Abstract
We performed a gene–smoking interaction analysis using families from an early-onset coronary artery disease cohort (GENECARD). This analysis was focused on validating and expanding results from previous studies implicating single nucleotide polymorphisms (SNPs) on chromosome 3 in smoking-mediated coronary artery disease. We analyzed 430 SNPs on chromosome 3 and identified 16 SNPs that showed a gene–smoking interaction at P < 0.05 using association in the presence of linkage—ordered subset analysis, a method that uses permutations of the data to empirically estimate the strength of the association signal. Seven of the 16 SNPs were in the Rho-GTPase pathway indicating a 1.87-fold enrichment for this pathway. A meta-analysis of gene–smoking interactions in three independent studies revealed that rs9289231 in KALRN had a Fisher’s combined P value of 0.0017 for the interaction with smoking. In a gene-based meta-analysis KALRN had a P value of 0.026. Finally, a pathway-based analysis of the association results using WebGestalt revealed several enriched pathways including the regulation of the actin cytoskeleton pathway as defined by the Kyoto Encyclopedia of Genes and Genomes.
Introduction
Coronary artery disease (CAD) is the leading cause of death in the United States (Lopez et al. 2006). Despite the many studies using cohort, family-based, and case–control designs to address the genetic etiology of CAD (Franchini and Mannucci 2008; Musunuru and Kathiresan 2010; Samani et al. 2007; Schunkert et al. 2011) and common CAD risk factors (Heard-Costa et al. 2009; Lemaitre et al. 2011; Schunkert et al. 2011; Smith et al. 2010), there remains lack of knowledge concerning how genetic variation mediates and/or modifies the links between CAD and its common risk factors like hyperlipidemia, obesity, and smoking. Further work is needed in this area.
We previously reported a genetic linkage peak on chromosome 3q13 for CAD in families with an early-age-of-onset (EOCAD) (Hauser et al. 2004), which was subsequently replicated in the Diabetes Heart Study (DHS) (Bowden et al. 2006). Fine-mapping of this linkage peak in a case–control sample from the CATHGEN study revealed several associations between CAD and single nucleotide polymorphisms (SNPs) in this region (Wang et al. 2007). Replication of the 11 most significant SNPs in the case– control series from the Intermountain Heart Collaborative Study (IHCS) was only possible when the analysis was restricted to smokers (Horne et al. 2009). Further examination revealed a difference between the prevalence of smoking in the two datasets with smoking more prevalent in CATHGEN than in IHCS (41 vs. 11 % in controls, 74 vs. 29 % in cases). Significant gene–smoking interactions were found for SNPs in KALRN and ARHGAP31/CDGAP, and a second analysis in the DHS noted additional gene–smoking interactions in KALRN (Rudock et al. 2011). Given that smoking is a well-established risk factor for CAD (Ambrose and Barua 2004; Kannel et al. 1987) discovering genetic variants differentially associated with CAD in smokers may uncover mechanistic factors connecting genes, smoking, and CAD. We hypothesized that using APL-OSA as a test of gene–smoking interactions would reveal several SNPs that are significantly more associated with CAD in families with greater smoking exposure, measured by mean pack-years. We also hypothesized that the Rho-GTPase pathway would be enriched among the significant associations, based on its previously noted importance (Wang et al. 2007).
Methods
Subjects
The Genetics of Early Onset Cardiovascular Disease Study (GENECARD) is comprised of families with at least two siblings diagnosed with early onset CAD. Early onset CAD was defined on the basis of a medically documented acute coronary syndrome (unstable angina or myocardial infarction), a revascularization procedure, or a positive functional imaging study at or before the age of 50 years in men or 55 years in women. Risk factor history was collected via in person interviews in a location convenient for the subject. A blood sample was also collected during the interview (Hauser et al. 2003). Pack-years were calculated as pack of cigarettes smoked per day multiplied by total years spent smoking. A pack of cigarettes was defined as 20 cigarettes. People with zero pack-years were defined as those responding “NO” when asked if they smoked at least 100 cigarettes. We used pack-years because it is a measure of lifetime smoking and is well documented in the GENECARD dataset. A total of 2,434 individuals had sufficient information to calculate pack-years; the rest were considered as missing and did not contribute to family specific means.
Genotypes were generated in the Center for Human Genetics Molecular Genetics Core. All studies conformed to the same laboratory-wide quality control (QC) standards (Crosslin et al. 2009; Shah et al. 2009; Wang et al. 2007). Genotyping was done using either Taqman from Applied Biosystems or Golden Gate assay platform from Illumina. Applied Biosystems genotyping was done using the 7900HT Taqman SNP genotyping system and Illumina BeadStation genotyping was done on the 500G system. Six quality control samples were included on each quadrant of a 384-well plate. All SNPs were successfully genotyped for 95 % of the individuals and checked for the absence of any Mendelian inconsistencies and deviations from HWE both in their respective studies and the combined set of studies. Error rate estimates for SNPs meeting QC benchmarks were <0.2 %. Any SNP typed on fewer than 100 individuals was removed from analysis; a total of 3,516 individuals were sampled during the GENECARD study, of which 3,061 contributed genotypes these analyses. A total of 430 SNPs were analyzed across the chromosome 3q13 region in 1,360 nuclear families. All 11 SNPs analyzed by Horne et al. (2009) were genotyped in these families.
Statistical methods
APL-OSA
There are several family-based association tests each with its own strengths and weaknesses (Chung et al. 2006; Horvath et al. 2001; Martin et al. 2003; Spielman et al. 1993). We used the APL-OSA method. APL-OSA is a method that implements the APL test to detect increased evidence for association in the tail of a covariate distribution (Chung et al. 2008). Simulation studies show that APL has the same, or better, power as other tests without type I error inflation (Martin et al. 2003), even in regions with strong linkage (Chung et al. 2007). APL-OSA detects increased association in the tails of a covariate distribution, and is an effective test for SNP-smoking interactions. If there is truly an interaction between a SNP and smoking, we expect people with both the risk allele and higher cigarette smoking exposure to be at much greater risk of developing CAD, thus increasing the strength of the association in the upper extreme of the smoking distribution. We used pack-years as a measure of lifetime smoking and a dominant genetic model as the inheritance pattern for the putative CAD disease locus. The APL test is based on a test statistic, Z, which compares the number of alleles observed in affected sib-pairs with the number expected under the null hypothesis of no association conditioned on the parental genotypes.
has mean 0 and variance 1 under the null hypothesis, s is the number of families in the dataset, and Ti is the difference between the observed number of alleles in affected sib-pairs and the expected number of alleles given parental genotypes. In APL-OSA the APL test is calculated iteratively as each family is entered into the analysis. The analysis procedure is described in the following paragraph.
First the mean pack-years, xi, for each family, i, is calculated. For this calculation only the affected siblings are used. As the GENECARD study is an affected sibling pair design many families only have affected siblings and pack-years is well measured amongst the affected individuals within GENECARD. Thus we chose to only use the pack-years for the affected siblings to estimate the family-specific means of pack-years. Additionally, the family-specific associations are calculated based on the observed versus expected alleles in affected individuals; thus it is their exposure that will determine any gene-environment interactions. Since smoking increases the risk of CAD, the families are ordered by xi starting with the highest value of average pack-years. For exposures that decrease risk we would order the families low-to-high according to xi. Starting with a small subset of families c (c = 20 for our analyses), where c ≤ the total number of families (S), the APL test statistic, Z, is calculated as indicated above, and proceeding in order of xi each family is added into the set and the APL test statistic is recalculated. If two or more families have the same xi value they are added at the same time. Once all families have been added in, we define the maximum subset as the subset of families that contribute to the max |Z|, Z*. The absolute value is taken because Z can be significant in either the positive or negative direction, depending on the direction of association. The APL-OSA P value for Z is calculated by permuting the xi and repeating the procedure k times obtaining a Z*j for each permutation j. Finally, the P value is calculated as , where I is the indicator function with I(X) = 1 if X true and 0 otherwise.
Since the P value is computed via permutation, to reduce computing time for the initial analysis 500 permutations (k = 500) are used. Then SNPs with a P value less than 0.15 were selected for a second stringent analysis. In this analysis the number of permutations was increased, k = 1,000, to provide a more precise P value estimate. While the use of a permutation procedure increases computational demands, it also provides a natural correction for multiple testing by comparing the observed test statistic to the empirical null distribution of the data. The number of bootstrap iterations used to calculate is also varied between the initial and stringent analysis: 200 initially and 400 in the more stringent analysis. Despite the fact that the original linkage peak was found in GENECARD, using APL and APL-OSA to detect associations does not increase the type I error under the null hypothesis of no association (Chung et al. 2007).
Meta-analysis
We performed a meta-analysis by examining gene–smoking interaction signals amongst SNPs in Rho-GTPase pathway genes that were analyzed in GENECARD, IHCS, or DHS. We excluded the study by Wang et al. (2007) using CATHGEN because they did not report on any direct gene–smoking interactions, and we only report results for SNPs that were at least analyzed in GENECARD (the current study). Fischer’s method was used to compute the combined P value for those SNPs that appear in two or more studies. Fischer’s method computes the meta-analysis P value (PM) from S independent studies as .Pi is the P value of the ith study and PM is drawn from a Chi-squared distribution with 2S degrees of freedom. For the gene-based meta-analysis each gene was represented by the minimum P value observed across all genetic variants within the gene after correcting for the number of SNPs in the gene using the Sidak (1967) combination test (Peng et al. 2010). After collapsing across the genetic variants within a gene the gene-based P values were combined using Fischer’s method.
Pathway-based analysis
We used WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/) (Zhang et al. 2005) to perform the pathway-based gene set association analysis. We applied the hypergeometric test as implemented in WebGestalt to allow for the potential non-independence of the gene sets and set of SNPs chosen for genotyping. We used the 430 SNPs analyzed as the reference gene set. For significance we used a Bonferroni corrected cutoff of 0.05 and required that any gene sets called significant contain at least two genes from our list to avoid spurious associations caused by small gene sets that are “enriched” by only a single gene.
Results
GENECARD individuals have the clinical characteristics and risk factor distributions expected in an early-onset CAD cohort including a younger age of onset, more males than females, and high proportion of people diagnosed with hypertension and dyslipidemia (Table 1). At 33.8 % our percentage of smokers is higher than what was seen by Horne et al. (2009) (18.8 % for affected and controls combined). The various studies that have examined associations in the chromosome 3q13 region have used different definitions of smoking leading us to be careful when making comparisons between them. When the smoking definition used by Horne et al. current smoker or >10 pack-years, was applied to the CATHGEN cohort analyzed by Wang et al. (2007) the percentage of smokers in the datasets was 60.5, 55.2, and 54.2 % for the White Initial Dataset, White Validation Dataset, and African-American Dataset, respectively. All three of these datasets had a lower proportion of smokers than GENECARD (61.4 %) when an identical definition of smokers was applied.
Table 1.
Clinical covariates
| Characteristic, (N)a | Mean (SD)/percentage |
|---|---|
| Sex (% female), (3,061) | 37.3 % |
| Age of onset, (2,010) | 45.2 (7.67) |
| BMI, (2,387) | 29.7 (6.48) |
| Pack-years, (2,434) | 25.6 (27.1) |
| Diabetes (%), (2,435) | 19.5 % |
| Hypertension (%), (2,436) | 53.3 % |
| Dyslipoproteinemia (%), (2,438) | 72.4 % |
The clinical covariates for the GENECARD cohort analyzed here. Only those individuals who contributed genotypes are included in the table. Other individuals would have been included in the analysis to complete the family structure
3,061 participants provided genotypes for analysis
N is the total number of people used to calculate the mean/percentage
Counts done among individuals who provided genotype data to the study
APL results
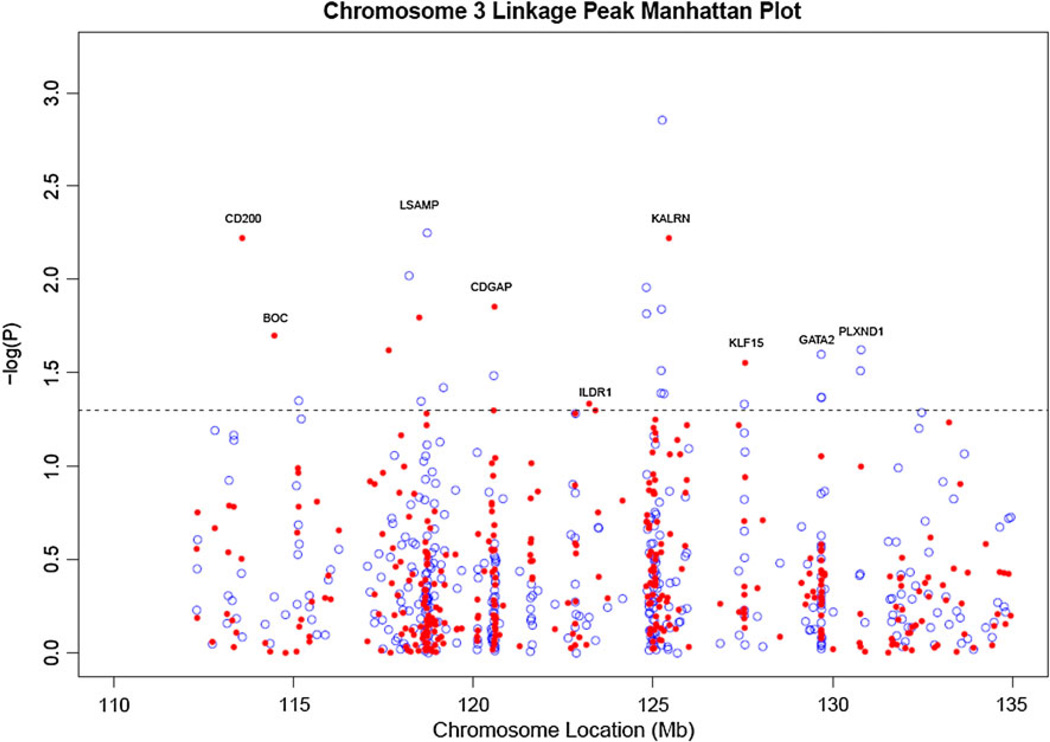
We performed the standard APL test (Martin et al. 2003) for all 430 SNPs to allow comparison with the APL-OSA results from the initial analysis with 200 bootstrap iterations and 500 permutations (Fig. 1). As in the analysis by Wang et al. (2007), rs9289231 was associated with EO-CAD (P < 0.000048), and was the only significant SNP after Bonferroni correction for 430 tests (P < 0.00012). Rs9289231 has previously been shown to be associated with high-density lipoproteins (Shah et al. 2006) in addition to early onset CAD. Twenty-three additional SNPs had unadjusted APL P values less than 0.05. A comparison of all 430 SNPs in the APL-OSA (initial run with 500 permutations, 200 bootstrap iterations) and the APL analysis is presented in Supplementary Table 1.
Figure 1.
APL and APL-OSA results. APL (blue open circles) and APL-OSA (red filled circles) results within the linkage peak region on chromosome 3. The y-axis is −log(P value) and the horizontal line is the 0.05 significance line. The x-axis is the location of each SNP along chromosome 3 in mega-bases (Mb). Only the area under the chromosome 3 linkage peak found in (Hauser et al. 2004) is shown (color figure online)
APL-OSA results
Table 2 contains the APL-OSA association results. Of the 430 SNPs, 16 had an APL-OSA P value < 0.05 after completing the second stringent analysis (Table 2). Of these, 12 were associated with a gene and four were intergenic SNPs. Though no results met a Bonferroni corrected P value of P < 0.00012, this cutoff may be overly conservative given the linkage disequilibrium (LD) in the region and the permutation procedure used to calculate empirical P values. Of the 16 SNPs with P < 0.05, six were in genes associated with the Rho-GTPase pathway, rs870995 (PIK3CA), rs2272486 (KALRN), rs4234218 (KALRN), rs11707609 (MYLK), rs1343700 (MYLK), rs10934651 (MYLK), rs6766899 (ARHGAP31/CDGAP). All of these genes fell under the chromosome 3 linkage peak (Hauser et al. 2004); however, there was low to moderate LD between the SNPs under this peak (Fig. 2). One SNP of particular interest is rs2272486 (P = 0.033, KALRN). Rs2272486 is a synonymous variant (His → His) and was the most significant SNP in the initial cohort analysis by Wang et al. (2007) (P = 0.0005).
Table 2.
APL-OSA results
| Marker | Gene | BP location | MAF | Z* | Z* P value | APL-OSA P value |
N, maximum subset (pack-years cutoff) |
|---|---|---|---|---|---|---|---|
| rs9289186 | ILDR1 | 121740304 | 0.07 | 2.75 | 0.003 | 0.006 | 232 (41.8) |
| rs6766899 | ARHGAP31 | 119107251 | 0.19 | 2.95 | 0.0016 | 0.008 | 200 (45.0) |
| rs1875111 | BOC homolog (mouse) | 112978764 | 0.26 | 3.23 | 0.0006 | 0.011 | 349 (9.0) |
| rs2084385 | PAK2 | 196553867 | 0.12 | 3.62 | 0.0001 | 0.014 | 684 (16.0) |
| rs11708769 | Intergenic | 150075547 | 0.13 | 2.68 | 0.0037 | 0.017 | 88 (42.0) |
| rs4234218 | KALRN | 123961210 | 0.36 | 2.91 | 0.0018 | 0.018 | 249 (40.5) |
| rs1513172 | Intergenic | 117011888 | 0.33 | 3.54 | 0.0002 | 0.019 | 232 (42.0) |
| rs870995 | PIK3CA | 178913005 | 0.42 | 3.00 | 0.0014 | 0.031 | 358 (6.25) |
| rs2272486 | KALRN | 123988038 | 0.36 | 2.72 | 0.0032 | 0.033 | 211 (44.0) |
| rs11707609 | MYLK | 123503423 | 0.42 | 2.41 | 0.008 | 0.034 | 319 (37.8) |
| rs1343700 | MYLK | 123571753 | 0.38 | 2.47 | 0.0067 | 0.041 | 59 (72.0) |
| rs1358087 | Intergenic | 126078890 | 0.37 | 2.35 | 0.0093 | 0.046 | 124 (56.0) |
| rs10934651 | MYLK | 123533208 | 0.05 | 2.55 | 0.0054 | 0.047 | 334 (35.0) |
| rs10511352 | LSAMP | 115643277 | 0.09 | 2.42 | 0.0078 | 0.047 | 653 (17.5) |
| rs1394041 | Intergenic | 147096847 | 0.10 | 2.55 | 0.0054 | 0.048 | 102 (58.0) |
| rs9822445 | LSAMP | 116161534 | 0.30 | 2.28 | 0.0114 | 0.048 | 24 (65.8) |
Results of the APL-OSA analysis with 1,000 permutations and 400 bootstrap iterations (for calculation of the variance)
Only those results with P < 0.05 are shown in the table
MAF minor allele frequency,
Z* is the association test statistic for the maximum subset
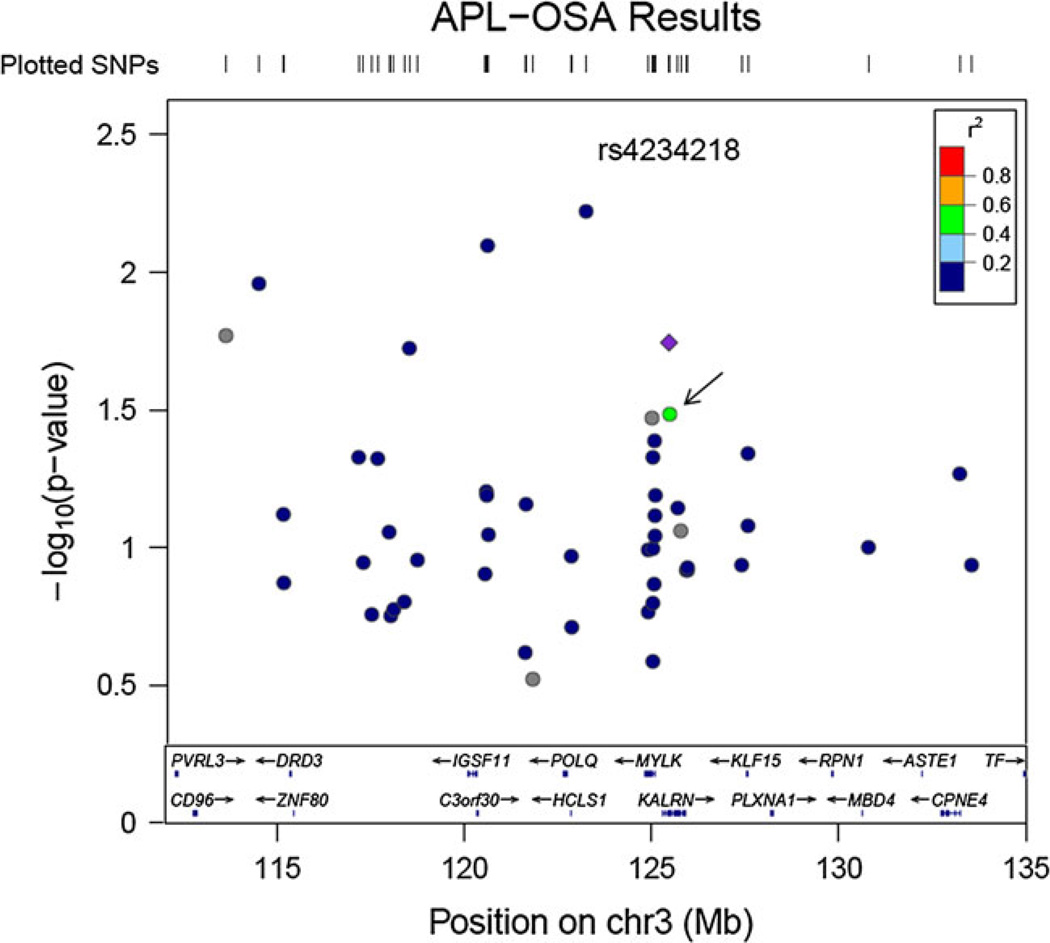
Figure 2.
APL-OSA associations. APL-OSA associations after restricting initial analyses (Fig. 1) based on a liberal P value (P < 0.15). It is these associations that are used throughout the paper as they represent our best estimation of the gene–smoking interactions. In the LocusZoom (Pruim et al. 2010) plot the −log10(P value) is given on the y-axis and the genomic location on the x-axis. Only the area under the chromosome 3 linkage peak (Hauser et al. 2004) is shown. Colors indicate the linkage disequilibrium with the most significant SNP in KALRN (rs4234218, shown as a diamond) and are based on LD within the CEU HapMap population. Only rs2272486 (arrow) shows even modest LD (color figure online)
Of the 11 SNPs analyzed by Horne et al. rs4234218 had an APL-OSA P < 0.05 (P = 0.018, Table 2). This SNP did not have a significant gene–smoking interaction P value in any of the datasets analyzed by Horne et al.; however, the maximum subset for rs4234218 contained 249 families each of which had an average pack-years of 40.5 or greater. The definition of smokers used by Horne et al. was pack-years >10. This heterogeneity in the definition of the “exposed” subgroup may account for the heterogeneity in association results and would indicate that a more severe smoking phenotype is needed to observe interactions with rs4234218.
A meta-analysis of three gene–smoking interaction studies in the 3q13 genomic region revealed two SNPs with evidence for effect modification via smoking. Rs12637456 (meta-analysis P = 0.044, KALRN) and rs9289231 (meta-analysis P = 0.0017) both showed evidence for association when information from independent cohorts was combined. As independent studies often have heterogeneity in the SNPs selected for analysis, a gene-based approach may be superior (Neale and Sham 2004). By using Sidak’s combination test (Peng et al. 2010) to combine P values across genes followed by a gene-based meta-analysis of three independent cohorts (GENECARD, IHCS, and DHS), we observed evidence for association between EOCAD and gene–smoking interactions in KALRN (meta-analysis P = 0.026) and ARH-GAP31 (meta-analysis P = 0.0056) (Table 3).
Table 3.
Gene–smoking interactions across three independent studies
| SNP | IHCS | GENECARD | DHS | Meta-analysis | Gene |
|---|---|---|---|---|---|
| rs9289231 | 0.07 | 0.282 | <0.001** | 0.0017 | KALRN |
| rs12637456 | 0.010 | 0.736 | 0.044 | KALRN | |
| rs6810298 | 0.75 | 0.564 | 0.012** | 0.10 | KALRN |
| rs4234218 | 1.00 | 0.018 | 0.515 | 0.15 | KALRN |
| rs10934490 | 0.019 | 0.156 | 0.20 | CDGAP/ARHGAP31 | |
| rs16834817 | 0.14 | 0.396 | 0.22 | MYLK | |
| rs1444768 | 0.54 | 0.504 | 0.63 | KALRN | |
| rs1444754 | 0.87 | 0.644 | 0.88 | KALRN |
SNPs in Table 3 are located in the Rho-GTPase pathway and analyzed in a gene–smoking interaction model in at least two of three studies, IHCS (Horne et al. 2009), DHS (Rudock et al. 2011), or the current analysis (GENECARD). Only SNPs appearing in two or more studies were included and the column “meta-analysis” is the Fisher’s combined P value across all the studies
For DHS P values listed as <0.001 converted to 0.001
P value for smoking interaction with carotid artery intima medial thickness as outcome
Pathway-based WebGestalt results
We performed a pathway-based gene set association analysis with WebGestalt (Zhang et al. 2005) using the 16 significant (P < 0.05) SNPs discovered from the APL-OSA analysis of the 430 SNPs across the chromosome 3q13 linkage region. We used the hypergeometric test as a test of enrichment, and after WebGestalt restricted our initial list of 16 SNPs to those mapping to known human genes we were left with 11 SNPs mapping to seven human genes (rs1875111 mapped to a homolog of the mouse gene BOC). We tested enrichment for both Gene Ontology (GO) (Ashburner et al. 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000) gene sets. The GO gene sets cytosol (P = 0.0095) and cellular projection (P = 0.046) and the KEGG pathways regulation of actin cytoskeleton (P = 0.028) and renal cell carcinoma (P = 0.043) were enriched in the pathway analysis.
Most of the GO and KEGG pathways are defined according to biological function and thus do not offer a direct test of the Rho-GTPase pathway as presented by Wang et al. (2007). Using the UCSC Table Browser (Karolchik et al. 2004) we extracted SNPs in the coding and 3′-UTR regions for the genes within the Rho-GTPase pathway as depicted in (Wang et al. 2007). We then further restricted to only those genes on chromosome 3 included in our peak-wide analysis. These restrictions left 6 genes (CDGAP/ARHGAP31, KALRN, MYLK, PIK3CA, PIK3CB, PIK3R4) comprising 4,912 SNPs. We used the 430 SNPs included in the analysis as the reference set and used the intersection of the 4,912 SNPs pulled from the UCSC Table Browser and our 430 analyzed SNPs as the SNP set of interest (97 SNPs). Of our 16 significant SNPs 7 were in the SNP set of interest, compared with 101 out of 430 when considering all genes. This 1.87-fold enrichment yielded a suggestive P value of 0.055 under the hyper-geometric test. In order to determine the degree to which smoking drives this enrichment we performed the same test but used the 23 SNPs significant in the association in the presence of linkage (APL) analysis. APL does not take into account the influence of smoking on the genetic associations, and instead simply tests for an overall association of SNP on CAD (Martin et al. 2003). In this analysis only 4 out of 23 significant SNPs were in the aforementioned Rho-GTPase SNP set, giving no evidence for enrichment (P = 0.855).
Discussion
This study used a family-based dataset, GENECARD, to validate smoking interactions in a linkage region, chromosome 3q13, associated with CAD (Hauser et al. 2004; Wang et al. 2007). We continued to observe strong evidence for association in this region, particularly in the gene KALRN. Previous evidence for association was more pronounced in smokers (Horne et al. 2009; Rudock et al. 2011) leading us to believe that a SNP–smoking interaction may be responsible for many of the initial associations. Using GENECARD, the EOCAD family-based dataset in which the original linkage was identified, we performed an APL-OSA analysis using pack-years as a measure of smoking history and found that 16 SNPs out of 430 analyzed had an APL-OSA P value <0.05 suggesting effect modification by smoking.
Chromosome 3q13 associations and LD
Among the 16 gene–smoking interactions identified via APL-OSA there were several SNPs not located in the Rho-GTPase pathway or associated with the identified KEGG or GO pathways. LSAMP, a gene previously implicated in CAD (Wang et al. 2008), was represented by two SNPs both located in introns, rs10511352 (P = 0.047) and rs9822445 (P = 0.048). Rs1394041 (P = 0.048) is an intergenic SNP previously found to be associated with several blood lipid phenotypes including small low-density lipoproteins, high-density lipoproteins, and triglycerides (Kathiresan et al. 2007).
To examine the correlation among the signals we identified in this study we looked at the LD of 26 SNPs, the 16 significant gene–smoking interactions and the ten additional SNPs analyzed by Horne et al. Across all 26 SNPs the LD was quite low, max r2 = 0.64 rs1444768 – rs1444754 (Fig. 3), indicating that these analyses are identifying independent loci that interact with smoking.
Figure 3.
Linkage disequilibrium across SNPs. Linkage disequilibrium (LD) plot of 11 SNPs from Horne et al. and 15 significant SNPs (APL-OSA P value <0.05) from across chromosome 3. Boxed SNPs are the 11 analyzed by Horne et al. (Horne BD). Underlined SNPs were also analyzed by Rudcock et al. (2011). The plot shows the pairwise r2 with darker cells having a higher r2 than lighter cells
Evidence for validation
To date analyses with four independent datasets (CATH-GEN, IHCS, DHS, and GENECARD) have identified EOCAD associations and/or gene–smoking interactions in the chromosome 3q13 region. Genome-wide analyses in GENECARD and the DHS were a family-based linkage analysis, both of which showed strong evidence of linkage in the 3q13 region (Bowden et al. 2006; Hauser et al. 2004). The more targeted candidate region analyses in CATHGEN identified specific genetic variants in KALRN and the Rho-GTPase pathway as associated with CAD (Wang et al. 2007), and validation studies in IHCS and the DHS (Horne et al. 2009; Rudock et al. 2011) found evidence for gene-smoking interactions in KALRN and selected Rho-GTPase genes. Our study provides a further confirmation of these gene–smoking interaction signals from Rho-GTPase pathway genes, in particular KALRN as it contained the only exonic gene–smoking interaction association (rs2272486).
Evidence for allelic heterogeneity in KALRN
At the SNP level there were multiple, potentially independent, associations within KALRN. Out of the 11 SNPs analyzed by Horne et al., we identified one gene–smoking interaction (rs4234218, P = 0.018). Rs4234218 was not one of the significant SNPs found by Horne et al. (2009). The most significant SNP found in the fully adjusted gene–smoking interaction model for young affected (most closely matching our EOCAD cohort) by Horne et al. was rs12637456 (P = 0.010), but this SNP did not have a significant APL-OSA P value, P = 0.736. Thus we do not have validation at the SNP level in GENECARD. However, rs4234218 and rs12637456 are located in KALRN yet not in LD in the GENECARD dataset, r2 = 0. Thus, KALRN may be important in the etiology and pathogenesis of CAD and we observe substantial allelic heterogeneity within the interaction associations between KALRN and CAD. Additional explanations for the observed heterogeneity are partial LD with an un-typed causal allele or a difference in the definition of smokers versus non-smokers. While APL-OSA allows the data to define the best cutoff, Horne et al. and Rudock et al. used >10 pack-years to define smokers. However, identical exposure definitions still resulted in allelic heterogeneity. For example, rs6810298 was significant in the Rudock et al. (2011) (P = 0.012), but not Horne et al. (2009) (P = 0.75). Despite this heterogeneity, the finding of multiple the gene–smoking interactions in multiple independent datasets in KALRN gives us confidence that these results are not due to random noise or confounding, but instead come from a true signal from these, or nearby, genetic variants. The meta-analysis can yield some clarity by showing those SNPs that had the most consistent signal. In meta-analysis we observed some association for replication at the SNP level with rs9289231. However, there was significant heterogeneity in the selection of SNPs for the three studies examined. Rudock et al. (2011) selected 28 SNPs within KALRN using a gene tagging approach, Horne et al. (2009) selected 11 SNPs based on their previous association with Rho-GTPase genes, and in our study the SNPs were selected based on their location on chromosome 3 and having been previously typed in the GENECARD cohort. Given this SNP selection heterogeneity a gene-based approach may better capture the consistent gene–smoking interactions. We see significant evidence for consistent associations between gene–smoking interactions in KALRN and EOCAD (P = 0.026).
Functional heterogeneity in KALRN
In addition to the aforementioned allelic heterogeneity a literature search revealed that KALRN exhibits significant functional heterogeneity. KALRN is a gene with multiple isoforms and multiple functional domains. The gene was originally identified in the brain and has been associated with schizophrenia (Kushima et al. 2012) and ADHD (Lesch et al. 2008). Among cardiovascular diseases, in addition to CAD, KALRN has been associated with ischemic stroke (Krug et al. 2010), but this result was not replicated in an independent data set (Olsson et al. 2011).
Smoking and KALRN
KALRN is primarily known as a neuronal gene and it has several functions related to the protection and growth of neurons (Rabiner et al. 2005). However, another important function of KALRN is its inhibition of inducible nitric oxide synthase (iNOS) activity through the prevention of iNOS homodimerization (Ratovitski et al. 1999). Research indicates that cigarette smoke-induced intima wall thickening in mice is markedly greater in wild-type than in iNOS-deficient mice (Anazawa et al. 2004), and long-term survival after ischemic stroke was associated with interactions between nitric oxide synthase genetic variants and cigarette smoke (Oksala et al. 2008). Given these relationships between cardiovascular phenotypes and iNOS, and the evidence that cigarette smoke decreases iNOS expression (Hoyt et al. 2003), it is possible that the functional link among KALRN, smoking, and CAD is the joint activity of KALRN and smoking on iNOS.
Rho-GTPase associations beyond KALRN and other pathway relationships
In addition to KALRN, other genes in the chromosome 3q13 region demonstrated gene–smoking interactions. MYLK was the most represented gene among the significant results having 3 SNPs with P < 0.05. All three SNPs, rs11707609 (P = 0.034), rs1343700 (P = 0.041), and rs10934651 (P = 0.047) are located in introns of MYLK. Rs6766899 (P = 0.008) is located in ARHGAP31, a Rho-GTPase pathway gene also noted as having a gene–smoking interaction by Horne et al. (2009) (rs10934490, P = 0.017). ARHGAP31 was also the most significant gene in the gene-based meta-analysis (P = 0.0056). Rs870995 had an APL-OSA P value of 0.031 and is located in PIK3CA. PIK3CA is the alpha subunit of phosphoinositide 3 kinase and was implicated in a meta-analysis of four genome-wide linkage studies for CAD (Chiodini and Lewis 2003). We compared the enrichment of Rho-GTPase pathway genes, as noted in (Wang et al. 2007), among the 16 significant SNPs from APL-OSA with those significant in APL. The 16 gene–smoking interactions were suggestive for enrichment (P = 0.055); however, the 23 significant SNPs from the APL analysis, which would indicate an overall association with CAD independent of smoking, were not enriched for Rho-GTPase pathway genes (P = 0.855). Thus, we concluded that the enrichment of the Rho-GTPase pathway is likely due to gene–smoking interactions rather than marginal genetic associations with CAD.
In addition to specifically examining the Rho-GTPase pathway genes we examined the enrichment of functional KEGG and GO pathways. The KEGG pathway regulation of the actin cytoskeleton was enriched (P = 0.028) in our analyses. Regulation of the actin cytoskeleton may be mechanistically involved in the pathogenesis of CAD through its role in the migration and morphology changes of smooth muscle cells. An important component of the association between smooth muscle cells and CAD is their differentiation from a contractile to a synthetic phenotype, where the cell can respond to vascular injury (Doran et al. 2008), and genetic variants associated with vascular disease are associated with smooth muscle cell differentiation (Milewicz et al. 2010). Smooth muscle cell differentiation involves extensive morphology reorganization, implicating the actin cytoskeleton as an important mediator of the process. The actin cytoskeleton also plays a role in cellular migration and proliferation, and proliferation of smooth muscle cells is proposed to be a key component in the development of atherosclerosis (Libby and Theroux 2005). While other members of the regulation of the actin cytoskeleton pathway may be targets for future analyses, we recognize that it will take careful functional studies in model systems to elucidate the causal actors. Furthermore, our study suggests that these analyses will need to be augmented with careful analysis of environmental risk factors to understand the relationship between pathways/genes/genetic variants and CAD.
Conclusion
Coronary artery disease is a complex disorder with a variety of genetic variants found to modify risk (Bis et al. 2011; Maouche and Schunkert 2012; Schunkert et al. 2011). Functional relationships are elusive, perhaps due to the presence of allelic heterogeneity, genetic heterogeneity, and gene–environment actions. While genome-wide association studies have great utility, unraveling epistasis or environmental interactions via this method can require sample sizes in the hundreds of thousands (Zuk et al. 2012). Targeted gene–environment interactions studies, along with higher level analyses at the pathway or functional level, are needed to elucidate causative links between genetic variants and CAD. This study reaffirms published gene–smoking interactions in KALRN while identifying a novel coding variant, highlights multiple gene–smoking interactions under the chromosome 3q13 linkage peak, and uses pathway analyses to integrate and better understand the gene–smoking interactions arising from this region. Each of these analyses implicates specific pathogenic mechanisms that can be followed up in functional studies. KALRN and smoking are potentially mechanistically linked via their inhibition of iNOS, the Rho-GTPase pathway has several members lying outside our chromosome 3 region of investigation that may present novel candidate genes for analysis, and our pathway analyses reveal global pathways that may be interrupted or jointly analyzed to further this work.
Supplementary Material
Acknowledgments
We thank all of the volunteers for their participation in the GENECARD study. We would also like to acknowledge the essential contributions of the following individuals: the GENE-CARD Investigators Network, and the faculty and staff at the Center for Human Genetics for their innumerable contributions to this manuscript. This work was supported by NIH grants HL073389 and MH595228 and an award from the Neurosciences Education and Research Foundation. This work was supported in part by an appointment to the Research Participation Program at the Office of Research and Development, US Environmental Protection Agency.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-013-1339-7) contains supplementary material, which is available to authorized users.
Contributor Information
Cavin Ward-Caviness, Email: cavin.wardcaviness@gmail.com, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA.
Carol Haynes, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA.
Colette Blach, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA.
Elaine Dowdy, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA.
Simon G. Gregory, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA
Svati H. Shah, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA Division of Cardiovascular Medicine, School of Medicine, Duke University Medical Center, Durham, NC 27710, USA.
Benjamin D. Horne, Cardiovascular Department, Intermountain Medical Center, University of Utah, 391 Chipeta Way, Suite D, Salt Lake City, UT 84122, USA
William E. Kraus, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA Division of Cardiovascular Medicine, School of Medicine, Duke University Medical Center, Durham, NC 27710, USA.
Elizabeth R. Hauser, Center for Human Genetics, School of Medicine, Duke University Medical Center, Box 3382, Durham, NC 27710, USA Epidemiologic Research and Information Center, Durham Veterans Affairs Medical Center, Durham, NC, USA.
References
- Ambrose JA, Barua RS. The pathophysiology of cigarette smoking and cardiovascular disease: an update. J Am Coll Cardiol. 2004;43:1731–1737. doi: 10.1016/j.jacc.2003.12.047. [DOI] [PubMed] [Google Scholar]
- Anazawa T, Dimayuga PC, Li H, Tani S, Bradfield J, Chyu K-Y, Kaul S, Shah PK, Cercek B. Effect of exposure to cigarette smoke on carotid artery intimal thickening: the role of inducible NO synthase. Arterioscler Thromb Vasc Biol. 2004;24:1652–1658. doi: 10.1161/01.ATV.0000139925.84444.ad. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis JC, Kavousi M, Franceschini N, Isaacs A, Abecasis GR, Schminke U, Post WS, Smith AV, Cupples LA, Markus HS, Schmidt R, Huffman JE, Lehtimaki T, Baumert J, Munzel T, Heckbert SR, Dehghan A, North K, Oostra B, Bevan S, Stoegerer E-M, Hayward C, Raitakari O, Meisinger C, Schillert A, Sanna S, Volzke H, Cheng Y-C, Thorsson B, Fox CS, Rice K, Rivadeneira F, Nambi V, Halperin E, Petrovic KE, Peltonen L, Wichmann HE, Schnabel RB, Dorr M, Parsa A, Aspelund T, Demissie S, Kathiresan S, Reilly MP, Taylor K, Uitterlinden A, Couper DJ, Sitzer M, Kahonen M, Illig T, Wild PS, Orru M, Ludemann J, Shuldiner AR, Eiriksdottir G, White CC, Rotter JI, Hofman A, Seissler J, Zeller T, Usala G, Ernst F, Launer LJ, D’Agostino RB, O’Leary DH, Ballantyne C, Thiery J, Ziegler A, Lakatta EG, Chilukoti RK, Harris TB, Wolf PA, Psaty BM, Polak JF, Li X, Rathmann W, Uda M, Boerwinkle E, Klopp N, Schmidt H, Wilson JF, Viikari J, Koenig W, Blankenberg S, Newman AB, Witteman J, Heiss G, Duijn Cv, Scuteri A, Homuth G, Mitchell BD, Gudnason V, O’Donnell CJ. Meta-analysis of genome-wide association studies from the CHARGE consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet. 2011;43:940–947. doi: 10.1038/ng.920. http://www.nature.com/ng/journal/v43/n10/abs/ng.920.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden DW, Rudock M, Ziegler J, Lehtinen AB, Xu J, Wagenknecht LE, Herrington D, Rich SS, Freedman BI, Carr JJ, Langefeld CD. Coincident linkage of type 2 diabetes, metabolic syndrome, and measures of cardiovascular disease in a genome scan of the diabetes heart study. Diabetes. 2006;55:1985–1994. doi: 10.2337/db06-0003. [DOI] [PubMed] [Google Scholar]
- Chiodini BD, Lewis CM. Meta-analysis of 4 coronary heart disease genome-wide linkage studies confirms a susceptibility locus on chromosome 3q. Arterioscler Thromb Vasc Biol. 2003;23:1863–1868. doi: 10.1161/01.ATV.0000093281.10213.DB. [DOI] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Chung R-H, Hauser ER, Martin ER. Interpretation of simultaneous linkage and family-based association tests in genome screens. Genet Epidemiol. 2007;31:134–142. doi: 10.1002/gepi.20196. [DOI] [PubMed] [Google Scholar]
- Chung R-H, Schmidt S, Martin ER, Hauser ER. Ordered-subset analysis (OSA) for family-based association mapping of complex traits. Genet Epidemiol. 2008;32:627–637. doi: 10.1002/gepi.20340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosslin DR, Shah SH, Nelson SC, Haynes CS, Connelly JJ, Gadson S, Goldschmidt-Clermont PJ, Vance JM, Rose J, Granger CB, Seo D, Gregory SG, Kraus WE, Hauser ER. Genetic effects in the leukotriene biosynthesis pathway and association with atherosclerosis. Hum Genet. 2009;125:217–229. doi: 10.1007/s00439-008-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini MPF, Mannucci PM. The genetic basis of coronary artery disease: from candidate genes to whole genome analysis. Trends Cardiovasc Med. 2008;18:5. doi: 10.1016/j.tcm.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Mooser V, Crossman DC, Haines JL, Jones CH, Winkelmann BR, Schmidt S, Scott WK, Roses AD, Pericak-Vance MA, Granger CB, Kraus WE. Design of the genetics of early onset cardiovascular disease (GENECARD) study. Am Heart J. 2003;145:602–613. doi: 10.1067/mhj.2003.13. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJH, Mooser V, McAdam B, Winkelmann BR, Wiseman AH, Muhlestein JB, Bartel AG, Dennis CA, Dowdy E, Estabrooks S, Eggleston K, Francis S, Roche K, Clevenger PW, Huang L, Pedersen B, Shah S, Schmidt S, Haynes C, West S, Asper D, Booze M, Sharma S, Sundseth S, Middleton L, Roses AD, Hauser MA, Vance JM, Pericak-Vance MA, Kraus WE. A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD study. Am J Hum Genet. 2004;75:436–447. doi: 10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard-Costa NL, Zillikens MC, Monda KL, Johansson Å, Harris TB, Fu M, Haritunians T, Feitosa MF, Aspelund T, Eiriksdottir G, Garcia M, Launer LJ, Smith AV, Mitchell BD, McArdle PF, Shuldiner AR, Bielinski SJ, Boerwinkle E, Brancati F, Demerath EW, Pankow JS, Arnold AM, Chen Y-DI, Glazer NL, McKnight B, Psaty BM, Rotter JI, Amin N, Campbell H, Gyllensten U, Pattaro C, Pramstaller PP, Rudan I, Struchalin M, Vitart V, Gao X, Kraja A, Province MA, Zhang Q, Atwood LD, Dupuis J, Hirschhorn JN, Jaquish CE, O’Donnell CJ, Vasan RS, White CC, Aulchenko YS, Estrada K, Hofman A, Rivadeneira F, Uitterlinden AG, Witteman JCM, Oostra BA, Kaplan RC, Gudnason V, O’Connell JR, Borecki IB, van Duijn CM, Cupples LA, Fox CS, North KE. NRXN3: is a novel locus for waist circumference: a genome-wide association study from the CHARGE consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne BD, Hauser ER, Wang L, Muhlestein JB, Anderson JL, Carlquist JF, Shah SH, Kraus WE. Validation study of genetic associations with coronary artery disease on chromosome 3q13-21 and potential effect modification by smoking. Ann Hum Genet. 2009;73:551–558. doi: 10.1111/j.1469-1809.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Hoyt JC, Robbins RA, Habib M, Springall DR, Buttery LDK, Polak JM, Barnes PJ. Cigarette smoke decreases inducible nitric oxide synthase in lung epithelial cells. Exp Lung Res. 2003;29:17–28. doi: 10.1080/01902140303759. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, D’Agostino RB, Belanger AJ. Fibrinogen, cigarette smoking, and risk of cardiovascular disease: insights from the Framingham study. Am Heart J. 1987;113:1006–1010. doi: 10.1016/0002-8703(87)90063-9. [DOI] [PubMed] [Google Scholar]
- Karolchik DHA, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:4. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Manning AK, Demissie S, D’Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, Arnett DK, Peloso GM, Ordovas JM, Cupples LA. A genome-wide association study for blood lipid phenotypes in the Framingham heart study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug TMH, Gouveia L, Sobral J, Xavier J, Gaspar G, Correia M, Viana-Baptista M, Simoes R, Pinto A, Taipa R, Ferreria C, Fontes J, Silva M, Gabriel J, Matos I, Lopes G, Ferro J, Vicente A, Oliveira S. Kalirin: a novel genetic risk factor for ischemic stroke. Hum Genet. 2010;127:11. doi: 10.1007/s00439-010-0790-y. [DOI] [PubMed] [Google Scholar]
- Kushima I, Nakamura Y, Aleksic B, Ikeda M, Ito Y, Shiino T, Okochi T, Fukuo Y, Ujike H, Suzuki M, Inada T, Hashimoto R, Takeda M, Kaibuchi K, Iwata N, Ozaki N. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schiophr Bull. 2012;38(3):552–560. doi: 10.1093/schbul/sbq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng L-C, Bhattacharya S, Bandinelli S, Bis JC, Rich SS, Jacobs DR, Jr, Cherubini A, McKnight B, Liang S, Gu X, Rice K, Laurie CC, Lumley T, Browning BL, Psaty BM, Chen Y-DI, Friedlander Y, Djousse L, Wu JHY, Siscovick DS, Uitterlinden AG, Arnett DK, Ferrucci L, Fornage M, Tsai MY, Mozaffarian D, Steffen LM. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch K-P, Timmesfeld N, Renner T, Halperin R, Röser C, Nguyen T, Craig D, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schäfer H, Walitza S, Reif A, Stephan D, Jacob C. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111:3481–3488. doi: 10.1161/CIRCULATIONAHA.105.537878. [DOI] [PubMed] [Google Scholar]
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJL. Global burden of disease and risk factors. New York: Oxford University Press; 2006. [PubMed] [Google Scholar]
- Maouche S, Schunkert H. Strategies beyond genome-wide association studies for atherosclerosis. Arterioscler Thromb Vasc Biol. 2012;32:170–181. doi: 10.1161/ATVBAHA.111.232652. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Hauser ER, Kaplan NL. Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet. 2003;73:1016–1026. doi: 10.1086/378779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewicz DM, Kwartler CS, Papke CL, Regalado ES, Cao J, Reid AJ. Genetic variants promoting smooth muscle cell proliferation can result in diffuse and diverse vascular diseases: evidence for a hyperplastic vasculomyopathy. Genet Med. 2010;12:196–203. doi: 10.1097/GIM.0b013e3181cdd687. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Kathiresan S. Genetics of coronary artery disease. Annu Rev Genomics Hum Genet. 2010;11:91–108. doi: 10.1146/annurev-genom-082509-141637. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksala N, Oksala A, Erkinjuntti T, Pohjasvaara T, Kunnas T, Vataja R, Kaste M, Karhunen P. Long-term survival after ischemic stroke in postmenopausal women is affected by an interaction between smoking and genetic variation in nitric oxide synthases. Cerebrovasc Dis. 2008;26:250–258. doi: 10.1159/000147452. [DOI] [PubMed] [Google Scholar]
- Olsson S, Jood K, Melander O, Sjögren M, Norrving B, Nilsson M, Lindgren A, Jern C. Lack of association between genetic variations in the KALRN region and ischemic stroke. Clin Biochem. 2011;44:1018–1020. doi: 10.1016/j.clinbiochem.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S, Zhao J, Zhou X, Reveille JD, Jin L, Amos CI, Xiong M. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur J Hum Genet. 2010;18:111–117. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner CA, Mains RE, Eipper BA. Kalirin: a dual rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neurosci. 2005;11:148–160. doi: 10.1177/1073858404271250. [DOI] [PubMed] [Google Scholar]
- Ratovitski EA, Alam MR, Quick RA, McMillan A, Bao C, Kozlovsky C, Hand TA, Johnson RC, Mains RE, Eipper BA, Lowenstein CJ. Kalirin inhibition of inducible nitric-oxide synthase. J Biol Chem. 1999;274:993–999. doi: 10.1074/jbc.274.2.993. [DOI] [PubMed] [Google Scholar]
- Rudock M, Cox AJ, Ziegler J, Lehtinen A, Connelly J, Freedman B, Carr JJ, Langefeld C, Hauser E, Horne B, Bowden D. Cigarette smoking status has a modifying effect on the association between polymorphisms in KALRN and measures of cardiovascular risk in the diabetes heart study. Genes Genomics. 2011;33:483–490. [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AFR, Barbalic M, Gieger C, Absher D, Aherrahrou Z, Allayee H, Altshuler D, Anand SS, Andersen K, Anderson JL, Ardissino D, Ball SG, Balmforth AJ, Barnes TA, Becker DM, Becker LC, Berger K, Bis JC, Boekholdt SM, Boerwinkle E, Braund PS, Brown MJ, Burnett MS, Buysschaert I, Carlquist JF, Chen L, Cichon S, Codd V, Davies RW, Dedoussis G, Dehghan A, Demissie S, Devaney JM, Diemert P, Do R, Doering A, Eifert S, Mokhtari NEE, Ellis SG, Elosua R, Engert JC, Epstein SE, de Faire U, Fischer M, Folsom AR, Freyer J, Gigante B, Girelli D, Gretarsdottir S, Gudnason V, Gulcher JR, Halperin E, Hammond N, Hazen SL, Hofman A, Horne BD, Illig T, Iribarren C, Jones GT, Jukema JW, Kaiser MA, Kaplan LM, Kastelein JJP, Khaw K-T, Knowles JW, Kolovou G, Kong A, Laaksonen R, Lambrechts D, Leander K, Lettre G, Li M, Lieb W, Loley C, Lotery AJ, Mannucci PM, Maouche S, Martinelli N, McKeown PP, Meisinger C, Meitinger T, Melander O, Merlini PA, Mooser V, Morgan T, Muhleisen TW, Muhlestein JB, Munzel T, Musunuru K, Nahrstaedt J, Nelson CP, Nothen MM, Olivieri O, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. http://www.nature.com/ng/journal/v43/n4/abs/ng.784.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Kraus WE, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, Huang L, Haynes C, Dowdy E, Vega GL, Grundy SM, Vance JM, Hauser ER. Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q. Ann Hum Genet. 2006;70:738–748. doi: 10.1111/j.1469-1809.2006.00288.x. [DOI] [PubMed] [Google Scholar]
- Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, Wenner BR, Dowdy ZE, Granger CB, Ginsburg GS, Newgard CB, Kraus WE. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5:258. doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidak Z. Rectangular confidence regions for the means of multivariate normal distributions. J Am Stat Assoc. 1967;62:626–633. [Google Scholar]
- Smith EN, Chen W, Kähönen M, Kettunen J, Lehtimäki T, Peltonen L, Raitakari OT, Salem RM, Schork NJ, Shaw M, Srinivasan SR, Topol EJ, Viikari JS, Berenson GS, Murray SS. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet. 2010;6:e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, Harris M, Nelson S, Hale AB, Granger CB, Haines JL, Jones CJH, Crossman D, Seo D, Gregory SG, Kraus WE, Goldschmidt-Clermont PJ, Vance JM. Peakwide mapping on chromosome 3q13 identifies the Kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Seo D, Sivashanmugam P, Exum ST, Gregory SG, Granger CB, Haines JL, Jones CJH, Crossman D, Haynes C, Kraus WE, Freedman NJ, Pericak-Vance MA, Goldschmidt-Clermont PJ, Vance JM. Polymorphisms of the tumor suppressor gene LSAMP are associated with left main coronary artery disease. Ann Hum Genet. 2008;72:443–453. doi: 10.1111/j.1469-1809.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.