Abstract
Activated hepatic stellate cells (HSC) are the primary source of extracellular matrix proteins found in liver fibrosis/cirrhosis patients. Therefore, the prevention of HSC activation is an important strategy for treating severe liver injury. This study examined the effects of KR62776, a new peroxisome proliferator-activated receptor γ (PPARγ) agonist, on the rate of cell proliferation and expression of α-smooth muscle actin (α-SMA) in rat hepatic stellate HSC-T6 cells. In addition, its effects on the liver damage induced by carbon tetrachloride were investigated. KR62776 caused the apoptosis of activated HSC-T6 cells with the concomitant decrease in the α-smooth muscle actin levels in a time- and concentration-dependent manner. However, KR62776 did not cause the apoptosis of human HepG2 and rat McARH7777 hepatoma cells, suggesting that KR62776 has a specific effect on stellate cells. KR62776 increased the levels of Gadd45, p27, p21 and PPARγ proteins but decreased the cell cycle-related proteins, such as cdk2, cyclin B and cyclin D1. These changes were reversed by BADGE, a specific PPARγ antagonist, indicating that the effects of KR62776 are, at least in part, PPARγ-dependent. In addition, KR62776 administration showed some protection against carbon tetrachloride-induced hepatocellular damage in rats. Overall, these results suggest that KR62776 may have potential in the chemoprevention of liver fibrosis/cirrhosis.
Keywords: PPARγ, Stellate cells, Collagen α1(I), α-smooth muscle actin, KR62776
INTRODUCTION
Hepatic fibrosis is a clinically serious problem with chronic liver diseases caused by a variety of insults including viral infection, autoimmune liver diseases, and sustained alcohol abuse (Friedman, 2000). It is well-established that activated hepatic stellate cells (HSCs) play an important role in the development of liver fibrosis/cirrhosis (Eng et al., 2000; Reeves and Friedman, 2002). HSCs are derived from mesenchymal cells in the subendothelial space of an embryonic liver and are located behind the endothelial cells in the space of Disse. In a normal liver, HSCs exhibit quiescent phenotypes and contain vitamin A-rich lipid droplets. However, after liver injury, the quiescent HSC undergo differentiation into myofibroblast-like phenotypes with subsequent proliferation, leading to a significant increase in the synthesis of collagen α1(I) and α-smooth muscle actin (α-SMA) (Milani et al., 1990; Geerts et al., 1991; Blazejewski et al., 1995; Baroni et al., 1996).
Recent reports have shown that partial or full recovery from the experimental models of liver fibrosis can occur through the apoptosis of activated HSC, resulting in reduced collagen α1(I) and α-SMA levels (Zhao et al., 2003). Therefore, it is important to identify or discover certain agents that may directly promote the apoptosis of HSCs with the concomitant suppression of HSC activation. Recent studies demonstrated a beneficial role of peroxisome proliferator-activated receptors (PPARs) belonging to a family of ligand-activated transcription factors in HSC proliferation during liver injury (Dubuquoy et al., 2000). Among the three major types of PPARs (namely, PPARα, γ, and δ), PPARγ mRNA is expressed in the HSC of a normal rat liver but its expression is significantly decreased in HSC during liver fibrosis (Dubuquoy et al., 2000; Marra et al., 2000; Miyahara et al., 2000; Galli et al., 2002).
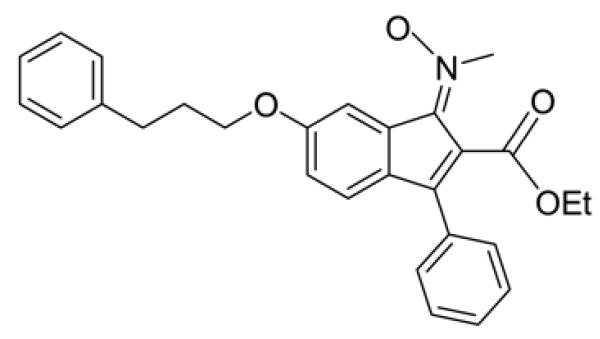
Accordingly, a few of ligands for PPARγ were reported to inhibit the proliferation of HSC and suppress the levels of α-SMA and collagen type I (Marra et al., 2000; Miyahara et al., 2000), possibly through the up-regulation of PPARγ expression. In addition, some PPARγ agonists were shown to induce the apoptosis of HSC (Kon et al., 2002; Enomoto et al., 2003; Xu et al., 2003; Ohata et al., 2004; Kawaguchi et al., 2004). Based on these reports, it was hypothesized that KR62776 (Fig. 1, Patent WO2005100303), a novel selective ligand of PPARγ, may suppress the rate of cell proliferation in the rat hepatic stellate cell line, HSC-T6 (Vogel et al., 2000), and prevent the liver fibrosis caused by carbon tetrachloride. These results showed that KR62776 promoted the apoptosis of activated stellate HSC-T6 cells without affecting the two different hepatoma cells. The apoptosis of HSC-T6 was determined to be mediated by PPARγ. These results further demonstrated that KR62776 can offer some protection against carbon tetrachloride-induced liver damage in rats, suggesting a new chemopreventive candidate against liver fibrosis.
Fig. 1.
Structure of KR62776. 1-(trans-methylimino-N-oxy)-3-phenyl-6-(3-phenylpropoxy)-1H-indene-2-carboxylic acid ethyl ester
MATERIALS AND METHODS
Materials
Unless specified otherwise, all chemicals used in this study were purchased from Sigma Chemicals. KR62776, 1-(trans-methylimino-N-oxy)-3-phenyl-6-(3-phenylpropoxy)-1H-indene-2-carboxylic acid ethyl ester, was synthesized at the Korea Research Institute of Chemical Technology. All tissue culture media and related reagents including fetal bovine serum were obtained from Invitrogen. The specific antibodies to the target protein analyzed were supplied by Santa Cruz Biotechnologies.
Cell culture and determination of cell viability
HSC-T6 cells, which were used as a convenient in vitro surrogate model to examine the mechanisms of the progressive development of hepatic fibrosis (Vogel et al., 2000), were kindly provided by Dr. Scott L. Friedman (Mount Sinai Medical School, New York). The HepG2 human hepatoma cells and McARH-7777 rat hepatoma cells were purchased from the American Type Culture Collection. The HSC-T6 cells were grown in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. The HSC-T6, HepG2 and McARH7777 cells, which were grown in 96-well microtiter plates (1×104 cells/well) for 1 day, were incubated with various concentrations of KR62776 (diluted in DMSO at 0.05% final concentration) for 48 h. The cell viability and cytotoxicity were measured by using commercial kits for the MTT and LDH assays, as described elsewhere (Bae et al., 2001).
Determination of apoptotic DNA fragments
The apoptotic DNA fragmentation of HSC-T6 cells treated with KR62776 was determined as previously elsewhere (Bae et al., 2001). Briefly, the supernatant of the cell lysates was incubated for 2 h at 37°C in the presence of RNase A, proteinase K and SDS (final 1%). The DNA fragments were then precipitated with 2.5 volumes of cold 100% ethanol in the presence of 0.5 M ammonium acetate and air-dried. The DNA sample was dissolved in 10 mM Tris-HCl and 1 mM EDTA buffer (pH 8.0), separated on 1.8% agarose gels and visualized by ethidium bromide staining under UV exposure.
RNA extraction and real time RT-PCR
The total RNA was extracted using an RNA easy kit (Promega) according to the manufacturer’s instruction. A reverse transcription-polymerase chain reaction (RT-PCR) was performed using a Platinum PCR Supermix kit (Invitrogen) after the total RNA (1 μg) had been reverse-transcribed in a 20 μL reaction volume by incubating with murine leukemia virus reverse transcriptase for 1 h at 42°C. Real time RT-PCR was performed using the following sets of specific primers: PPARγ1 (sense, 5′-GGCTGCGGCCTTAGT-ACATGTCCT-3′; antisense, 5′-TTCTGACAGGACTGT-GTGACAG-3′), PPARγ2 (sense, 5′-ATAAGGTGGAGA-TGCAGGTTC-3′; antisense, 5′-ATAAGGTGGAGA-TGCAGGTTC-3′), α-SMA (sense, 5′-TGTGCTGGACTC-TGGAGATG-3′; antisense, 5′-GATCACCTGCCCATC-AGG-3′), collagen α1 (I) (sense, 5′-CGATGGATTCCC-GTTCGAGTAC-3′; antisense, 5′-GTCCACAACCCT-GTAGGTG-3′), CD36 (sense, 5′-GGAGGCATTCTC-ATGCCGGTTGGAG-3′; antisense, 5′-TGAGAACTG-TGAAGTTGTCATTCTC-3′), and GAPDH (sense, 5′-ACCACAGTCCATGCCATCAC-3′; antisense, 5′-TCC-ACCACCCTGTTGCTGTA-3′). Each PCR mixture contained 5 μL of a Master SYBR®Green solution (Qiagen) and primers (1 μM). The instrument settings were as follows: holding at 94°C for 10 min; denaturing at 94°C for 10 s, annealing at 56°C for 30 s, and elongation at 72°C for 30 s for collagen α1 (I), PPAR γ1 and γ2; denaturing at 94°C for 10 s, annealing at 60°C for 30 s, and elongation at 72°C for 20 s for CD36; and denaturing at 94°C for 10 s, annealing at 63°C for 10 s, and elongation at 72°C for 20 s for α-SMA. All data was normalized to the the amount of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which was used as the internal control. The specificity of the PCR product for each tested gene was confirmed by gel electrophoresis.
Immunoblot analyses
The cellular lysates were prepared as previously described (Bae and Song, 2003), and equal amounts of the cell lysate protein (20 μg/well) were separated on 10% SDS polyacrylamide gels and transferred electrically to nitrocellulose membranes (Millipore Co.). Immunoblot analyses were performed by incubation with the primary antibodies specific to the target protein followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Enhanced chemiluminescence (ECL) was used to finally visualize the target protein.
Flow cytometric cell cycle analysis
After a treatment with 20 μM KR62776 for up to 72 h, both the adherent and floating HSC cells were combined, washed twice with cold PBS, fixed in 70% ice-cold ethanol with vortexing and finally stored at −20°C for at least 4 h. After two additional washes with PBS, the cell pellets were stained with a fluorescent probe solution containing 50 μg/mL propidium iodide, 0.1% Triton X-100, and 0.5 mg/mL RNaseA in PBS for 1 h at room temperature in the dark. The cells were then analyzed using a FACS Calibur cytometer (Becton Dickinson) (excitation at 488 nm; emission at 620 nm). The percentage of cells undergoing apoptosis was calculated from the percentage of cells in the distinct sub-diploid region of the DNA distribution histograms.
Effects of KR62776 on the expression of α-SMA in rat models of liver damage
Male Wistar rats, weighing 200 to 260 g, were obtained from Orient company (9 weeks of age, Orient Co., n = 5 or 6 per group). All rats were kept in specially prepared cages in a room with daylight for 12 h. The animal study was carried out in accordance with the NIH guidelines for the experimental animal studies. All Wistar rats were divided randomly into the following four groups: (1) normal control (olive oil), (2) liver damage control (1.0 mL/kg of CCl4), (3) liver damage (1.0 mL/kg of CCl4) + KR62776 (10 mg/kg), (4) liver damage (1.0 mL/kg of CCl4) + KR62776 (50 mg/kg). Throughout the experiment, all animals were given access to water and laboratory chow ad libitum. After 1 week acclimatization, each rat received a subcutaneous injection of an initial dose of 40% (v/v) carbon tetrachloride (1.0 mL/kg of CCl4), in olive oil twice per week for 3 weeks, as described previously (Zhao et al., 2003). KR62776, dissolved in 20% (v/v) Tween80, was administered orally at 10 or 50 mg/kg three times per week for 4 weeks. Seventy two hours after the final CCl4 dose, the animals were briefly anesthetized with pentobarbital and sacrificed by exsanguination from the inferior vena cava. The liver and serum samples were frozen immediately in liquid nitrogen and stored at −80°C until analyzed. The serum ALT and AST activities were analyzed by standard spectrophotomeric methods using commercial test reagents. Relative degree of liver damage was determined by a histological evaluation of hematoxylin and eosin (H&E)-stained slides and immunohistochemical staining with the anti-α-SMA antibody (Abcam Co.) after fixing the liver specimens in 10% buffered formaldehyde and embedding them in paraffin.
Statistical analysis
Each experiment was repeated three times, and the results are expressed as the mean ± S.D. To determine the effects of the KR62776 treatment, the data was evaluated by one-way ANOVA, followed by a nonparametric Turkey’s posthoc test. The mean values were compared using a Student’s t-test. A p-value < 0.05 was considered significant.
RESULTS
Effects of KR62776 on the cell proliferation rates of HSC-T6 cells
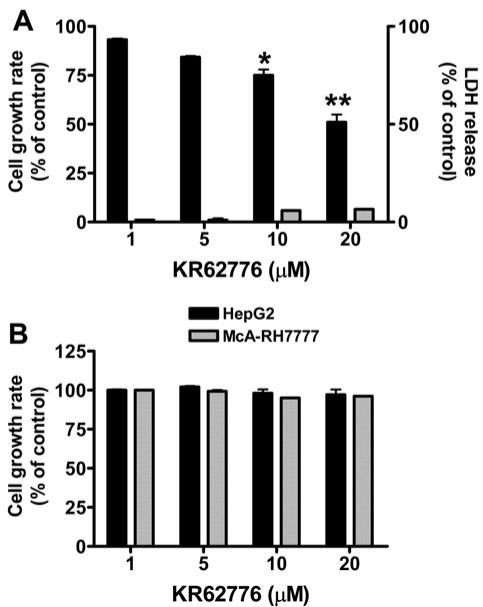
The effect of KR62776 on hepatic stellate cells was examined by treating the semi-confluent HSC-T6 cells with various concentrations of KR62776 for 48 h and measuring the rate of cell proliferation using a MTT reduction assay. As shown in Fig. 2A, KR62776 significantly decreased the growth rates of HSC-T6 cells in a concentration-dependent manner. Approximately 50% growth inhibition was detected at 20 μM KR62776, whereas no significant change was observed in the DMSO-treated control group. In contrast, KR62776 at the same concentration for up to 48 h did not significantly inhibit the growth of hepatoma HepG2 or McARH7777 cells, suggesting the specific action of KR62776 on stellate cells (Fig. 2B).
Fig. 2.
Effects of KR62776 on the cell growth rates o stellate HSC-T6 cells and hepatoma cells. (A) HSC-T6 cells (1×104 cells/well), grown in 96-well microtiter plates, were treated with different concentrations of KR62776 for 48 h. The cell viability and cytotoxicity were then measured sing a commercial kit for MTT (■) or LDH ( ) assay. The results represent the mean ± S.D. from three replicate experiments. *p < 0.05, **p < 0.01 versus control. (B) HepG2 hepatoma cells or McARH7777 hepatoma cells (1×104 cells/well), which were grown in 96-well plates, were treated with different concentrations of KR62776 for 48 h. The cell death rate was determined by a MTT reduction assay. The results represent the mean ± S.D. from three replicate experiments.
) assay. The results represent the mean ± S.D. from three replicate experiments. *p < 0.05, **p < 0.01 versus control. (B) HepG2 hepatoma cells or McARH7777 hepatoma cells (1×104 cells/well), which were grown in 96-well plates, were treated with different concentrations of KR62776 for 48 h. The cell death rate was determined by a MTT reduction assay. The results represent the mean ± S.D. from three replicate experiments.
Based on the antiproliferative effects of KR62776 on cultured HSC-T6 cells, it was hypothesized that the compound might cause the apoptosis of HSC-T6 cells. The morphology of the KR62776-treated HSC-T6 cells changed considerably into the round and lobulated appearances of typical apoptotic cells, while the DMSO-treated control cells remained the same (data not shown).
Effects of KR62776 on apoptosis of HSC-T6 cells
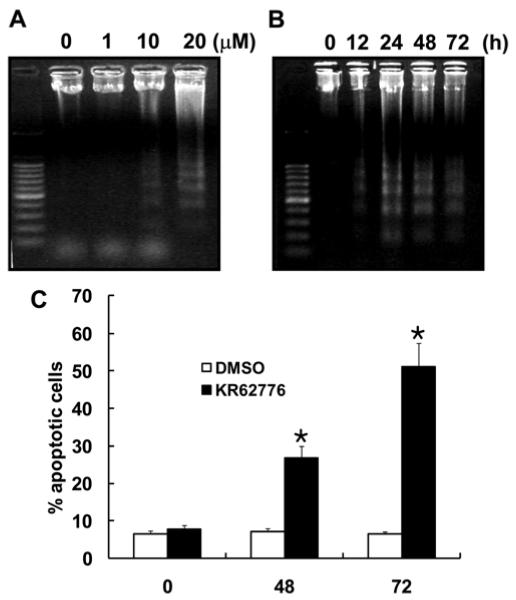
In order to obtain further evidence of apoptosis, therelative degree of DNA fragmentation, a hallmark of apoptosis, was determined after exposing the HSC-T6 cells to various concentrations of KR62776 for different times. As shown in Fig. 3A and B, KR62776 markedly induced DNA fragmentation in a time- and concentration-dependent manner, respectively. In addition, TUNEL staining, which is another marker of apoptosis, revealed the apoptotic effect of KR62776 on activated HSC-T6 cells (data not shown). This shows that the inhibition of HSC-T6 cell growth by KR62776 is mediated mainly by apoptosis.
Fig. 3.
Analysis of apoptosis in HSC-T6 cells by DN fragmentation and flow cytometry. Exponentially growing HSC-T6 cells were collected to assess the level of DNA fragmentation after exposure to different concentrations of KR62776 for 48 h (A) or for different times after exposure to 20 μM KR62776 (B). DNA samples were separated on 1.8% agarose gels and visualized by ethidium bromide staining. These results represent the typical examples of 3 independent determinations. The results represent one typical result of similar data from three independent experiments. Relative levels of the sub-G1 peak (apoptosis) was determined by using flow cytometric analysis after treating the HSC-T6 cells were treated with DMSO or 20 μM KR62776 for the indicated times. *p < 0.05 versus control (C).
The G1 phase transition state of the cell cycle is a critical point at which the cell assesses whether it should enter another full round of division, stop growing or die. Therefore, transition of the cell populations from the G1/G0 to subG1 phase is a frequent sign of apoptosis. As shown in Fig. 3C, the application of 20 μM KR62776 for 72 h significantly increased the accumulation of cells in the subG1 phase from 6.6 ± 0.8 (DMSO-vehicle control) to 51.2 ± 6.21%.
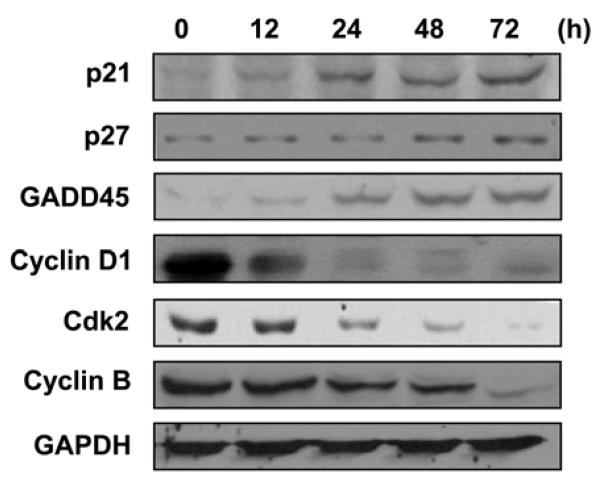
Because of the increased cell population in the subG1 phase after exposure to KR62776, the levels of cell cycle regulatory proteins, such as p21Waf1/cif1 (p21), p27kip1 (p27) and Gadd45, were also measured using the specific antibodies against each target protein. Immunoblot analyses showed that the KR62776 treatment gradually increased the levels of the cell cycle inhibitory proteins, such as p21, p27 and Gadd45, in a time-dependent manner (Fig. 4). In contrast, KR62776 markedly decreased the levels of the cell cycle-related proteins, including cyclin D1, cdk2 and cyclin B (Fig. 4) in a time-dependent manner.
Fig. 4.
Effects of KR62776 on the levels of the cell cyclerelated proteins in HSC-T6 cells. Cell lysate proteins (20 μg/lane) were separated on 10% SDS-polyacrylamide gels and subjected to immunoblot analysis with the specific antibody against p21, p27, GADD45, cyclin D1, cdk2, cyclin B, or GAPDH, followed by incubation with a horseradish peroxi-dase-conjugated secondary antibody. Detection of each antigen was performed using the ECL detection kit. The results shown are representative blots of three independent experiments.
Effects of KR62776 on the levels of marker genes for activated stellate cells
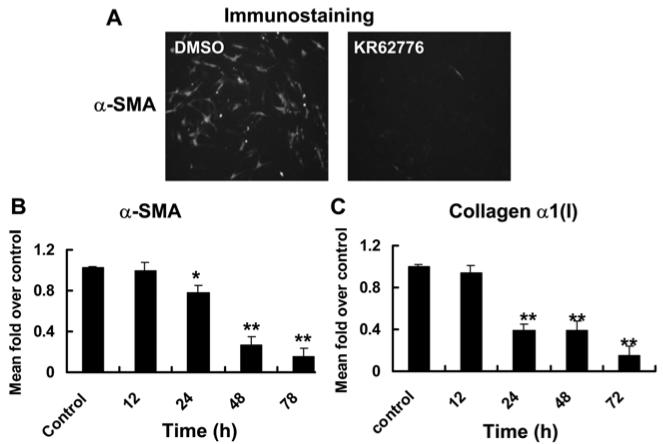
To determine the inhibitory effect of KR62776 on activated HSC-T6 cells, the levels of α-SMA protein expression was also examined by immunofluorescence staining using the specific anti-α-SMA antibody. As shown in Fig. 5A, KR62776 (20 μM) potently inhibited the expression of α-SMA protein. In addition, the levels of collagen α1(I) and α-SMA mRNAs, as surrogate markers of activated stellate cells, were measured to confirm the consequences of the inhibition of activated HSC-T6 cells by KR62776. As shown in Fig. 5B and C, considerable amounts of collagen α1(I) and α-SMA mRNA transcripts were detected in the DMSO-treated control HSC-T6 cells. However, KR62776 decreased the mRNA levels of collagen α1(I) and α-SMA significantly in a time-dependent manner, beginning from 24 h after treatment. These results suggest that KR62776 caused the apoptosis of HSC-T6 cells accompanied by the pretranslational suppression of the surrogate marker genes of the activated stellate cells.
Fig. 5.
Effects of KR62776 on the expression of α-SMA collagen α1(I). (A) The relative expression of α-SMA in HSC-T6 cells treated with DMSO or 20 μM KR62776 for 48 h was determined by immunofluorescence staining using the specific anti-α-SMA antibody. These slides represent the typical examples of 3 independent determinations. The total RNA was isolated from the HSC-T6 cells treated with 20 μM KR62776 for the indicated times, and the relative levels of α-SMA (B) and collagen α1(I) (C) mRNA transcripts were measured by real time RT-PCR. The levels of each transcript were calculate using the DDCT method and compared with the control. The results represent the mean ± S.D. from three replicate experiments. *p < 0.05, **p < 0.01 versus the control.
Specific induction of PPARγ1 and CD36 by KR62776 in HSC-T6 cells
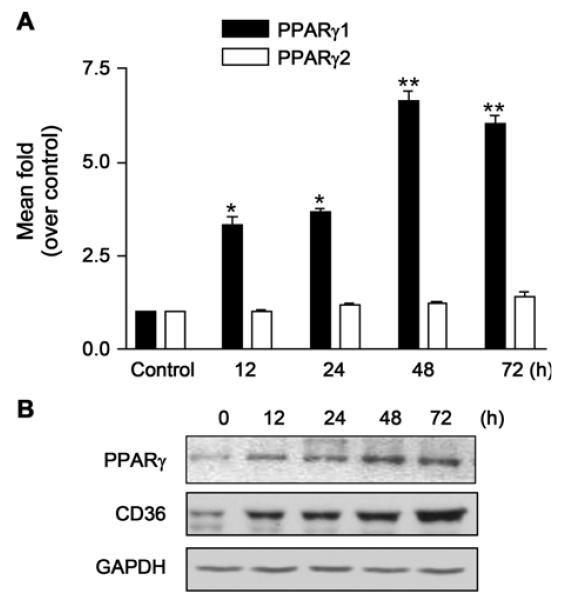
Since the expression of PPARγ1 appears to be inversely associated with the activation of stellate cells (Miyahara et al., 2000; Galli et al., 2002), this study examined whether KR62776 directly affects the expression of PPARγ1 and PPARγ2. Compared with the control, KR62776 significantly increased the levels of PPARγ1 mRNA (Fig. 6A) in a time-dependent manner with little effect on PPARγ2 expression. Consistent with the up-regulation of the PPARγ protein, the amount of CD36, a scavenger receptor containing a PPAR response element (PPRE) in its promoter region (Sato et al., 2002), was also increased by the KR62776 treatment (Fig. 6B).
Fig. 6.
Effects of KR62776 on the expression of PPAR mRNA and proteins. (A) The total RNA was isolated from the HSC-T6 cells treated with 20 μM KR62776 for the indicated times. The mRNA levels of PPARγ1 and PPARγ2 were determined by real time RT-PCR, and the relative levels of PPARγ mRNAs over the control were calculated using the DDCT method. The results represent the means ± S.D. from three replicate experiments. *p < 0.05, **p < 0.01 versus control (0 h). (B) The cell lysate proteins (20 μg/lane) were separated on 10% SDS-polyacrylamide gels and subjected to immunoblot analysis with the specific antibody against PPARγ or CD36 after the HSC-T6 cells were treated with 20 μM KR62776 for the indicated times. Detection of each antigen was performed using an ECL detection kit. The results shown are representative blots of three independent experiments.
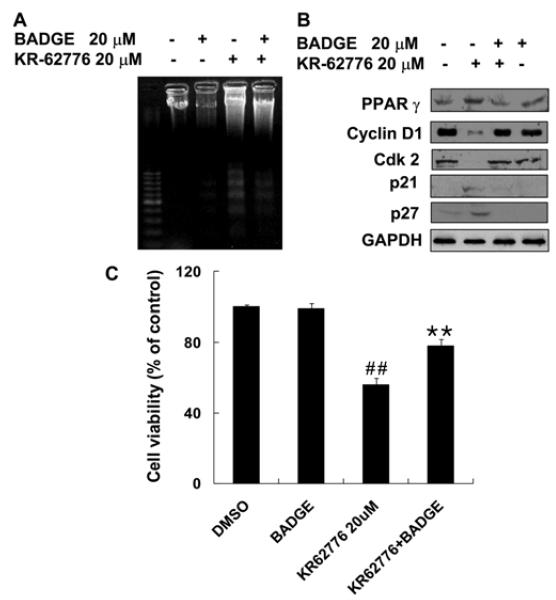
The involvement of PPARγ in the KR62776-mediated effects was investigated by examining the effect of a specific PPARγ inhibitor, BADGE, on KR62776-induced anti-proliferation in HSC-T6 cells. As shown in Fig. 7A and C, 20 μM BADGE blocked the KR62776-induced DNA fragmentation and apoptotic activity. Consistent with these results, immunoblot analyses showed that a pretreatment with BADGE effectively prevented the KR62776-mediated increase in the PPARγ protein (Fig. 7B). In addition, BADGE reversed the KR62776-mediated changes in the cell cycle regulatory proteins (cyclin D1, cdk2, p21 and p27). These results show that the antiproliferation action of KR62776 in HSC-T6 cells is mediated, at least partially, by PPARγ activation or the up-regulation of its expression.
Fig. 7.
Effects of BADGE on the KR62776-induced DN fragmentation, cell viability, and amounts of cell cyclerelated proteins. (A and C) After HSC-T6 cells were pre-incubated with 20 μM BADGE for 1 h, the cells were exposed to 20 μM KR62776 for an additional 48 h. The degree of DNA fragmentation and the cell death rates were determined, as described in Materials and Methods. The results represent the means ± S.D. from three replicate experiments. **p < 0.01 versus KR62776-treated sample. ##p < 0.01 versus control. (B) Another set of HSC-T6 cells, which were treated with KR62776 in the absence or presence of BADGE for 48 h, was harvested. Equal amounts of the cell lysate proteins (20 μg/lane) were separated on 10% SDS-polyacrylamide gels and subjected to immunoblot analysis. Each membrane was probed with the specific antibody against PPARγ, cyclin D1, cdk2, p21, p27, or GAPDH, followed by incubation with the horseradish per-oxidase-conjugated secondary antibody. Detection of each protein was performed using an ECL Western blotting detection system. The results shown are representative blots of three independent experiments.
In vivo effects of KR62776 in a rat model of liver fibrosis
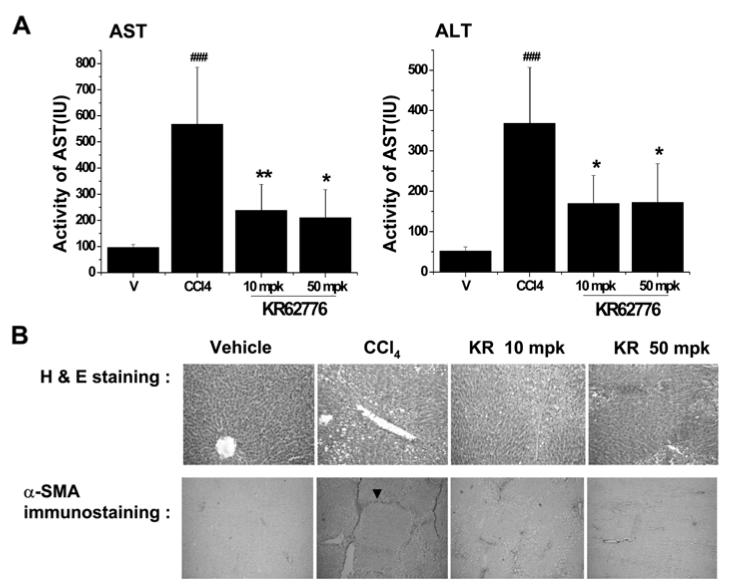
To determine the beneficial effects of KR62776 against liver damage in an animal model, Wistar rats were treated with repeated injections of carbon tetrachloride with or without the simultaneous administration of KR62776 (10 and 50 mg/kg, p.o). The average liver index (the ratio of liver weight to body weight) in the CCl4-treated Wistar rats was significantly higher than that of the controls (data not shown). However, the liver index in the KR62776-treated group (50 mg/kg) was similar to the control group. In addition, serum aspartate aminotransferase (AST) and alanine amino-transferase (ALT) levels were significantly higher in the CCl4-treated rats than the corn-oil treated control group (Fig. 8A). The increased levels of serum AST and ALT observed in the CCl4-treated group were significantly lower in the KR62776-treated groups. In addition, Fig. 8B (upper panel) shows the typical patterns of the liver histology stained with H&E. Normal hepatic architectures were observed in cornoil (vehicle) treated control rats. However, many in-filtrated inflammatory cells were detected in addition to mild or moderate liver damage in the pericentral regions of the CCl4-treated rat livers (Fig. 8B, upper panel). This liver damage caused by chronic CCl4 administration was prevented by a concomitant KR62776 treatment (Fig. 8B, upper panel). The hepatic architecture appeared greatly improved with lower levels of α-SMA at a higher dose of KR62776 (50 mg/kg) (Fig. 8B, upper panel).
Fig. 8.
In vivo effects of KR62776 on the serum transaminase activities, liver histology and expression of α-SMA in rat. (A and B) in the vehicle control, CCL4 and KR62776 treated rats. Male Wistar rats (n = 5-6/group) were treated with repeate injections of CCl4 in the absence or presence of KR62776 (10 and 50 mg/kg). (A) Serum transminase activities in the Vehicle, CCl4 and KR62776 treated groups. The data is reported as the mean ± S.D. (n = 5-6). One-way ANOVA was used for statistical analyses. *p < 0.05, **p < 0.01 versus CCl4-treated sample. ###p < 0.001 versus control. (B) Small pieces from each rat liver from different groups, as indicated, were fixed immediately with 10% formalin and subjected to H&E staining (upper panel). The fixed tissue paraffin blocks from each rat liver were cut into 5 micron-wide sections, placed onto glass slides and subjected to immunohistochemical detection for α-SMA expression using the specific antibody (bottom panel). These slides represent typical examples of 5 or 6 independent determinations.
The changes in the amounts of α-SMA expressed in the rat livers were also evaluated by immunohistochemical staining (Fig. 8B, bottom panel). As expected, the amounts and intensities of α-SMA expressed in the CCl4-treated rat livers were significantly higher than those in the corn-oil treated animals. The majority of intensely stained α-SMA was also observed in the pericentral regions. However, the increased amounts of α-SMA in the CCl4-treated rats were suppressed by KR62776. There were few changes in H&E staining and immunostaining for α-SMA in the rat livers after treatment with KR62776 alone (data not shown), which is similar to those of the corn-oil treated control rats. Comparable protective effects of KR62776 against CCl4-mediated liver injury were observed in mice (data not shown). These results suggest that KR62776 can prevent the hepatic damage caused by a chronic treatment with CCl4 in an in vivo model.
DISCUSSION
Liver fibrosis/cirrhosis can result from damaged hepatic parenchymal cells with the concomitant activation of stellate cells after chronic and multiple exposures to high doses of alcohol, hepatic viruses, autoimmune liver diseases, or potentially toxic compounds, including some drugs used in clinics. During liver fibrosis, the quiescent HSCs become activated with the excess production of extracellular matrix components, particularly collagen α1(I) and α-SMA. On the other hand, the apoptosis or inhibition of activated stellate cells reduced the accumulation of collagen α1(I) and α-SMA (Zhao et al., 2003), resulting in the partial or full recovery from the experimental models of fibrosis.
Many compounds have been developed to prevent or treat fibrosis and cirrhosis. These chemicals include HOE-077 (Sakaida et al., 1996), halofuginone (Pines et al., 1997; Bruck et al., 2001) and pirfenidone (Tada et al., 2001; Garcia et al., 2002), and appear to act by inhibiting collagen α1(I) biosynthesis. PPARγ, a nuclear receptor with transcriptional activity, has been shown to play an important role in the cellular physiology and metabolism for modulating the glucose and lipid metabolism/homeostasis (Berger and Moller, 2002; Rangwala and Lazar, 2004). Recently, the involvement of PPARγ in the protective effects against hepatic fibrosis was reported but the results are controversial. Li et al. (2001) reported that 15-deoxy-Δ12, 24-PG J2, an endogeneous PPARγ ligand, causes the apoptosis of hepatic myofibroblasts, which are similar to fully differentiated stellate cells, via a PPARγ-independent mechanism. In contrast, the majority of other reports (Galli et al., 2002; Kon et al., 2002; Enomoto et al., 2003; Xu et al., 2003; Ohata et al., 2004; Kawaguchi et al., 2004; Promrat et al., 2004) suggested that pioglitazone and rosiglitazone prevent experimental fibrosis mainly through the activation of PPARγ.
This study further examined the protective role of PPARγ against hepatic fibrosis using a new PPARγ agonist, KR62776, which is a structurally different compound from the existing PPARγ agonists with a canonical thiazolidinedione structure. These results showed that KR62776 had antiproliferative and apoptotic effects on the activated stellate cells. The apoptotic effects appear to be associated with the inhibition of the cell cycle related proteins. Previous studies suggested that cyclin D1 may play key roles in cell cycle progression, particularly at the early G0/G1 phase (Morgan, 1996; Sherr and Roberts, 1999). The inhibition of cyclin D1 with specific antibodies or anti-sense cyclin D1 prevented the target cells from entering the S phase. In this study, KR62776 markedly altered the expression of the cell cycle-regulatory proteins, such as cyclin D1, cyclin B and cdk2. These results are consistent with the previous observations that PPARγ activation by PGJ2, PGD2, BRL-49653 or troglitazone selectively inhibited the expression of cyclin D1 (Wang et al., 2001). The mechanism for the modulation of the cell cycle-related proteins by KR62776 is unknown and warrants further study.
The level of the PPARγ protein in cultured HSCs decreases upon activation, whereas PPARγ up-regulation by its ligands inhibits the proliferation of HSC and the expression of extracellular matrix components. For example, a recent report showed that PPARγ1 mRNA is expressed in normal HSCs, while it is not detected in the HSCs isolated from livers with cholestatic fibrosis induced by bile duct ligation, suggesting an inverse relationship between PPARγ1 and fibrosis (i.e., stellate cell activation) (Miyahara et al., 2000). The present results showing the specific effect of KR62776 on the apoptosis of HSCs without affecting two different hepatoma cells (Fig. 2) indicate that KR62776 exerts its beneficial effect through the PPARγ1 isoform expressed in stellate cells. Indeed, the levels of PPARγ1 mRNA and protein in HSC were increased by KR62776 in this study. Therefore, it isreasonable to hypothesize that the PPARγ1 pathway is important for maintaining the quiescent phenotypes of HSCs. This study further demonstrated that KR62776 inhibits the proliferation of activated HSC-T6 cells by promoting apoptosis, leading to a decrease in the levels of collagen α1(I) and α-SMA mRNAs. The regulatory effects of KR62776 on the α-SMA and collagen α1(I) expression could result from transcriptional activation, even though the presence of PPRE in the promoter region of α-SMA and collagen α1(I) is unknown. Nevertheless, the action of KR62776 in stellate cells appears to be, at least partially, mediated by PPARγ1 activation because BADGE, a specific PPARγ antagonist, effectively blunted the KR62776-induced apoptosis and antiproliferative action.
Recently, Date et al. (2003) reported that 15-deoxy-Δ12, 24-PGJ2 induces apoptosis in human hepatoma cells, such as Huh-7 and HepG2. However, they did not examine or mention the role of PPARγ in directly promoting apoptosis of these cells. In addition, Bae et al. (2003) demonstrated that TRO induced apoptosis in human hepatoma cell lines via a PPARγ-independent pathway by activating the cell-death related stress-activated protein kinases. Previous studies showed that KR62776 binds to the ligand binding domain of PPARγ in a different mode from that of rosiglitazone, which is a well known PPARγ ligand with a thiazolidinedione structure (Ahn et al., 2006). Since KR62776 showed no or little effect on human hepatoma cells, it is believed that the novel structure of KR62776 might be important for exhibiting the selective inhibitory action on stellate cells without affecting the cells with a hepatic origin.
It was reported that a liver injury can be induced either by ligation of the common bile duct or by the chronic administration of hepatotoxins, such as CCl4 (Wu et al., 1996). Therefore, a model of rat liver injury and liver damage by repeated treatment with CCl4 was used in this study to evaluate the protective effect of KR62776 against hepatic damage. KR62776 not only improved the hepatic architecture, as determined by H&E staining, but also reduced the levels of α-SMA detected by immunohistochemical staining. The suppression of α-SMA by KR62776 in animal studies is consistent with the in vitro result from rat HSC-T6 cells.
In conclusion, PPARγ activation by KR62776, a novel agonist, inhibited the proliferation of HSC-T6 cells by promoting apoptosis and cell cycle arrest. Furthermore, KR62776 had beneficial effects in a rodent model of hepatic injury caused by the repeated administration of carbon tetrachloride. Overall, KR62776 may have a therapeutic potential for the prevention and treatment of liver fibrosis/cirrhosis.
ACKNOWLEDGEMENTS
This study was supported by grant from the Center for Biological Modulators of the 21th Century Frontier R&D Program, The Ministry of Science and Technology, Korea.
REFERENCES
- Ahn JH, Shin MS, Jung SH, Kang SK, Kim KR, Rhee SD, Jung WH, Yang SD, Kim SJ, Woo JR, Lee JH, Cheon HG, Kim SS. Indene derivatives: A novel template for peroxosome proliferator-activated receptor γ (PPARγ) agonists. J. Med. Chem. 2006;49:4781–4784. doi: 10.1021/jm060389m. [DOI] [PubMed] [Google Scholar]
- Bae MA, Pie JE, Song BJ. Acetaminophen induces apoptosis of C6 glioma cells by activating the c-Jun NH2-terminal protein kinase-related cell death pathway. Mol. Pharmacol. 2001;60:847–856. [PubMed] [Google Scholar]
- Bae MA, Song BJ. Critical role of c-Jun N-terminal protein kinase activation in troglitazone-induced apoptosis of human HepG2 hepatoma cells. Mol. Pharmacol. 2003;63:401–408. doi: 10.1124/mol.63.2.401. [DOI] [PubMed] [Google Scholar]
- Baroni GS, D’Ambrosio L, Curto P, Casini A, Mancini R, Jezequel AM, Benedetti A. Interferon gamma decreases hepatic stellate cell activation and extracellular matrix deposition in rat liver fibrosis. Hepatology. 1996;23:1189–1199. doi: 10.1002/hep.510230538. [DOI] [PubMed] [Google Scholar]
- Berger J, Moller DE. The mechanisms of action of PPARs. Annu. Rev. Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- Blazejewski S, Preaux AM, Mallat A, Brocherion I, Mavier P, Dhumeaux D, Hartmann D, Schuppan D, Rosenbaum J. Human myofibroblast-like cells obtained by outgrowth are representative of the fibrogenic cells in the liver. Hepatology. 1995;22:788–797. [PubMed] [Google Scholar]
- Bruck R, Genina O, Aeed H, Alexiev R, Nagler A, Avni Y, Pines M. Halofuginone to prevent and treat thio-acetamide-induced liver fibrosis in rats. Hepatology. 2001;33:379–386. doi: 10.1053/jhep.2001.21408. [DOI] [PubMed] [Google Scholar]
- Date M, Fukuchi K, Morita S, Takahashi H, Ohura K. 15-Deoxy-delta12,14-prostaglandin J2, a ligand for peroxisome proliferators-activated receptor-gamma, induces apoptosis in human hepatoma cells. Liver Int. 2003;23:460–466. doi: 10.1111/j.1478-3231.2003.00877.x. [DOI] [PubMed] [Google Scholar]
- Dubuquoy L, Dharancy S, Nutten S, Petterson S, Auwerx J, Desreumaux P. Role of peroxisome proliferator-activated receptor and retinoid × receptor heterodimer in hepatogastroenterological diseases. Lancet. 2000;360:1410–1418. doi: 10.1016/S0140-6736(02)11395-X. [DOI] [PubMed] [Google Scholar]
- Enomoto N, Takei Y, Hirose M, Konno A, Shibuya T, Matsuyama S, Suzuki S, Kitamura KI, Sato N. Prevention of ethanol-induced liver injury in rats by an agonist of peroxisome proliferator-activated receptor-gamma, pioglitazone. J. Pharmacol. Exp. Ther. 2003;306:846–854. doi: 10.1124/jpet.102.047217. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- Galli A, Crabb DW, Ceni E, Salzano R, Mello T, Svegliati-Baroni G, Ridolfi F, Trozzi L, Surrenti C, Casini A. Antidiabetic thiazolidinediones inhibit collagen synthesis and hepatic stellate cell activation in vivo and in vitro. Gastroenterology. 2002;122:1924–1940. doi: 10.1053/gast.2002.33666. [DOI] [PubMed] [Google Scholar]
- Garcia L, Hernandez I, Sandoval A, Salazar A, Garcia J, Vera J, Grijalva G, Muriel P, Margolin S, Armendariz-Borunda J. Pirfenidone effectively reverses experimental liver fibrosis. J. Hepatol. 2002;37:797–805. doi: 10.1016/s0168-8278(02)00272-6. [DOI] [PubMed] [Google Scholar]
- Geerts A, Lazou JM, De Bleser P, Wisse E. Tissue distribution, quantitation and proliferation kinetics of fatstoring cells in carbon tetrachloride-injured rat liver. Hepatology. 1991;13:1193–1202. [PubMed] [Google Scholar]
- Kawaguchi K, Sakaida I, Tsuchiya M, Omori K, Takami T, Okita K. Pioglitazone prevents hepatic steatosis, fibrosis, and enzyme-altered lesions in rat liver cirrhosis induced by a choline-deficient L-amino acid-defined diet. Biochem. Biophys. Res. Commun. 2004;315:187–195. doi: 10.1016/j.bbrc.2004.01.038. [DOI] [PubMed] [Google Scholar]
- Kon K, Ikejima K, Hirose M, Yoshikawa M, Enomoto N, Kitamura T, Takei Y, Sato N. Pioglitazone prevents early-phase hepatic fibrogenesis caused by carbon tetrachloride. Biochem. Biophys. Res. Commun. 2002;291:55–61. doi: 10.1006/bbrc.2002.6385. [DOI] [PubMed] [Google Scholar]
- Li L, Tao J, Davaille J, Feral C, Mallat A, Rieusset J, Vidal H, Lotersztajn S. 15-Deoxy-Δ12,14-prostag-landin J2 induces apoptosis of human hepatic myofibroblasts. A pathway involving oxidative stress independently of peroxisome-proliferator-activated receptors. J. Biol. Chem. 2001;276:38152–38158. doi: 10.1074/jbc.M101980200. [DOI] [PubMed] [Google Scholar]
- Marra F, Efsen E, Romanelli RG, Caligiuri A, Pastacaldi S, Batignani G, Bonacchi A, Caporale R, Laffi G, Pinzani M, Gentilini P. Ligands of peroxisome proliferator-activated receptor g modulate profibrogenic and proinflammatory actions in hepatic stellate cells. Gastroenterology. 2000;119:466–478. doi: 10.1053/gast.2000.9365. [DOI] [PubMed] [Google Scholar]
- Milani S, Herbst H, Schuppan D, Kim KY, Riecken EO, Stein H. Procollagen expression by nonparenchymal rat liver cells in experimental biliary fibrosis. Gastroenterology. 1990;98:175–184. doi: 10.1016/0016-5085(90)91307-r. [DOI] [PubMed] [Google Scholar]
- Miyahara T, Schrum L, Rippe R, Xiong S, Yee HF, Jr, Motomura K, Anania FA, Willson TM, Tsukamoto H. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J. Biol. Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1996;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Ohata M, Suzuki H, Sakamoto K, Hashimoto K, Nakajima H, Yamauchi M, Hokkyo K, Yamada H, Toda G. Pioglitazone prevents acute liver injury induced by ethanol and lipopolysaccharide through the suppression of tumor necrosis factor-a. Alcohol. Clin. Exp. Res. 2004;28:139S–144S. doi: 10.1097/01.alc.0000134412.38510.f7. [DOI] [PubMed] [Google Scholar]
- Pines M, Knopov V, Genina O, Lavelin I, Nagler A. Halofuginone, a specific inhibitor of collagen type I synthesis, prevents dimethylnitrosamine-induced liver cirrhosis. J. Hepatol. 1997;27:391–398. doi: 10.1016/s0168-8278(97)80186-9. [DOI] [PubMed] [Google Scholar]
- Promrat K, Lutchman G, Uwaifo GI, Freedman RJ, Soza A, Heller T, Doo E, Ghany M, Premkumar A, Park Y, Liang TJ, Yanovski JA, Kleiner DE, Hoofnagle JH. A pilot study of pioglitazone treatment for nonalcoholic steatohepatitis. Hepatology. 2004;39:188–196. doi: 10.1002/hep.20012. [DOI] [PubMed] [Google Scholar]
- Rangwala SM, Lazar MA. Peroxisome proliferator-activated receptor gamma in diabetes and metabolism. Trends Pharmacol. Sci. 2004;25:331–336. doi: 10.1016/j.tips.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Reeves HL, Friedman SL. Activation of hepatic stellate cells-a key issue in liver fibrosis. Front Biosci. 2002;7:808–826. doi: 10.2741/reeves. [DOI] [PubMed] [Google Scholar]
- Sakaida I, Matsumura Y, Kubota M, Kayano K, Takenaka K, Okita K. The prolyl 4-hydroxylase inhibitor HOE-077 prevents activation of Ito Cells, reducing procollagen gene expression in rat liver fibrosis induced by choline-deficient L-amino acid-defined diet. Hepatology. 1996;23:755–763. doi: 10.1053/jhep.1996.v23.pm0008666329. [DOI] [PubMed] [Google Scholar]
- Sato O, Kuriki C, Fukui Y, Motojima K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J. Biol. Chem. 2002;277:15703–15711. doi: 10.1074/jbc.M110158200. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1 phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Tada S, Nakamuta M, Enjoji M, Sugimoto R, Iwamoto H, Kato M, Nakashima Y, Nawata H. Pirfenidone inhibits dimethylnitrosamine-induced hepatic fibrosis in rats. Clin Exp. Pharmacol. Physiol. 2001;28:522–527. doi: 10.1046/j.1440-1681.2001.03481.x. [DOI] [PubMed] [Google Scholar]
- Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, Blaner WS. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J. Lipid Res. 2000;41:882–893. [PubMed] [Google Scholar]
- Wang C, Fu M, D’Amico M, Albanese C, Zhou JN, Brownlee M, Lisanti MP, Chatterjee VK, Lazar MA, Pestell RG. Inhibition of cellular proliferation through IkappaB kinase-independent and peroxisome proliferator-activated receptor gamma-dependent repression of cyclin D1. Mol. Cell. Biol. 2001;21:3057–3070. doi: 10.1128/MCB.21.9.3057-3070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Norton PA. Animal models of liver fibrosis. Scand. J. Gastroenterol. 1996;31:1137–1143. doi: 10.3109/00365529609036901. [DOI] [PubMed] [Google Scholar]
- Xu J, Fu Y, Chen A. Activation of peroxisome proliferator-activated receptor-gamma contributes to the inhibitory effects of curcumin on rat hepatic stellate cell growth. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:20–30. doi: 10.1152/ajpgi.00474.2002. [DOI] [PubMed] [Google Scholar]
- Zhao WX, Zhao J, Liang CL, Zhao B, Pang RQ, Pan XH. Effect of caffeic acid phenethyl ester on proliferation and apoptosis of hepatic stellate cells in vitro. World J. Gastroenterol. 2003;9:1278–1281. doi: 10.3748/wjg.v9.i6.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]