers of the head, neck, and anogenital region; MDS, AML |
| Dyskeratosis Congenita |
Triad of nail dysplasia, lacy skin pigmentation abnormalities, and oral leukoplakia; BMF, pulmonary fibrosis, stenosis of esophagus, lacrimal ducts, or urethra, liver cirrhosis or fibrosis |
Very short telomeres, macrocytosis, elevated HbF |
Telomer e biology |
XLR: DKC1 AD: TERT, TERC, TINF2, RTEL1 AR: NOP10, NHP2, WRAP53, RTEL1, TERT, CTC1 |
MDS, AML, Head and neck squamous cell cancers |
| Hoyeraal Hreidarsson Syndrome (DC variant) |
Features of DC and developmental delay,intra-uterine growth retardation, immunodeficiency, enteropathy |
Very short telomeres, macrocytosis, elevated HbF |
Telomer e biology |
XLR:DKC1, AD: TINF2,RTEL1 AR: TERT,RTEL1 |
|
| Revesz Syndrome (DC variant) |
Features of DC and bilateral exudative retinopathy, IUGR, intracranial calcifications, developmental delay, fine sparse hair |
Very short telomeres, macrocytosis, elevated HbF |
Telomer e biology |
AD: TINF2 | |
| Coat’s plus or Cerebroretinal microangiopathy with calcifications and cysts (CRMCC) |
Bilateral exudative retinopathy, retinal telangiectasias, growth retardation, intracranial calcifications, neurological symptoms, bone abnormalities, gastrointestinal vascular ectasias, dystrophic nails, sparse or graying hair, and anemia |
Very short telomeres, macrocytosis, elevated HbF |
Telomer e biology |
AR: CTC1 | |
| Diamond-Blackfan Anemia |
Severe anemia, typically in infancy, ~25% with birth defects |
Elevated RBC ADA, macrocytosis, elevated HbF |
Ribosom e biogenes is |
AD: RPS19, RPS17, RPS24, RPL35A, RPL5, RPL11,RPS7, RPS26, RPS10 XLR: GATA1 |
MDS, AML, osteosarcoma, colon cancer |
| Thrombocytopenia absent radii |
Absent radii with thumbs present, petechiae or hemorrhage in infancy |
Thrombocytopenia in infancy/childhood, usually improves with time |
Not yet elucidate d |
AR: 1q21.1 deletion and RBM8A variant |
AML, ALL |
Abbreviations: DC, dyskeratosis congenita; MDS, myelodysplastic syndrome; AML, acute myeloid leukemia; ALL, acute lymphocytic leukemia; AD, autosomal dominant; AR, autosomal recessive; XLR, X-linked recessive; RBC, red blood cell; ADA, adenosine deaminase; HbF, fetal hemoglobin; BMF, bone marrow failure
BMF in FA typically occurs in the first decade of life and may initially present as a single or bilineage cytopenia. Patients may also have macrocytosis and elevated fetal hemoglobin levels (1). The severity of BMF may progress and require medical intervention such as regular blood product transfusions or hematopoietic stem cell transplantation (HSCT). Patients with severe BMF unable to undergo HSCT, due to medical or personal reasons, have been treated with androgens such as oxymetholone with an estimated 50% response rate (1). FA patients who are treated with androgens are at an increased risk of developing liver tumors (25).
Defective DNA repair confers a propensity for specific cancers in patients with FA. In addition to BMF, patients with FA are at increased risk of myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) (26). FA also confers a higher risk of developing solid tumors, particularly squamous cell cancers (SCC) of the head, neck, skin, GI tract and genital tract. Patients with FA subtype D1, caused by biallelic mutations in BRCA2, have an extremely high risk of early onset AML, brain tumors, or Wilms tumor (26, 27). These risk factors make the correct diagnosis of FA essential for proper management as well as genetic counseling.
Genetics and Pathophysiology of FA
All the known FA subtypes are caused by germline mutations in key components in the DNA repair pathway. They are inherited in an autosomal recessive (AR) pattern, except for subtype B, which is X-linked recessive (XLR). The first causative FA gene was discovered over 20 years ago, and there are now16 known FA subtypes (A, B, C, D1 (BRCA2), D2, E, F, G, I, J (BRIP1, BACH1), L, M, N (PALB2), O (RAD51C), P (SLX4), and Q (ERCC4)) (17, 28) (Figure 1). At least 95% of FA cases are caused by germline mutations in one of these 16 genes (2). In AR FA, the mutations are often biallelic and may include the combination of a single base substitution and a partial or full gene deletion (28). The heterozygote carrier frequency of FA is estimated to be 1 in 181 in the United States (29); it may be higher in populations with founder effects, such as in Ashkenazi Jewish populations that have an estimated carrier frequency of 1 in 89 for the IVS2(+4)A>T mutation in FANCC (30).
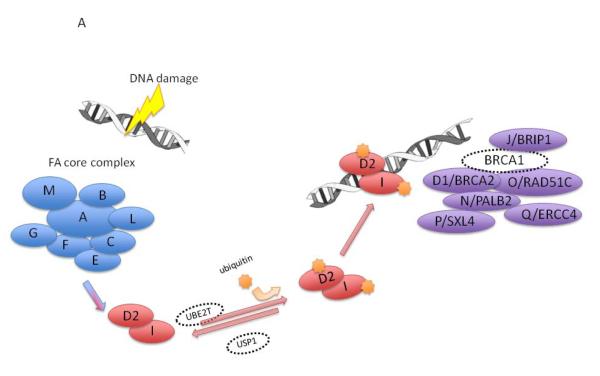
Figure 1. Fanconi anemia (FA) genetics and biologic pathway.
A)FA/BRCA DNA damage response pathway. Following DNA damage, the proteins represented by A, B, C, E, F, G, L, and M form the core complex which is required for ubiquitination of the I and D2 proteins, which are in turn, required for the downstream complex of D2-ubi, I-ubi, and D1/BRCA1, N/PALB2, and J/BACH1/BRIP1 to form foci for DNA repair. Only BRCA1 is not yet known to be a FA gene. Adapted from Shimamura and Alter Blood Reviews 2010;24(3):101-22
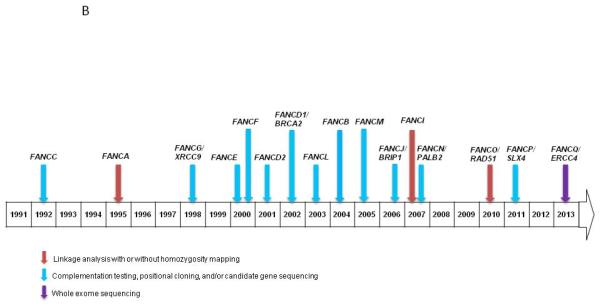
B) Approximate timeline and methods of gene discovery in FA
The genes mutated in FA encode proteins that work together to resolve DNA interstrand cross-links during cellular replication (Figure 1A). The FA protein core complex is a large nuclear E3 ubiquitin ligase complex consisting of FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM. The core complex interacts with FANCD2 and FANCI, which then interact with FANCD1 (known as BRCA2), FANCN (known as PALB2), FANCJ (known as BRIP1 or BACH1), FANCP (known as SLX4) and FANCO (known as RAD51C), as well as NBS1, BRCA1, and FAN1 to preserve genome integrity (28). Germline mutations in any of the FA genes result in markedly reduced or absent protein function and deficient DNA repair.
NGS in FA
The first gene associated with FA, FANCA, was discovered by linkage analysis in 1995 (31) (Figure 1A). Subsequent mutation discovery efforts utilized a complementation testing approach in which defective cells were transfected in an attempt to rescue the chromosome breakage phenotype (32). The gene encoding the protein that results in the rescue can then be sequenced to identify causative mutations (2). However, complementation testing for FA is costly and time consuming. It requires a highly specialized technique that is not widely available. Genetic linkage and positional cloning have continued to be valuable in identifying mutations in FA-associated genes.
Although the genetic cause of FA is known in approximately 95% of patients, WES has accelerated the identification of mutations in the rare FA patients without a known genetic etiology. For example, ERCC4, the DNA-repair endonuclease XPF, was discovered as a cause of FA through WES in a patient with clinical FA (17). Notably ERCC4 mutations have previously been associated with xeroderma pigmentosum type F (XPF) or XFE progeroid syndrome (33, 34) but interestingly, in this case, the patient lacked the dermatologic manifestations of XPF or XFE. Homozygous truncating mutations in XRCC2, a gene critical for DNA repair by homologous recombination, have also been implicated as a cause of FA after autozygome analysis failed to reveal a genetic cause in a consanguineous family (35).
Targeted NGS has the potential to be cost-effective in identifying a patient’s FA-associated mutation in newly diagnosed individuals. For example, as proof of principle, Ameziane et al the sequenced germline DNA of 11 patients with known FA mutations on a custom NGS sequencing panel of eight FA genes (FANCA, FANCB, FANCC, FANCD1, FANCE, FANCG, FANCI and FANCN) (19). This approach identified not only the known mutations but a novel deletion. This study also noted that it is critical to carefully evaluate the potential confounding effect of pseudogenes, especially for FANCD2, and deletions in all FA-associated genes. This study also suggested that NGS is useful in uncovering somatic mosaicism by comparing different tissue types.
A multi-platform approach was recently used to successfully identify the genetic cause of FA in 27 patients (21). The combination of custom array CGH (aCGH) to detect deletions and duplications of FA genes, WES to evaluate mutations, and RNA-Seq to evaluate gene expression changes resulting from these mutations was shown to be an efficient for characterization of both the complementation group and the germline mutations.
WES has been used to successfully identify germline mutations in four patients with unknown complementation groups (18). Sanger sequencing confirmed the point mutations and small indels present in FANCD1, FANCD2, and FANCJ. Another study used WES to detect a rare, yet clinically significant germline mutation in SLX4/FANCP in a young adult patient clinically diagnosed with FA in early childhood and an elevated chromosome breakage with MMC (20).
DYSKERATOSIS CONGENITA (DC)
Clinical features and diagnosis of DC
DC is an IBMFS caused by germline defects in telomere biology and is diagnosed by the presence of the classic triad of nail dysplasia, lacy skin pigmentation, and oral leukoplakia (Table 1). Not all patients present with this diagnostic triad, but they are at high risk of trilineage BMF, pulmonary fibrosis, liver disease (cirrhosis and fibrosis), and malignancy. Additional medical problems may include avascular necrosis of the femoral or humeral heads, stenosis of the esophagus, urethra, or lacrimal ducts, and developmental delay (1, 36, 37). Patients with DC are at increased risk for many of the same cancers as patients with FA, namely, SCC of the head and neck, and anogenital region, MDS, and AML. The actuarial risk of cancer in patients with DC is 40% by 50 years of age, with an 1100-fold increased risk for tongue cancer, 2500-fold increased risk for MDS and 200-fold for AML (26, 38).
BMF can develop in up to 90% patients with DC and is often life-threatening (36, 39, 40). However, the rate of BMF in individuals who have a germline mutation in a DC-associated gene but lack clinical features at the time of evaluation (e.g., silent carriers) is not known. BMF can present as single or multi-lineage cytopenia that progress to severe BMF. Hematopoietic stem cell transplant (HSCT) is the only current modality for cure of BMF associated with DC. As in FA, treatment with androgen therapy is often considered for BMF in individuals who cannot, or do not wish to undergo HSCT (36, 37). Between 50-70% of DC patients treated with androgens respond and no longer be dependent on red blood cell and platelet transfusions (41, 42).
The clinical diagnosis of DC can be challenging due to phenotypic heterogeneity, variable age of onset of the mucocutaneous triad, and presence of several clinical variants of DC. Table 1 describes the known clinical variants of DC, namely Hoyeraal Hreidarsson syndrome (HH), Revesz syndrome (RS), and Coats plus disease or cerebroretinal microangiopathy with calcification and cysts (CRMCC). The unifying feature of this complex set of clinical problems is the presence of very short telomeres, the result of germline mutations in key telomere biology genes. Telomere length less than the first percentile for age in leukocyte subsets measured by flow cytometry with in situ hybridization (flow FISH) is highly sensitive and specific for DC (43, 44)
Genetics and Pathophysiology of DC
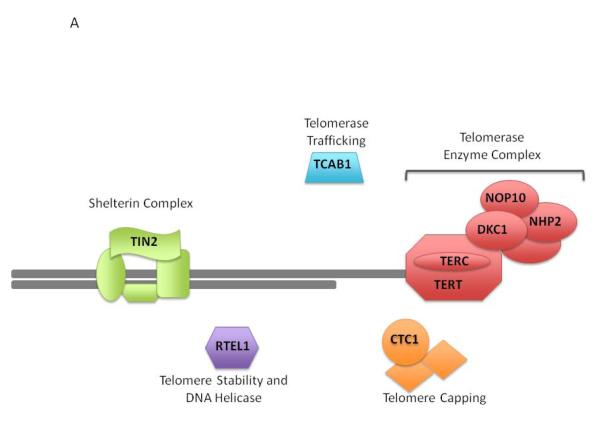
DC can be inherited in one of three forms, X-linked, autosomal dominant (AD), or AR. De novo germline mutations are also relatively frequent in DC and to date, about 70% DC patients have an identifiable germline mutation (36, 45). These mutations occur in genes responsible for the functioning and maintenance of telomeres (Figure 2A). Currently, there are nine known DC-associated genes (DKC1, TERT, TERC, TINF2, WRAP53, NOP10, NHP2, CTC1, and RTEL1) (36, 45).
Figure 2. Dyskeratosis Congenita genetics and biologic pathway.
A) Schematic of the telomere and functions of the proteins affected in dyskeratosis congenita and the related telomere biology disorders. Protein names are shown. Abbreviations: TCAB1, telomere Cajal body associated protein 1 (gene name: WRAP53); TIN2, TRF1-interacting nuclear factor 2 (TINF2); NOP10, NOP10 ribonucleoprotein (NOP10); NHP2, NHP2 ribonucleoprotein (NHP2); DKC1, dyskerin (DKC1); TERC, telomerase RNA component (TERC); TERT, telomerase (TERT); RTEL1, regulator of telomere elongation helicase 1 (RTEL1); CTC1, CTS telomere maintenance complex component 1 (CTC1)
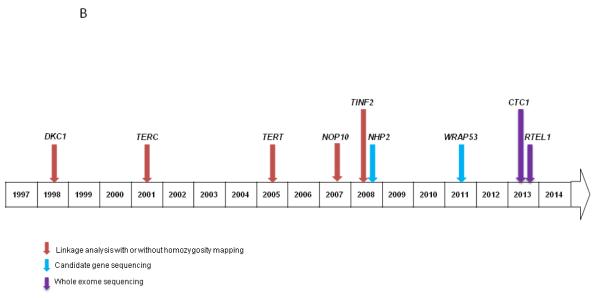
B) Approximate timeline and methods of gene discovery in DC
The first DC-associated gene, X-linked DKC1, was discovered by linkage analysis in 1998 (46) (Figure 2B). AD mutations in TERC, the RNA component of telomerase, were identified through linkage analysis of a large family (47). Subsequently, a combination of linkage (48) and candidate gene sequencing found mutations in TERT, as a cause of DC as well as in cases of isolated aplastic anemia and pulmonary fibrosis (49-52). These findings united the seemingly disparate diseases under the umbrella of DC-associated Telomere Biology Disorders (TBDs) (36, 39).
Linkage mapping was also used to identify AD mutations in TINF2, a key component of the shelterin telomere protection complex (53). Homozygosity mapping led to the discovery of AR NOP10 mutation in a consanguineous family with DC (54). That finding led investigators to follow-up with candidate gene sequencing of related genes and the detection of NHP2 mutations in two families (55). Candidate gene sequencing also led to the discovery of AR mutations in WRAP53 (TCAB1) as a cause of DC (56).
Lastly, mutations in two additional genes have been reported in DC, although, their connection with telomere biology is less straightforward. These include an intronic splice variant in Apollo (encoded by DCLRE1B) in a patient with HH and normal telomeres (57). On account of this finding, the role of Apollo in telomere biology is currently under investigation. Linkage analysis led to the identification of mutations in C16orf57, a gene with unknown function now called USB1 (58). C16orf57 mutations were reported in patients with DC and normal telomeres, but also in individuals with Rothmund Thomson syndrome and Poikiloderma with Neutropenia, suggesting an overlapping clinical spectrum (58).
NGS in DC
Prior to the use of NGS, the genetic cause of DC was known in less than one-half of patients (45). It is notable that mutational analyses have connected Coats plus (CRMCC) and DC based on WES discoveries that compound heterozygous mutations in the telomere capping protein encoded by CTC1 can cause either disorder (Table 1). Patients with these mutations have short telomeres and features that phenotypically overlapped with DC (9, 59, 60). Subsequently, candidate gene sequence analysis has led to the discovery of AR CTC1 mutations in a patient with DC (61).
Several groups have independently identified RTEL1 mutations using WES in families with DC (10, 11, 62). The RTEL1 protein regulates telomere length, may interact with PCNA (proliferating cell nuclear antigen), and also plays a role in DNA repair (10, 11). Most of the RTEL1 mutations appear to be AR, but AD mutations have been reported (10). It is estimated that the germline genetic cause of DC is now known in about 70% of families with DC due addition of CTC1 and RTEL1 to the list of DC-associated genes.
DIAMOND BLACKFAN ANEMIA (DBA)
Clinical features and diagnosis
DBA is a rare disorder of erythroid hypoplasia, characterized by macrocytic anemia, usually with normal WBC and platelet counts (Table 1). Patients with DBA are typically diagnosed at birth or within the first year of life (1); it is also associated with congenital anomalies, such as triphalangeal, bifid, or subluxed thumbs, or subtle flattening of the thenar eminence, with a normal radius. Genitoitourinary and heart defects, webbed neck, Klippel-Feil anomaly (fusion of cervical vertebrae), and Sprengel deformity (congenital asymmetric high scapula) have also been reported in DBA (3, 63). Like the other IBMFS, DBA is associated with an increased risk of certain malignancies, including MDS, AML, colon carcinoma, female genital cancers and osteosarcoma (64). The phenotypic spectrum of DBA is very broad, even within families. Some individuals with germline mutations may be silent carriers, or have only mild anemia whereas others are very severely affected. This observation suggests the importance of genetic modifiers, which have yet to be defined.
The diagnosis of DBA is primarily based upon the early onset, in infancy or childhood of persistent severe anemia with reticulocytopenia in the absence of other bone marrow abnormalities (1, 65). The bone marrow of patients with DBA shows erythroblastopenia with normal myeloid and megakaryocytic lineages. Overall, the bone marrow cellularity is usually normal or slightly reduced. An elevated erythrocyte adenosine deaminase (eADA) level in pretransfusion samples supports the diagnosis of DBA, whereas normal eADA levels do not exclude the diagnosis (66, 67).
Red blood cell transfusions are the primary treatment modality for severe anemia until the diagnosis is firmly established (1, 68). Oral corticosteroids may mitigate the need for regular transfusions in some DBA patients; however side effects must be carefully balanced with response (65). In those who do not respond to steroid therapy and require RBC transfusions, iron overload is a major clinical concern and should be treated early.
Genetics and Pathophysiology of DBA
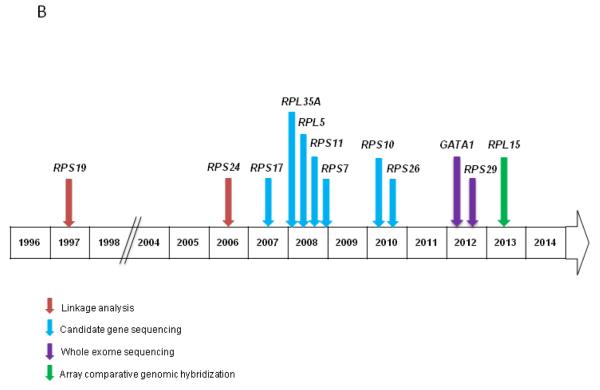
DBA is AD inherited disease and is usually caused by heterozygous germline mutations in genes encoding key components of small 40S or large 60S ribosomal subunits (Figure 3A) (65, 69). AD germline mutations in RPS19 were the first known genetic cause of DBA, providing a critical link between DBA and ribosomal biogenesis (70). RPS19 germline mutations account for approximately 25% of DBA cases. Although there are now nine DBA-associated genes (RPS19, RPS17, RPS24, RPS26, RPS10, RPS7, RPL35A, RPL5, and RPL11), at least three genes are suspected to be associate with DBA, RPL36, RPS15, and RPS27A (63), but approximately 50% of DBA patients have no known genetic mutation (3). De novo mutations and/or variable disease penetrance can also account for DBA in a subset of patients.
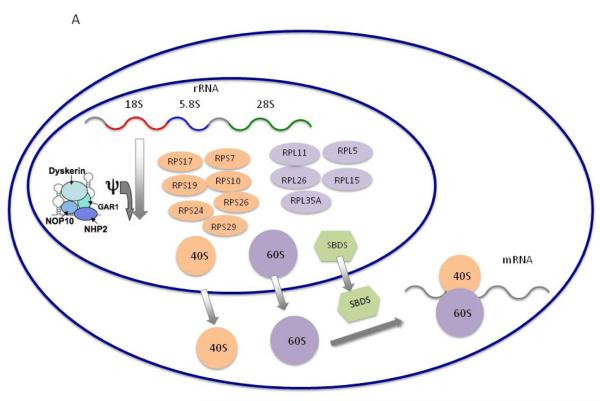
Figure 3. Schematic of the ribosomal biogenesis pathway associated with DBA.
A) Ribosomes consist of a small 40S subunit and a large 60S subunit and catalyze protein synthesis. Small and large subunits are composed of four RNA species and approximately 80 structurally distinct proteins. The DBA-associated proteins are in both the small 40S and large 60S ribosomal subunit. They are encoded by RPS19, RPS24, RPS1, RPS15, RPS27A, RPS10, RPS29 and RPS26 which belong to the small ribosomal subunit, and by RPL5, RPL11, RPL35A, RPL15 and RPL36 which are components of the large ribosomal subunit. The DKC1 gene encodes the dyskerin protein, which has been implicated in ribosomal RNA pseudouridylation (Φ). The SBDS protein appears to be involved in the joining of the 40S and 60S ribosomal subunits to form the mature 80S ribosome. AR mutations in SBDS, a key component of ribosomal assembly cause Shwachman-Diamond syndrome (SDS), a disorder of neutropenia and exocrine pancreatic insufficiency. SDS is not discussed herein because NGS studies have not been published in this disorder. Adapted from Shimamura and Alter Blood Reviews 2010;24(3):101-22
B) Approximate timeline and methods of gene discovery in DBA
Most DBA-associated germline mutations result in abnormal assembly of ribosomal proteins (Figure 3). While ribosome assembly is a highly regulated process, the reason these defects cause a specific defect in erythropoiesis is still not known. Aberrant ribosomal biogenesis due to germline ribosomal protein gene mutations activates cellular stress signaling pathways, such as p53 (71). This change in the balance of the p53 tumor suppressor pathway could disrupt cellular homeostasis and result in increased cancer risk, but further work is required to explain the underlying pathophysiology of the genetic mutations.
NGS in DBA
An NGS panel of 79 genes encoding ribosomal proteins, including the known DBA-associated genes, has been used to define the genetic cause in some patients and to evaluate potentially novel DBA-associated genes (63, 72-77).
WES has expanded the biological mechanism of DBA and expanded the clinical phenotype. Two families with DBA were found to have germline X-linked GATA1 mutations by WES. Previously, GATA1 mutations were only associated with X-linked congenital dyserythropoietic anemia and thrombocytopenia (12, 78). GATA1 encodes a key component of the GATA family of transcription factors that is important in erythroid development. Notably, the red cell ADA levels were normal in the DBA patients with GATA1 mutations (13), suggestive of an interaction or a different biological mechanism that is yet to be determined.
As expected based on the other DBA-associated genes, WES has also uncovered mutations in a gene encoding a ribosomal protein, RPS29 (79). RPS29 is a component of the 40S ribosomal subunit and critical for normal hematopoiesis in a zebrafish model (80). More recently, aCGH also identified deletions in RPL15 as a novel cause for DBA (14). Similar to RPS26, RPL15 is important in early synthesis of the 60S ribosomal subunit and in the cleavage of the internal transcribed spacer 1 (14).
THROMBOCYTOPENIA ABSENT RADII (TAR)
Clinical features and diagnosis
TAR is typically diagnosed in infancy due to the constellation of thrombocytopenia with bilateral absence of radii with the presence of thumbs, albeit abnormal (1, 81, 82). Patients can also have additional bony abnormalities of the ulna or humerus. Occasionally, hip and/or patellar dislocation and other non-specific bony abnormalities are present. The thumbs are always present in TAR, in contrast to FA where the radial ray abnormality results in missing thumbs if radii are absent. Cardiac, gastrointestinal and genitourinary system abnormalities have also been described in TAR (81).
Thrombocytopenia in TAR may be congenital or occur within the first few weeks to months of life (81). The majority of patients present with thrombocytopenia with platelet counts less than 50,000/ul. This is usually transient and significantly improves with time, but usually does not reach normal levels. Cow’s milk allergy is frequent in patients with TAR and can exacerbate thrombocytopenia, however the underlying pathophysiology of this relationship is not completely understood (81). Transient leukemoid reactions have also been reported in TAR, with white blood cell counts exceeding 35,000 cells/mm3 (15). There have been four reports of leukemia in patients with TAR, which include either AML or ALL (83, 84).
The management of TAR is mainly supportive care of thrombocytopenia with platelet transfusions as needed, and orthopedic treatment of bony abnormalities to improve function of the upper limbs (81, 85).
TAR Genetics and NGS in TAR
The inheritance of TAR typically fits into either an AR or de novo pattern (86). The molecular etiology of TAR was unknown until 2007 when a combination of chromosome GTG-banding and submegabase-resolution whole-genome tiling array CGH was used to identify a microdeletion at chromosome 1q21.1in TAR patients (15). This was further confirmed and fine-mapped to a common deleted region in all 30 individuals with TAR studied. Notably, the deletion was de novo in only 25% of affected individuals; an unaffected parent was a carrier of the same deletion in the other families. A subsequent study of 14 patients confirmed the presence of the 1q21.1 deletion in affected individuals and confirmed that unaffected parents also carried the deletion (87). However, the etiology of TAR was not yet been fully defined because unaffected parents can also be carriers of the 1q21.1 deletion. The deleted region of 1q21.1 contains 10 protein-coding genes but the initial candidate gene sequencing across this region failed to identify TAR-associated mutations. However, TAR cases were found to have low-frequency SNPs in regulatory regions of the RBM8A gene in the 1q21.1 deleted region (16, 82). The parent without the 1q21.1 deletion was the carrier of the RBM8A SNP. Thus, TAR can be caused by the biallelic inheritance of the 1q21.1 deletion from one parent and a rare SNP in RBM8A from the other. Notably, the inheritance of two hypomorphic variants in RBM8A also appears to cause TAR (16, 82). RBM8A encodes the conserved Y14 subunit of the exon-junction complex (EJC) that is essential for RNA processing and expressed in all hematopoietic lineages. These findings suggest that TAR is caused by the loss or significant reduction of RBMBA expression (82). This comprehensive NGS approach led to the first report of EJC defects associated with human disease.
UNCOVERING THE GENETIC ETIOLOGY OF UNCLASSIFIED IBMFS
In some instances patients may present with BMF and select features, such as congenital anomalies or family history, suggestive of an inherited disorder but not consistent with a well defined constellation of features consistent with a known IBMFS. Additionally, atypical presentations of known disorders may also complicate the diagnosis. NGS methods make it possible to uncover the genetic etiology of many previously unclassified patients (88-93) (Table 2). For example, a linkage study did not reveal the genetic cause of BMF and congenital nerve deafness in a family of three siblings and their mother but WES of the four affected individuals led to the discovery of germline mutations in SRP72 in this family (89). Targeted SRP72 sequencing identified an additional mother-child pair with BMF and a missense mutation. SRP72 encodes a component of a signal recognition particle responsible for protein translocation and processing that had not previously been implicated in BMF.
TABLE 2.
Examples of studies using whole exome sequencing to uncover the genetic etiology of unclassified IBMFS.
| Disorder/Clinical Features | Inheritance: Gene |
Biological pathway | Reference |
|---|---|---|---|
| Primary neutropenia and thrombocytopenia, oculocutaneous albinism, inflammatory bowel disease |
AR: SLC45A2 AR: G6PC3 |
SLC45A2: Transcription factor involved in trafficking of melanocyte-specific proteins to melanosomes G6PC3: Encodes enzyme involved in glucose-6-phosphate hydrolysis, maintenance of neutrophil viability and regulation of spontaneous neutrophil apoptosis |
Culliane et al. (2011)(88) |
| MonoMAC syndrome: Monocytopenia, severe infections with nontuberculous Mycobacteria, pulmonary alveolar proteinosis; Emberger syndrome; familial MDS/AML |
AD: GATA2 | Transcription factor involved in development and proliferation of hematopoietic and endocrine cell lineages |
Hsu et al. (2011)(91) |
| Bone marrow failure, congenital nerve deafness, MDS |
AD: SRP72 | Transcription factor involved in intracellular translocation of proteins by endoplasmic reticulum |
Kirwan et al. (2012)(89) |
| Familial aplastic anemia presenting as undiagnosed congenital amegakaryocytic thrombocytopenia |
AR: MPL | Encodes TPO receptor involved in megakaryopoiesis and HSC maintenance |
Walne et al. (2012)(90) |
| Congenital neutropenia, primary myelofibrosis and bone marrow failure in infancy, bony abnormalities, nephromegaly |
AR: VPS45 | Transcription factor involved in regulation of endosomal system |
Stephensky et al. (2013)(92) |
| Thrombocytopenia, fair hair and skin |
AR: SBF2 | Biological pathway unknown. | Abuzenadah et al. (2013)(93) |
Abbreviations: MDS, myelodysplastic syndrome; AD, autosomal dominant; AR, autosomal recessive; HSC, hematopoietic stem cell; WES, whole exome sequencing
WES of one affected child and the father was used to uncover homozygous mutations in SBF2 that are likely associated with disease in a family with two children with early onset thrombocytopenia and fair skin (93).The SBF2 gene encodes a pseudophosphatase, which is a member of the myotubularin-related protein family. Already germline mutations of SBF2 have been associated with Charcot-Marie-Tooth Diease, type 4B2. Its role in megakaryopoiesis is not understood and the connection between these findings is not known.
Similarly, germline mutations in VPS45, a component of congnate syntaxin Tlg2 which is required for membrane traffic through the endosomal system, have been linked to congenital neutropenia and myelofibrosis by WES and homozygosity mapping (92). Cells with the VPS45 mutation appeared to have defects in the endosomal-lysosomal pathways, a novel finding in IBMFS.
In addition to discovering novel causes of disease, WES and other NGS platforms can uncover mutations in genes associated with other phenotypes. Recently, AR germline mutations in MPL, which encodes the thrombopoietin receptor and are primarily responsible for congenital amegakaryocytic thrombocytopenia (CAMT), were linked to childhood onset BMF through exome sequencing of a consanguineous family (90). Evaluation of an additional 33 patients with childhood, but not neonatal BMF, identified another sibling pair with MPL mutations. CAMT typically presents during infancy with thrombocytopenia in the absence of physical anomalies but it can also present as BMF without a specific history of thrombocytopenia (94). These findings suggest screening for MPL mutations is warranted in individuals with BMF.
Occasionally, an individual with a previously undiagnosed disorder may be found to have two rare syndromes as illustrated in the case of a woman with oculocutaneous albinism, bleeding diathesis, and neutropenia without findings consistent with Hermansky-Pudlak syndrome. Her parents were consanguineous and the combination of WES with homozygosity mapping revealed AR mutations in both G6PC3, a congenital neutropenia gene, and SLC45A2, a gene associated with oculocutaneous albinism type 4 (88).
TRANSLATIONAL UTILITY OF GENOMICS IN THE IBMFS
Mutation identification in individuals with a clinically diagnosed IBMFS and their family members
It is important to identify the germline mutation in individuals with an IBMFS to confirm the diagnosis and tailor medical management accordingly. Genetic testing of a single IBMFS can be expensive and time consuming since there are often many genes that can underlie the main types of IBMFS (Table 1). Logical, sequential testing of specific targeted genes is complicated because data on genotype-phenotype relationships in the IBMFS are limited by sample size and clinical heterogeneity. In many instances, BMF may be the presenting sign for any of the IBMFS described above. For example, patients with FA may not have congenital anomalies, and patients with DC may not have the diagnostic mucocutaneous triad. Importantly, an IBMFS-related cancer, such as head and neck squamous cell cancer, may be the initial manifestation of disorders such as FA and DC. Since many patients with an IBMFS will need an HSCT, knowledge of the causative mutation allows for testing of potential related hematopoietic stem cell donors. This is critical because across the IBMFS spectrum, there is variable penetrance and expressivity of the clinical features. For example, use of a clinically silent carrier of a TERC mutation as a HSCT donor for a DC relative led to delayed engraftment and death from infection (95). In another example, HSCT of a DC patient using a sibling, who was a silent carrier of a TERC mutation, resulted in non-mobilization of the hematopoietic stem cells and the subsequent need for androgen therapy to maintain blood counts (95). Molecular testing may also help in distinguishing post-HSCT complications, such as chronic graft versus host disease, from late manifestations of DC (96).
NGS multi-gene platforms, specific for certain IBMFS, have been developed and currently are used on a research basis only (18, 19, 21, 63). These panels have the potential to supplant other modalities for establishing the diagnosis while identifying the genetic etiology of IBMFS and other disorders. However, they will not likely replace molecular diagnostics in IBMFS, but will be used to aid in the diagnostic work-up. For example, increased clastogen-induced chromosome breakage is diagnostic of FA and should continue to be used as a screening test. In FA it is possible that the complementation analysis performed to identify the defective protein could be replaced by NGS gene panels. In this instance, NGS gene panels may prove more cost-effective in identifying the genetic etiology of FA, and possibly other IBMFS.
Characterization of individuals with a mutation-negative or unclassified IBMFS
The causative mutation is not known in a large number of patients that meet clinical criteria for specific IBMFS. Although aberrations in the ribosomal biogenesis pathway are well known to cause DBA, a mutation has been identified in only 50% of patients with clinical DBA (3). Similarly, in DC the list of associated telomere biology genes is now up to nine, but only about 70% of patients have a mutation in one of those genes (36). Ongoing NGS efforts, including WES, whole genome sequencing, and comprehensive searches for insertions/deletions, will be essential in uncovering the genetic etiology of these disorders.
In some instances, patients may not meet diagnostic criteria for a specific IBMFS but the clinical suspicion of an inherited syndrome remains high. This may include patients with BMF who failed to respond to immunosuppressive therapy, those with a family history of features seen in other IBMFS (e.g., early onset cancers, BMF, pulmonary fibrosis), or a personal history of BMF and other clinical features consistent with, but non-diagnostic of a classic IBMFS (e.g., BMF and nail dysplasia with non-diagnostic telomere lengths). WES, comprehensive gene panels, or the judicious use of other NGS technologies can be extremely valuable in understanding the genetic cause of disease in these patients. It is extremely important to comprehensively evaluate the clinical features of family members of such patients in order to determine whether subtle signs of a familial disorder exist. Sequence analysis should include the affected and unaffected relatives whenever possible. This leads to improved efficiency in filtering of both common and rare genetic variants.
Genetic Variants in IBMFS Genes May Be Associated with Other Disorders
For the most part, the IBMFS-associated germline mutations are highly penetrant, resulting in clinically significant disease. In AD disorders, variable clinical penetrance of mutations in key telomere biology genes can complicate the diagnosis and genetic counseling. As noted above, AD germline mutations in TERT or TERC can result in classic DC, but may also result in a TBD lacking the typical DC-associated features (e.g., aplastic anemia, pulmonary fibrosis, or liver disease) (36, 97). Clinically silent carriers of mutations other DC genes and in DBA genes have also been reported (1, 39, 53).
Hereditary breast cancer is associated with monoallelic germline mutations in some of the same DNA repair genes that cause FA in the presence of biallelic mutations. For example, AR inheritance of mutation in BRCA2 (FANCD1) causes FA whereas AD inheritance of a single mutation can result in a significant predisposition to breast and ovarian cancer (98). The FA-associated BRCA2 mutations are located in distinct regions from the breast and ovarian cancer susceptibility mutations. Mono-allelic germline mutations in other FA-associated genes, BRIP1/FANCJ, PALB2/FANCN, RAD51C/FANCO, and SLX4/FANCP, were identified as breast cancer susceptibility genes through candidate gene sequencing (99, 100). WES in breast cancer families found rare, deleterious mutations in FANCC, BLM, and XRCC2 were associated with breast cancer (101, 102).
As NGS technologies are applied to a broader spectrum of clinical disorders, it is likely that expanded clinical phenotypes will be defined, based, in part, on the underlying molecular pathogenesis. Throughout all of these studies, it will be important to differentiate the highly-penetrant, clinically-significant germline mutations from those that result in a modest or even relatively minor increased risk of cancer or other disorder.
GENETIC COUNSELING CONSIDERATIONS
Genetic Education and Counseling Are Crucial
With the discovery of a compendium of disease mutations comes the responsibility of appropriate genetic counseling of the patients and families. All patients and their family members should receive education and counseling related to the genetics of the disorder for which they are being tested. This includes ensuring their understanding of the concepts of the inheritance of traits and disease, as well as of DNA and how DNA is transcribed to RNA then translated into protein. The molecular consequences of common and very rare, disease-associated DNA nucleotide changes should be described and understood by all individuals undergoing genetic testing. For young children, the parents should be educated and counseled. If the child is of the age of assent (usually around 11 or 12 years of age, depending on institutional policies), age-specific education and counseling should be performed with appropriate assent of the minor child. All individuals undergoing genetic testing should also understand that genetic testing has implications for the entire family. Healthy individuals may be found to be “silent carriers” of a mutation that causes clinically significant disease in their relative.
Couples from IBMFS families are now using pre-implantation genetic diagnosis (PGD) to attempt to have a healthy, unaffected child (103). PGD can only be successful if the causative mutation is known in the family. For example, sequence analysis of FANCA in an FA patient with positive FANCA complementation testing will uncover the specific mutations in the patient; the parents are then tested for the specific FANCA mutations. However, in mutation-negative families, a comprehensive research-based approach may be required prior to clinical testing. This approach could include WES that leads to the discovery of a new disease-associated gene (e.g., RTEL1 in DC).
Return of NGS Results
NGS technologies have been introduced into the clinical diagnosis and management of IBMFS, providing new insights. It is likely that, in the near future, we will know the genetic cause of most of the classic Mendelian disorders. NGS platforms, by the sheer volume of sequence data they generate, identify genetic variants that may or may not be related to the disease being studied. The clinical genetics community agrees that there is some duty to disclose incidental findings in clinical exome and genome sequencing (104). The American College of Medical Genetics and Genomics (ACMG) policy statement recommended a list of conditions, genes, and variants for return of incidental findings to patients. This list includes genes associated with cancer-predisposition disorders, connective tissue diseases, cardiac-related phenotypes, and susceptibility to malignant hyperthermia.
Currently, there appears to be a lack of consensus concerning the return of findings in the research setting. This is especially challenging because many times the investigators seeking to identify the cause of an inherited disorder with a specific phenotype do not have the expertise to identify and prioritize variants in genes important for different phenotypes. This problem is further compounded in studies of pediatric patients (105). Current recommendations include consideration of whether the findings have a known, urgent clinical significance. This includes careful consideration of the balance between knowing about a potential genetic disorder versus the potential risks of anxiety or other psychosocial harm that could result from the knowledge (105). The ACMG statement acknowledged a lack of data on the consequences of return of incidental findings to participants in NGS studies and recommended future longitudinal studies (104).
SUMMARY
NGS technologies have already had a substantial impact on advancing our knowledge of the etiology of the IBMFS. Ongoing NGS efforts will likely uncover the genetic etiology of the majority of the clinically diagnosed IBMFS. We can also expect to learn more about the pathophysiology of BMF through the study of patients and families with currently unclassified BMF disorders. These findings will advance our understanding of the underlying genetic defect. Follow-up with detailed functional studies will lead to important advances in understanding the underlying pathways and perhaps consider novel approaches towards therapy in the distant future. Patients and their family members need to be involved in the mutation discovery process through genetic education and counseling.
ACKNOWLEDGEMENTS
This work was supported by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI), National Institutes of Health. We thank Drs. Blanche P. Alter, Bari J. Ballew, and Neelam Giri, NCI, for their helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Shimamura A, Alter BP. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–22. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter BP, Kupfer G. Fanconi Anemia. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. University of Washington, Seattle; Seattle (WA): 1993. Updated 2013 Feb 7. [Google Scholar]
- 3.Clinton C, Gazda HT. Diamond-Blackfan Anemia. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. University of Washington, Seattle; Seattle (WA): 1993. [Google Scholar]
- 4.Gullapalli RR, Lyons-Weiler M, Petrosko P, Dhir R, Becich MJ, LaFramboise WA. Clinical integration of next-generation sequencing technology. Clinl Lab Med. 2012;32:585–99. doi: 10.1016/j.cll.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.H C. Rare Genetic Disorders: Learning About Genetic Disease Through Gene Mapping, SNPs, and Microarray Data. Nature Education. 2008;1(1) [Google Scholar]
- 6.Mutz KO, Heilkenbrinker A, Lönne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Current Opinions Biotech. 2013;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Shendure J, Lieberman Aiden E. The expanding scope of DNA sequencing. Nature Biotechnol. 2012 Nov;30(11):1084–94. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massie CE, Mills IG. Mapping protein-DNA interactions using ChIP-sequencing. Methods Molec Biol. 2012;809:157–73. doi: 10.1007/978-1-61779-376-9_11. [DOI] [PubMed] [Google Scholar]
- 9.Anderson BH, Kasher PR, Mayer J, et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nature Genetics. 2012;44:338–42. doi: 10.1038/ng.1084. [DOI] [PubMed] [Google Scholar]
- 10.Ballew BJ, Yeager M, Jacobs K, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Human Genetics. 2013;132:473–80. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe Dyskeratosis congenita. American Journal of Human Genetics. 2013;92:448–53. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kacena MA, Chou ST, Weiss MJ, Raskind WH. GATA1-Related X-Linked Cytopenia. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. University of Washington, Seattle; Seattle (WA): 1993. [Google Scholar]
- 13.Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Inves. 2012;122:2439–43. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landowski M, O’Donohue MF, Buros C, et al. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Human Genetics. 2013 Jun; doi: 10.1007/s00439-013-1326-z. (PMID: 23812780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klopocki E, Schulze H, Strauss G, et al. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Human Genetics. 2007;80:232–40. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albers CA, Paul DS, Schulze H, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nature Genetics. 2012;44:435–9. S1–2. doi: 10.1038/ng.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogliolo M, Schuster B, Stoepker C, et al. Mutations in ERCC4, Encoding the DNA-Repair Endonuclease XPF, Cause Fanconi Anemia. Am J Human Genetics. 2013;92:800–6. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knies K, Schuster B, Ameziane N, et al. Genotyping of Fanconi anemia patients by whole exome sequencing: advantages and challenges. PloS ONE. 2012;7:e52648. doi: 10.1371/journal.pone.0052648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameziane N, Sie D, Dentro S, et al. Diagnosis of Fanconi anemia: mutation analysis by next-generation sequencing. Anemia. 2012:132856. doi: 10.1155/2012/132856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuster B, Knies K, Stoepker C, et al. Whole exome sequencing reveals uncommon mutations in the recently identified Fanconi anemia gene SLX4/FANCP. Human mutation. 2013;34:93–6. doi: 10.1002/humu.22221. [DOI] [PubMed] [Google Scholar]
- 21.Chandrasekharappa SC, Lach FP, Kimble DC, et al. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood. 2013;121:e138–48. doi: 10.1182/blood-2012-12-474585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gille JJ, Floor K, Kerkhoven L, Ameziane N, Joenje H, de Winter JP. Diagnosis of Fanconi Anemia: Mutation Analysis by Multiplex Ligation-Dependent Probe Amplification and PCR-Based Sanger Sequencing. Anemia. doi: 10.1155/2012/603253. 2012603253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soulier J. Fanconi anemia. Hematology / the Education Program of the American Society of HematologyAmerican Society of HematologyEducation Program. 2011;2011:492–7. doi: 10.1182/asheducation-2011.1.492. [DOI] [PubMed] [Google Scholar]
- 24.Alter BP, Rosenberg PS. VACTERL-H Association and Fanconi Anemia. Molec Syndrom. 2013;4:87–93. doi: 10.1159/000346035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velazquez I, Alter BP. Androgens and liver tumors: Fanconi’s anemia and non-Fanconi’s conditions. Am J Hematol. 2004;77:257–67. doi: 10.1002/ajh.20183. [DOI] [PubMed] [Google Scholar]
- 26.Alter BP, Giri N, Savage SA, et al. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol. 2010;150:179–88. doi: 10.1111/j.1365-2141.2010.08212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genetics. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitao H, Takata M. Fanconi anemia: a disorder defective in the DNA damage response. Intern J Hematol. 2011;93:417–24. doi: 10.1007/s12185-011-0777-z. [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genetics A. 2011;155A:1877–83. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verlander PC, Kaporis A, Liu Q, Zhang Q, Seligsohn U, Auerbach AD. Carrier frequency of the IVS4 + 4 A-->T mutation of the Fanconi anemia gene FAC in the Ashkenazi Jewish population. Blood. 1995;86:4034–8. [PubMed] [Google Scholar]
- 31.Pronk JC, Gibson RA, Savoia A, et al. Localisation of the Fanconi anaemia complementation group A gene to chromosome 16q24.3. Nature Genetics. 1995;11:338–40. doi: 10.1038/ng1195-338. [DOI] [PubMed] [Google Scholar]
- 32.Chandra S, Levran O, Jurickova I, et al. A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Molecular Therapeutics. 2005;12:976–84. doi: 10.1016/j.ymthe.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 33.Sijbers AM, de Laat WL, Ariza RR, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–22. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 34.Niedernhofer LJ, Garinis GA, Raams A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–43. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 35.Shamseldin HE, Elfaki M, Alkuraya FS. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. J Med Genetics. 2012;49:184–6. doi: 10.1136/jmedgenet-2011-100585. [DOI] [PubMed] [Google Scholar]
- 36.Ballew BJ, Savage SA. Updates on the biology and management of dyskeratosis congenita and related telomere biology disorders. Expert Rev Hemat. 2013;6:327–37. doi: 10.1586/ehm.13.23. [DOI] [PubMed] [Google Scholar]
- 37.Savage SA, Alter BP. Dyskeratosis congenita. Hematology/oncology clinics of North America. 2009;23(2):215–31. doi: 10.1016/j.hoc.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113:6549–57. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Savage SA, Bertuch AA. The genetics and clinical manifestations of telomere biology disorders. Genetics in medicine. 2010;12:753–64. doi: 10.1097/GIM.0b013e3181f415b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vulliamy T, Dokal I. Dyskeratosis congenita. Seminars in Hematology. 2006;43:157–66. doi: 10.1053/j.seminhematol.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Khincha P, Wentzensen I, Giri N, Alter BP, Savage SA. Response to Androgen Therapy and Side Effects in Patients with Dyskeratosis Congenita. American Society of Hematology Annual Meeting. 2012 [Google Scholar]
- 42.Islam A, Rafiq S, Kirwan M, et al. Haematological recovery in dyskeratosis congenita patients treated with danazol. Br J Haematol. 2013 doi: 10.1111/bjh.12432. DOI: 10.1111/bjh.12432. [DOI] [PubMed] [Google Scholar]
- 43.Alter BP, Baerlocher GM, Savage SA, et al. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–47. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alter BP, Rosenberg PS, Giri N, Baerlocher GM, Lansdorp PM, Savage SA. Telomere length is associated with disease severity and declines with age in dyskeratosis congenita. Haematologica. 2012;97:353–9. doi: 10.3324/haematol.2011.055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savage SA. Dyskeratosis Congenita. In: Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. GeneReviews. University of Washington, Seattle; Seattle (WA): 1993. [Google Scholar]
- 46.Knight SW, Heiss NS, Vulliamy TJ, et al. Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br J Haematol. 1999;107:335–9. doi: 10.1046/j.1365-2141.1999.01690.x. [DOI] [PubMed] [Google Scholar]
- 47.Vulliamy T, Marrone A, Goldman F, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–5. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 48.Armanios M, Chen JL, Chang YP, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Nat Ac Sci U S A. 2005;102:15960–4. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diaz de Leon A, Cronkhite JT, Katzenstein AL, et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PloS ONE. 2010;5:e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armanios MY, Chen JJ, Cogan JD, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. New Engl J Med. 2007;356:1317–26. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 51.Vulliamy TJ, Walne A, Baskaradas A, Mason PJ, Marrone A, Dokal I. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Molecules Diseases. 2005;34:257–63. doi: 10.1016/j.bcmd.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi H, Calado RT, Ly H, et al. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. New EnglJl Med. 2005;352:1413–24. doi: 10.1056/NEJMoa042980. [DOI] [PubMed] [Google Scholar]
- 53.Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Human Genetics. 2008;82:501–9. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walne AJ, Vulliamy T, Marrone A, et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Human Molecr Genetics. 2007;16:1619–29. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vulliamy T, Beswick R, Kirwan M, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Nat Ac Sci U S A. 2008;105:8073–8. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong F, Savage SA, Shkreli M, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Developm. 2011;25:11–6. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Touzot F, Callebaut I, Soulier J, et al. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc Nat Ac Sci U S A. 2010;107:10097–102. doi: 10.1073/pnas.0914918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Human Molecular Genetics. 2010;19(22):4453–61. doi: 10.1093/hmg/ddq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Polvi A, Linnankivi T, Kivela T, et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. American Journal of Human Genetics. 2012;90(3):540–9. doi: 10.1016/j.ajhg.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savage SA. Connecting complex disorders through biology. Nature Genetics. 2012;44(3):238–40. doi: 10.1038/ng.2206. [DOI] [PubMed] [Google Scholar]
- 61.Keller RB, Gagne KE, Usmani GN, et al. CTC1 Mutations in a patient with dyskeratosis congenita. Pediatric Blood & Cancer. 2012;59(2):311–4. doi: 10.1002/pbc.24193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Guen T, Jullien L, Touzot F, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Human Molecular Genetics. 2013 doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 63.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. American Journal of Human Genetics. 2008;83(6):769–80. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119(16):3815–9. doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lipton JM, Ellis SR. Diamond-Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematology/oncology clinics of North America. 2009;23(2):261–82. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glader BE, Backer K, Diamond LK. Elevated erythrocyte adenosine deaminase activity in congenital hypoplastic anemia. The New England Journal of Medicine. 1983;309(24):1486–90. doi: 10.1056/NEJM198312153092404. [DOI] [PubMed] [Google Scholar]
- 67.Fargo JH, Kratz CP, Giri N, et al. Erythrocyte adenosine deaminase: diagnostic value for Diamond-Blackfan anaemia. British Journal of Haematology. 2013;160(4):547–54. doi: 10.1111/bjh.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vlachos A, Ball S, Dahl N, et al. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. British Journal of Haematology. 2008;142(6):859–76. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Current Topics in Developmental Biology. 2008;82:217–41. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 70.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature Genetics. 1999;21(2):169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 71.Horos R, von Lindern M. Molecular mechanisms of pathology and treatment in Diamond Blackfan Anaemia. British Journal of Haematology. 2012;159(5):514–27. doi: 10.1111/bjh.12058. [DOI] [PubMed] [Google Scholar]
- 72.Boria I, Quarello P, Avondo F, et al. A new database for ribosomal protein genes which are mutated in Diamond-Blackfan Anemia. Human Mutation. 2008;29(11):E263–70. doi: 10.1002/humu.20864. [DOI] [PubMed] [Google Scholar]
- 73.Boria I, Garelli E, Gazda HT, et al. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Human Mutation. 2010;31(12):1269–79. doi: 10.1002/humu.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doherty L, Sheen MR, Vlachos A, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. American Journal of Human Genetics. 2010;86(2):222–8. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gazda HT, Preti M, Sheen MR, et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Human Mutation. 2012;33(7):1037–44. doi: 10.1002/humu.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farrar JE, Nater M, Caywood E, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–92. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Human Mutation. 2007 Dec;28(12):1178–82. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 78.Weiss MJ, Mason PJ, Bessler M. What’s in a name? The Journal of Clinical Investigation. 2012;122(7):2346–9. doi: 10.1172/JCI63989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mirabello L, Ballew BJ, Giri N, et al. RPS29 is Mutated in a Multi-Case Diamond Blackfan Anemia Family. American Society of Hematology Annual Meeting. 2012 [Google Scholar]
- 80.Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, Zon LI. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Experimental Hematology. 2012;40(3):228–37.e5. doi: 10.1016/j.exphem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toriello HV. Thrombocytopenia-absent radius syndrome. Seminars in Thrombosis and Hemostasis. 2011;37(6):707–12. doi: 10.1055/s-0031-1291381. [DOI] [PubMed] [Google Scholar]
- 82.Albers CA, Newbury-Ecob R, Ouwehand WH, Ghevaert C. New insights into the genetic basis of TAR (thrombocytopenia-absent radii) syndrome. Current Opinion in Genetics & Development. 2013 doi: 10.1016/j.gde.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 83.Go RS, Johnston KL. Acute myelogenous leukemia in an adult with thrombocytopenia with absent radii syndrome. European Journal of Haematology. 2003;70(4):246–8. doi: 10.1034/j.1600-0609.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 84.Fadoo Z, Naqvi SM. Acute myeloid leukemia in a patient with thrombocytopenia with absent radii syndrome. Journal of Pediatric Hematology/Oncology. 2002;24(2):134–5. doi: 10.1097/00043426-200202000-00015. [DOI] [PubMed] [Google Scholar]
- 85.Coccia P, Ruggiero A, Mastrangelo S, et al. Management of children with thrombocytopenia-absent radius syndrome: an institutional experience. Journal of Paediatrics and Child Health. 2012;48(2):166–9. doi: 10.1111/j.1440-1754.2011.02069.x. [DOI] [PubMed] [Google Scholar]
- 86.Ward RE, Bixler D, Provisor AJ, Bader P. Parent to child transmission of the thrombocytopenia absent radius (TAR) syndrome. American Journal of Medical Genetics Supplement. 1986;2:207–14. doi: 10.1002/ajmg.1320250625. [DOI] [PubMed] [Google Scholar]
- 87.Houeijeh A, Andrieux J, Saugier-Veber P, et al. Thrombocytopenia-absent radius (TAR) syndrome: a clinical genetic series of 14 further cases. impact of the associated 1q21.1 deletion on the genetic counselling. European Journal of Medical Genetics. 2011;54(5):e471–7. doi: 10.1016/j.ejmg.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Cullinane AR, Vilboux T, O’Brien K, et al. Homozygosity mapping and whole-exome sequencing to detect SLC45A2 and G6PC3 mutations in a single patient with oculocutaneous albinism and neutropenia. Journal of Investigational Dermatology. 2011 Oct;131(10):2017–25. doi: 10.1038/jid.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kirwan M, Walne AJ, Plagnol V, et al. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. American Journal of Human Genetics. 2012 May;90(5):888–92. doi: 10.1016/j.ajhg.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walne AJ, Dokal A, Plagnol V, et al. Exome sequencing identifies MPL as a causative gene in familial aplastic anemia. Haematologica. 2012 Apr;97(4):524–8. doi: 10.3324/haematol.2011.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011 Sep;118(10):2653–5. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stepensky P, Saada A, Cowan M, et al. The Thr224Asn mutation in the VPS45 gene is associated with the congenital neutropenia and primary myelofibrosis of infancy. Blood. 2013 Jun;121(25):5078–87. doi: 10.1182/blood-2012-12-475566. [DOI] [PubMed] [Google Scholar]
- 93.Abuzenadah AM, Zaher GF, Dallol A, et al. Identification of a novel SBF2 missense mutation associated with a rare case of thrombocytopenia using whole-exome sequencing. Journal of Thrombosis and Thrombolysis. 2013 Jan; doi: 10.1007/s11239-012-0864-x. [DOI] [PubMed] [Google Scholar]
- 94.Geddis AE. Congenital amegakaryocytic thrombocytopenia. Pediatric Blood & Cancer. 2011 Aug;57(2):199–203. doi: 10.1002/pbc.22927. [DOI] [PubMed] [Google Scholar]
- 95.Fogarty PF, Yamaguchi H, Wiestner A, et al. Late presentation of dyskeratosis congenita as apparently acquired aplastic anaemia due to mutations in telomerase RNA. Lancet. 2003 Nov;362(9396):1628–30. doi: 10.1016/S0140-6736(03)14797-6. [DOI] [PubMed] [Google Scholar]
- 96.Ivker RA, Woosley J, Resnick SD. Dyskeratosis congenita or chronic graft-versus-host disease? A diagnostic dilemma in a child eight years after bone marrow transplantation for aplastic anemia. Pediatric Dermatology. 1993 Dec;10(4):362–5. doi: 10.1111/j.1525-1470.1993.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 97.Gramatges MM, Bertuch AA. Short telomeres: from dyskeratosis congenita to sporadic aplastic anemia and malignancy. Translational Research. 2013 Jun; doi: 10.1016/j.trsl.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biswas K, Das R, Alter BP, et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood. 2011 Sep;118(9):2430–42. doi: 10.1182/blood-2010-12-324541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shah S, Kim Y, Ostrovnaya I, et al. Assessment of SLX4 Mutations in Hereditary Breast Cancers. PLoS ONE. 2013;8(6):e66961. doi: 10.1371/journal.pone.0066961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Filippini1 SE, Vega A. Breast cancer genes: beyond BRCA1 and BRCA2. Frontiers in Bioscience. 2013;18:1358–72. doi: 10.2741/4185. [DOI] [PubMed] [Google Scholar]
- 101.Thompson ER, Doyle MA, Ryland GL, et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genetics. 2012 Sep;8(9):e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Park DJ, Lesueur F, Nguyen-Dumont T, et al. Rare mutations in XRCC2 increase the risk of breast cancer. American Journal of Human Genetics. 2012 Apr;90(4):734–9. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuliev A, Rechitsky S, Tur-Kaspa I, Verlinsky Y. Preimplantation genetics: Improving access to stem cell therapy. Annals of the New York Academy of Sciences. 2005;1054:223–7. doi: 10.1196/annals.1345.028. [DOI] [PubMed] [Google Scholar]
- 104.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in Medicine. 2013 Jun; doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Abdul-Karim R, Berkman BE, Wendler D, et al. Disclosure of incidental findings from next-generation sequencing in pediatric genomic research. Pediatrics. 2013 Mar;131(3):564–71. doi: 10.1542/peds.2012-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]