Abstract
Heat shock proteins (Hsps) are increasingly implicated in aging phenotypes and control of lifespan across species. They are targets of the conserved heat shock factor (HSF) and insulin/IGF1-like (IIS) signaling pathways that affect lifespan and aging phenotypes. Hsps are expressed in tissue-specific and disease-specific patterns during aging, and their level of expression and induction by stress both correlates with, and in some instances predicts, lifespan. In model organisms, Hsps have been shown to increase lifespan and ameliorate aging-associated proteotoxicity. Finally, Hsps have emerged as key components in regulating aging-related cellular phenotypes including cell senescence, apoptosis and cancer. The Hsps therefore provide a metric of individual stress and aging and are potential targets for interventions in aging and aging-related diseases.
Stress resistance links Hsps with aging
Aging in biological systems is more correctly termed senescence, although for the purposes of this review, the common term aging is used for convenience (Box 1). Unless otherwise indicated, “old” refers to animals or cells that are greater than the median lifespan for that species/genotype. This review discusses the heat shock proteins (Hsps) with respect to aging (senescence) and certain aging-related diseases, with a focus on research of the last five years.
Box 1. What is aging?
The process we refer to as “aging” in biological systems is more correctly termed senescence (see Glossary); however, the word aging is commonly used for convenience. There are a number of ways to define aging [78]. Most researchers would likely agree with the basic definition of aging as a deteriorative change that causes increased mortality. Additional definitions include a cumulative, irreversible process associated with decreased reproductive fitness. Numerous cellular and molecular correlates of aging have been identified in various species, the most general being the accumulation of macromolecular damage and mutations.
It should be recognized that lifespan is not the same thing as aging; however, lifespan is still generally the most accepted and reliable metric for aging rate. Notably, the search for additional or better metrics of aging rate (biomarkers) is an area of active research, and has identified the Hsps. Lifespan is a quantitative trait that can be described and compared in various ways. The number of individuals dying per unit time is the mortality rate, and an exponential increase in mortality rate with age has also been used as a definition of aging.
The heat shock proteins (Hsps) are increasingly implicated in aging phenotypes and control of lifespan across species. The Hsps are robustly induced in response to a variety of stresses (both intrinsic and extrinsic), and are key mediators of the organism’s resistance to stress [1] (Box 2). Several aging theories posit that lifespan is causally related to the ability of the organism to resist intrinsic and extrinsic stress, which predicts that lifespan should positively correlate with stress resistance [2, 3]. Indeed, lifespan has been found to positively correlate with stress resistance in various species and this correlation holds true when lifespan is experimentally increased using single-gene mutations, dietary restriction (DR), or genetic selection of populations.
Box 2. Proteotoxicity, Hsps and Aging.
Proteins can become denatured (unfolded) due to mutations, heat, oxidative damage, and a variety of chemical modifications. Denatured proteins are a basic trigger for increased Hsp expresion (Figure 1). Protein denaturation exposes hydrophobic amino acid residues that can interact inter-molecularly to cause protein aggregation and precipitation. These hydrophobic residues are also the binding sites for Hsps, which can prevent aggregation and facilitate refolding. Hsps can also promote the entry of abnormal proteins into degradation pathways.
Protein aggregates and precipitates are resistant to degradation and can be toxic to cells (proteotoxicity) [1, 71]. The possible mechanisms by which abnormal proteins, protein aggregates and precipitates are toxic to cells include generation of ROS, physical disruption of membranes, and activation of apoptosis. By preventing the formation of toxic protein species, sequestering such species, and/or facilitating their degradation, the Hsps promote cell survival and function during stress.
One of the first molecular markers of aging identified was the accumulation of abnormal (conformationally altered) proteins, specifically the accumulation of inactive enzymes in aging nematodes [10]. Since then, denatured and aggregated proteins, typically associated with Hsps, have been found to mark aging cells in virtually every species examined. In humans, numerous aging-related diseases involve the accumulation of abnormally processed and/or folded proteins (e.g, Alzheimer’s Disease, Parkinson’s Disease, Prion diseases), and these aggregates are typically associated with Hsps [1]. The ability of the Hsps to counter proteotoxicity might be one mechanism by which they promote longevity, consistent with the correlation between a robust stress response and long life. At the same time, during aging, chronic expression and accumulation of Hsps might become toxic in some contexts, perhaps due to disruption of signaling pathways.
Classes and functions of Hsps and their induction by stress
The Hsps are molecular chaperones and have in common the property of being able to affect the conformation of other proteins [1]. The Hsps include constitutively expressed and stress-inducible forms and function to facilitate correct folding of newly synthesized polypeptides, regulate assembly and disassembly of multi-protein complexes, and to regulate intracellular protein trafficking and translocation across membranes. Several families of chaperones were named based on their induction by heat stress (heat shock proteins or Hsps) and their molecular weights: Hsp100, Hsp90, Hsp70, Hsp60, Hsp40 and the small heat shock proteins (sHsps). The constitutively expressed forms of Hsp70 are often referred to as heat-shock-cognate 70 (Hsc70). The Hsps also function with co-factors (co-chaperones) that confer specificity for substrates and functions [1]. A common role for Hsp70, Hsp90 and co-chaperone complexes is in regulating transcription factors (including p53 and HSF; see Figure 1) and signal-transduction pathway components (including the oncogene product Ras, steroid aporeceptors, kinases and phosphatases), such that the clients are maintained in an inactive state that is poised for rapid conformational change and activation.
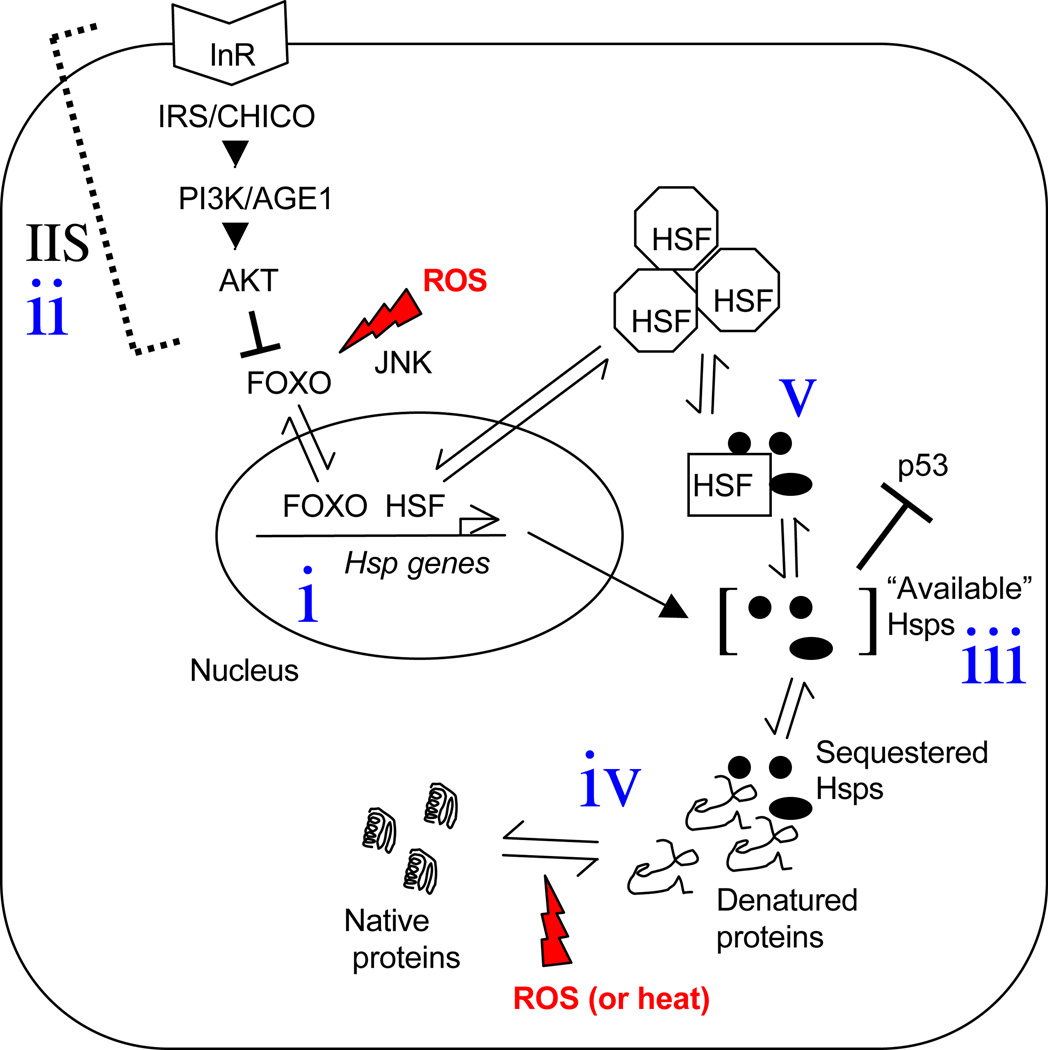
Figure 1.
Regulation of Hsp genes by HSF and FOXO. Three signaling pathways (HSF, IIS and JNK) converge upon the transcription factors FOXO and HSF to activate expression of Hsp genes. (i) FOXO and HSF transcription factors reside in the nucleus, bound to the promoter of an Hsp gene, where they activate expression of Hsps and other genes and promote stress resistance and longevity. ROS (Reactive Oxygen Species; shown in red) can activate the JNK stress-response pathway (and other factors) that in turn activate the FOXO transcription factor and facilitate its translocation into the nucleus. (ii) IIS acts to negatively regulate the activity of FOXO. The insulin-like receptor (InR in Drosophila; Daf2 in C. elegans) is activated by binding of insulin-like ligands (not shown). The InR initiates a signal transduction cascade involving the Insulin Receptor Substrate (called IRS in mammals and CHICO in Drosophila), PI3K (AGE1 in C. elegans) and the serine/threonine protein kinase AKT. AKT phosphorylates FOXO, which results in FOXO remaining sequestered in the cytoplasm. In this way, active signaling through the IIS pathway negatively regulates FOXO, Hsp expression, and lifespan. (iii) Although Hsps are abundant in cells, there appears to be very little excess, as partial reductions in specific Hsps using RNAi tends to have profound effects. Hsps that are “available” for binding to a client, such as a newly denatured protein, become so by dissociating from other clients. For example, the denaturation of cytosolic proteins by ROS or heat (iv) reveals hydrophobic amino acid residues that bind and sequester Hsps (Hsps indicated as black ovals). The Hsps are titrated away from other clients, including HSF. Dissociation of the Hsps from HSF monomers allows HSF to convert to the active trimeric form, translocate to the nucleus, and activate the expression of Hsp genes. The resultant high-level expression of the Hsp genes produces sufficient Hsp protein to again bind to and inactivate HSF (v), thereby providing a negative feedback to terminate the response. The Hsps bind to a wide variety of clients, including inhibitory interactions with the tumor-suppressor (and senescence-regulatory protein) p53. In this way, stresses such as heat or oxidative stress that cause the denaturation of proteins can coordinately affect a variety of pathways by sequestering the Hsps away from their networks of clients. HSF, heat shock factor; IIS, insulin/IGF1-like signaling pathway; InR, insulin receptor; PI3K, phosphatidyl inositol 3 kinase (AGE1 in C. elegans); AKT, serine/threonine protein kinase; JNK, Jun N-terminal kinase.
Key: Solid arrowheads indicate activation. T-bars indicate inhibition. Single-side arrowheads indicate partitioning between two subcellular compartments or states.
Increased expression and activity of Hsps upon heat stress is regulated primarily at the level of increased transcription by heat shock factor (HSF); however, RNA stability, translational regulation and altered Hsp activity and subcellular localization also play a role [1]. The transcriptional regulator HSF is highly conserved across metazoans. HSF is constitutively expressed in the cytoplasm and is maintained in an inactive state by association with Hsps, including Hsp70, Hsp90 and Hsp40 (see Figure 1) [4]. Stresses that cause protein unfolding in the cell, such as heat and oxidative stress, titrate the Hsps away from HSF, allowing HSF to undergo trimerization, activation, and translocation to the nucleus, where it binds to conserved heat shock elements (HSEs) in the promoters of Hsp genes and activates their transcription. Once Hsp levels have risen sufficiently and/or the stress is removed, the HSF is titrated back into the inactive monomeric form by association with Hsps, allowing for a feed-back loop that terminates the response (Figure 1). HSF is also regulated by posttranslational modifications such as phosphorylation, sumoylation and acetylation [1, 4]. Cellular stress that causes protein unfolding will therefore cause changes in the amount and availability of Hsps that will be transduced to their networks of clients, thereby coordinately altering signaling pathways, protein localization and stability, and affecting global changes in the cell’s physiology. In this way, chronic stress can lead to deleterious effects such as those associated with aging [5].
The insulin/IGF1-like signaling (IIS) pathway (Figure 1) has been found to be a key regulator of lifespan and aging phenotypes across species [6]. IIS acts through a conserved transcription factor (FOXO) whose targets include Hsps; therefore, Hsps might be key mediators of IIS effects on aging [7]. In Caenorhabditis elegans (C. elegans) the FOXO transcription factor acts in concert with HSF to regulate sHsp induction in response to stress and upon reduced IIS; also, consensus FOXO binding sites are present in most C. elegans sHsp gene promoters [6].
Additional pathways co-regulate stress resistance, lifespan, and Hsp expression. For example, the Drosophila JNK signaling pathway promotes oxidative stress resistance and expression of the Hsp70-class protein Hsp68, and both JNK signaling and Hsp68 over-expression were reported to extend fly life span [8]. JNK signaling also increases stress resistance and life span in C. elegans, by activating the FOXO transcription factor [9]. In summary, several stress response pathways, including HSF, IIS and JNK, converge upon the activation of Hsp gene expression, and can increase both stress resistance and life span.
Hsp expression during aging
Aging is associated with characteristic changes in gene expression, in particular the Hsps. During normal aging of an otherwise unstressed animal, expression of the Hsps increases in a variety of tissue-specific and disease-specific patterns, consistent with aging and disease as intrinsic stress states [10–12]. For example, up-regulated hsp22 and hsp70 gene expression during Drosophila aging requires functional HSE sites in the promoter. One likely mediator of increased Hsp expression during aging is increased oxidative stress [2, 10] (see Figure 1). Interestingly, the ability of Hsps to be further induced by an acute extrinsic stress (such as heat) declines with age, at least in certain mammalian cell types. Although no consensus mechanism for reduced Hsp induction in old cells has been identified, in liver cells from old rats and in senescent cells in culture, abundant HSF was present but had reduced DNA binding activity, suggesting an inhibitory posttranslational modification or damage to HSF. Another appealing mechanism for inhibition of the stress response in old cells is the elevated basal levels of Hsp70 and other Hsps, because these proteins can inhibit HSF and the heat shock response through the feedback loop described above [13](Figure 1).
Hsps as biomarkers of aging and life span
Only about 30% of the variability in lifespan is heritable in the Mendelian sense, in humans and in model organisms [14]. Differences in individual genotypes and environments can cause variation in lifespan, however even in isogenic C. elegans. populations, where genetic variance between individuals is almost zero, the worms are the same size, and the environmental conditions are as uniform as possible, the same variation in lifespan exists and this variation is almost as great as that in humans [15]. The causes of this inherent variation in lifespan are unknown, but could include differences in the amount of maternal gene products inherited, variable epigenetic states, variable cell number or cell size in critical tissues, and other possibilities [16, 17]. One of the goals in aging research is to identify biomarkers of aging that would predict some or all of this individual variability in lifespan and/or variability in other aging-related phenotypes such as aging-related functional losses or diseases [18–21]. Recently the Hsps have emerged as leading candidates for such biomarkers of aging.
C. elegans
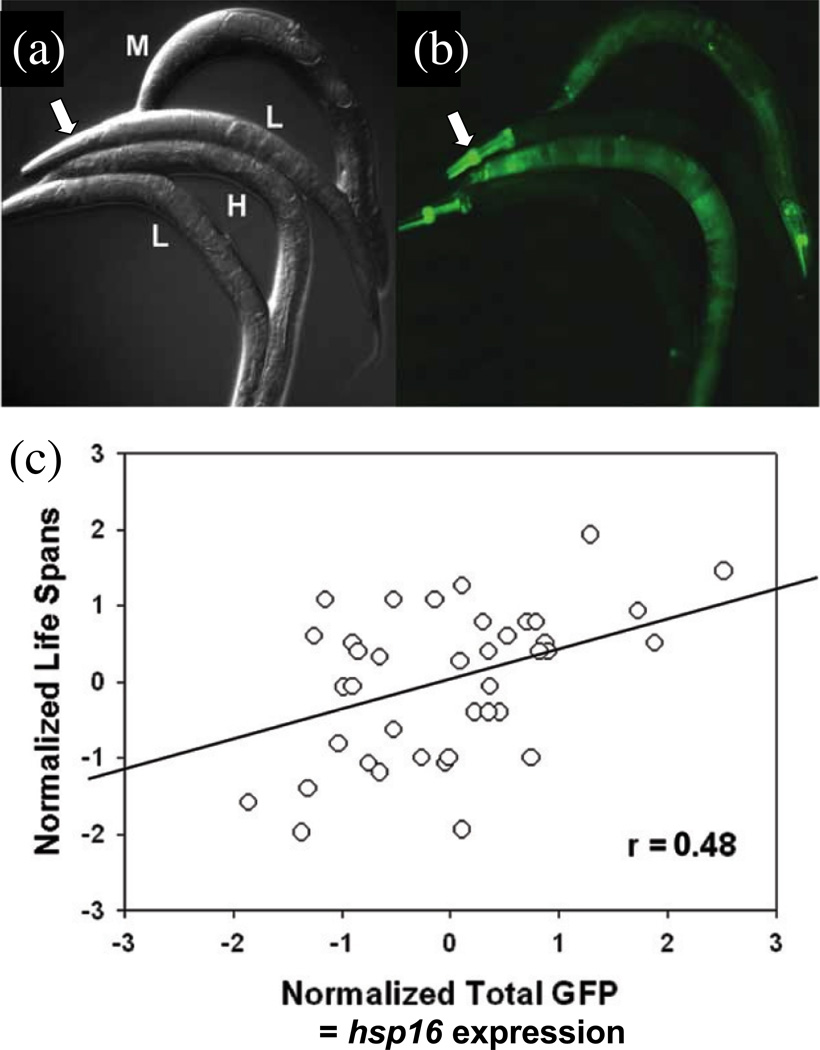
Rea et al found that the relative amount of expression of an sHsp transgene (Hsp16.2-GFP) in response to brief heat stress in a young worm is partially predictive of that individual’s remaining lifespan [15, 22] (Figure 2). Young worms with the most robust expression of the Hsp16.2-GFP reporter in response to heat pulse had the longest lifespans, and were relatively more resistant to subsequent toxic heat stresses. Interestingly the Hsp16.2-GFP transgenic worms varied not only in the level of expression, but also in which tissue expression was most prevalent. For example, a worm sorted into the low-expression group (“L”) based on overall expression levels had a relatively high level of expression limited to the pharynx region (marked with an arrow in Figure 2a & b). It is possible that expression in a particular tissue will turn out to be the best predictor of lifespan. These results indicate that whatever the mechanism or “state” of the worms that results in variability in individual lifespan, this state is present and quantifiable in the youngest worms and correlates with their ability to induce Hsp16.2 in response to heat stress. One possibility is that it is the ability of a particular worm to respond more robustly to stress that ultimately confers longer lifespan. For example, over-expression of Hsp16.2 itself can increase lifespan [23], and both the HSF and FOXO transcription factors that regulate induction of Hsp16.2 in response to stress can promote lifespan [6]. One interesting possibility for future research is to determine whether variable amounts of a regulator of the HSF or FOXO pathways is maternally supplied to the worms, and whether this regulator confers more robust stress response and greater lifespan [17, 24].
Figure 2.
Expression of Hsp16 in young C. elegans predicts subsequent survival. (a, b) Young worms (1 day old) bearing the Hsp16.2-GFP transgene were subjected to brief heat stress (35°C for two hours). Worms were sorted in to groups exhibiting low (L), medium (M) and high (H) levels of GFP expression, using an automated sorter, and representative animals are presented. (a) Nomarski image. (b) GFP image. Note that in addition to variation in overall level of GFP expression, there is tissue-specific variation in expression. For example, the white arrow in (b) indicates relatively abundant GFP expression in the pharynx of an animal with otherwise low expression. (c) Scatter plot showing correlation between Hsp16.2-GFP expression level in young worms and their ultimate lifespans. The data demonstrate that young worms possessing a more robust heat-stress response will live longer. TJ37-strain worms were grown synchronously from birth on NGM agar at 20°C and shifted to 31–32°C at three or four days of age, for two hours, resulting in a mild induction of the Hsp16.2-GFP transgene. Overall correlation coefficient was 0.48, p = 0.002. Data are reproduced from Rea et al [15].
Drosophila
Several Hsp genes, including hsp22 and hsp70, are up-regulated during normal Drosophila aging [10]. Recently, both hsp22 and hsp70 have been found to be predictive biomarkers of Drosophila lifespan and mortality: expression of hsp22-GFP and hsp70-GFP transgenes in young (~ 1 week old) flies was negatively correlated with remaining lifespan [25]; and during aging both genes were observed to spike in expression in the hours preceding death of the animal [26]. These data are consistent with model in which a robust stress response in young animals correlates with longevity, such as was demonstrated with the C. elegans Hsp16.2-GFP reporters (Figure 2); whereas chronic or basal-level expression of Hsp genes is indicative of aging and imminent mortality, as observed with Drosophila hsp22 and hsp70. The ability of Hsp transgenes to act as predictive biomarkers of aging should facilitate future research into basic mechanisms of aging and the role of the Hsps.
Interventions that alter Drosophila lifespan are also correlated with altered Hsp expression. For example, Drosophila lines genetically selected for increased lifespan had increased average message levels for hsp22 and hsp23 in young flies, perhaps indicative of a more robust stress response, whereas hsp70 levels appeared unchanged, or even reduced [10, 27].
Humans
Studies of centenarians indicates clustering of extreme longevity in families, suggesting it might be possible to identify specific human gene variants that confer longevity [28]. So far no natural variation in HSF genes has been correlated with life span in humans or model organisms [29], however analyses have been limited and this might be a useful area for future study. Mammals, including humans, have 17 Hsp70 genes, including three located within the major histocompatability locus (MHC). Preliminary studies suggest association of genetic markers in the MHC region and in specific Hsp70 genes with lifespan [30]. An Hsp70 gene polymorphism associated with shorter lifespan was also associated with a smaller induction of Hsp70 by heat in isolated blood cells, and offspring of centenarians were found to have significantly lower circulating Hsp70 levels than spousal controls [31]. These results are consistent with the general idea that longevity correlates with lower basal levels of Hsp gene expression and more robust heat shock response.
Effects of Hsps on life span
Prokaryotes
During division of E. coli cells, aggregated proteins persist at the older pole of daughter cells. These aging-associated aggregates can be visualized using a fluorescently tagged chaperone (IbpA), which is involved in protein aggregate processing. The cells inheriting these protein aggregates have reduced ability to replicate, a phenotype indicative of aging [32]. The co-localization of IbpA with the aggregates serves as a biomarker and suggests a possible direct role for the chaperone that will be interesting for further study.
Yeast
In budding yeast, the mother cell (older cell) preferentially retains protein aggregates and oxidatively damaged proteins [33]. This selective retention of damaged proteins requires the chaperone Hsp104 as well as Sir2p, and is required for normal lifespan of the daughter cell.
C. elegans
Reduced expression of HSF [6] or mitochondrial Hsp70 (Hsp-6 gene) [34] using RNAi caused a progeria (rapid aging-like) phenotype, consistent with a requirement for Hsps for normal lifespan. Conversely, over-expression of the Hsp regulators HSF and FOXO can each increase lifespan [6]. Creating a transgenic worm with extra copies of the Hsp16 gene increased lifespan in a FOXO(Daf16)-dependent manner [23], suggesting that sHsps are at least one mediator of the longevity-promoting effects of FOXO and HSF. Similarly, over-expression of the Mortalin-related gene Hsp70F was reported to increase worm lifespan [35].
Drosophila
Several studies implicate the sHsps in the control of lifespan in Drosophila. Hsp22, Hsp26 and Hsp27 are each reported to increase lifespan when over-expressed in flies using the Gal4/UAS system [36–38]. In contrast, ubiquitous over-expression of Hsp22 specifically in adult flies using a conditional (doxycyline-regulated) system decreased lifespan and stress resistance [39]. Presumably these differences in results reflect the different systems/promoters used for over-expression, suggesting that Hsp22 might have differential effects on stress resistance and lifespan depending upon the tissue and life stage for over-expression. Again these data are consistent with a general model in which a robust stress response in young animals (on and off) promotes longevity, whereas accumulation of certain Hsps to high levels during aging might be toxic.
Mammalian cell culture
Primary cells typically divide for only a limited number of times in vitro before they enter a non-dividing state called replicative senescence. It is possible that replicative senescence contributes to aging at the level of the whole animal, at least in species such as Drosophila and humans where adult tissues are maintained by dividing (stem) cell populations [40]. Mortalin is a mitochondrial Hsp that is related to Hsp70 but is not induced by heat stress. Mortalin can increase replicative lifespan when over-expressed in fibroblasts [41], likely related to the ability of Hsps to repress the senescence pathway, as discussed below.
Hormesis
In conditioning hormesis, a low dose of stress protects against a subsequent higher dose of that stress [42]. Strikingly, low dose stresses such as heat or irradiation typically administered in young animals can cause increased lifespan in a variety of models, including C. elegans and Drosophila [12, 43, 44]. For example, mild heat stress in young Drosophila increases lifespan, and this conditioning hormesis appears dependent upon functional HSF, but generally does not correlate with Hsp70 expression, suggesting that sHsps or some other heat-inducible factors are important [12, 45]. Significantly, the IIS pathway that regulates sHsp expression and lifespan in C. elegans is required for heat and oxidant-induced conditioning hormesis to increase lifespan, and this lifespan increase correlates with expression levels of Hsps, both the Hsp70 (Hsp-4) and sHsp (Hsp16.2) class [43, 46]. Strikingly, the pattern of gene expression changes associated with increased lifespan in IIS mutant worms and in flies where lifespan has been extended by over-expression of MnSOD show a statistically significant similarity to the gene expression changes normally observed in old animals, including up-regulation of Hsps [47, 48]. This is consistent with a conditioning-hormesis model wherein lifespan extension by reduced IIS or increased SOD results from early activation of an “aging-like” stress response involving Hsps, i.e. the early induction of this stress response would help protect the animal from the subsequent stresses associated with aging.
How Hsps affect life span possible mechanisms
The precise molecular mechanisms that cause aging and mortality are not yet known, however there are several possibilities for how Hsps might be directly involved in modulating these phenotypes.
Immune response
The HSF pathway, FOXO pathway, and specific Hsps (Hsp90 and sHsps), have each been found to function to promote an immune response [49–51]. However, evidence that immune function limits life span in the laboratory is limited and mixed. For C. elegans, the E. coli food source appears to limit lifespan in some instances [51], whereas in Drosophila, bacteria are found to affect lifespan under some conditions [52], but not all [53]. It seems likely that in the natural setting where pathogens are more prevalent, immune function will be more important for survival than in the laboratory setting; however, there is so far little evidence to suggest that immune function is an important modulator of aging rate.
Protein turnover and suppression of proteotoxicity
Numerous human aging-related diseases involve the misfolding, abnormal processing, modification and aggregation of proteins, for example, Parkinson’s Disease, ALS, prion diseases, Alzheimer’s Disease (AD), and CAG-repeat and polyQ expansion diseases including Huntington’s Disease [1] (Box 2). These protein conformation diseases have a strong age-dependence, and Hsps are typically found associated with the protein aggregates. Notably in animal models of these diseases, such as C. elegans and Drosophila engineered to express human disease genes, Hsps are often found to be modifiers, such that Hsp over-expression reduces the toxic effects. Consistent with this, the IIS and HSF pathways are potent modifiers of proteotoxicity. For example, inhibition of IIS (which activates FOXO and Hsp expression), and over-expression of HSF not only increases lifespan, but also suppresses the toxicity of polyQ and AD proteins expressed in C. elegans [6, 7]. Perhaps even during normal aging, the ability of Hsps to counteract proteotoxicity is a relevant pathway for their longevity-promoting effects.
Autophagy
Chaperone-mediated autophagy (CMA) is a lysosomal pathway for turnover of specific cytosolic proteins that is stimulated by stresses, including fasting and oxidative stress, and the activity of CMA declines during aging [54]. A complex in the cytoplasm including Hsp90, Hsc70, Hsp40 and cofactors including lysosomal Hsc70 unfolds target proteins to facilitate their translocation into the lysosome. Autophagy is implicated in lifespan control in several ways, including its ability to counteract proteotoxicity [54]. In C. elegans, autophagy has been reported to be required for lifespan extension in response to each of three lifespan pathways: reduced IIS [55], DR [56, 57], and reduced mitochondrial gene function [58]. In Drosophila, when autophagy gene activity is reduced during both development and adulthood, flies are short-lived, sometimes associated with reduced protein aggregate clearance and degeneration in the adult nervous system [58–60]. Moreover, over-expression of the autophagy gene Atg8a in Drosophila was reported to increase lifespan [59]. In contrast, conditional inactivation of autophagy genes specifically in adult flies did not affect lifespan [61]. Taken together, the data from C. elegans and Drosophila suggest a somewhat counter-intuitive model in which autophagy in adults might not limit lifespan under control conditions, but might be required for certain interventions to increase lifespan. The role of CMA in aging-related diseases and lifespan promises to be an active and productive area for future research.
Cellular senescence
Cellular senescence was first identified by the limited number of cell divisions that normal cells can undergo in vitro (called replicative senescence or the “Hayflick limit”) [40]. The cells exhibit changes in chromatin structure, undergo characteristic changes in gene expression and cell morphology, can no longer divide, and are resistant to apoptosis. It is now recognized that cellular senescence can be triggered by several stresses, including telomere shortening, oxidative stress, DNA damage, and expression of activated oncogenes such as Ras, Her-2 and PTEN. These stresses activate the tumor suppressor proteins p53 and pRb, which in turn result in accumulation of cell cycle inhibitors such as p16 and p21. It appears clear that cellular senescence pathways normally function as potent anti-tumor mechanisms, because mutations in the tumor suppressor genes p53 and Rb cause increased cancer incidence [40]. In addition, recent results suggest that cellular senescence can play a beneficial role in limiting damage to tissues under stress [62]. One way cancer cells can escape senescence is by increased expression of Hsps [63]. For example, numerous human cancer cell lines are found to over-express Hsp70 family members such as Hsp70-2, Hsp72, and the major mitochondrial chaperone, Mortalin (aka Grp75). These Hsp70-class proteins bind to and inhibit p53, thereby preventing senescence. Similarly, in certain cancer cell lines, the sHsps Hsp27 and Clusterin (aka vcp/p97, aka apolipoprotein J) are over-expressed and inhibit p53 and the senescence pathway. Another way Hsps might promote tumor cell growth is by suppressing missense mutations that have accumulated during transformation [64]. The over-expression of Hsps in cancer cells might result from over-expression of HSF or activation of HSF by AKT phosphorylation, or other mechanisms [63]. Cellular senescence is likely to be more relevant to aging in organisms such as humans and Drosophila, where specific tissues are maintained by dividing stem cell populations, as opposed to C. elegans where all tissues except the gonads are post-mitotic.
Apoptosis
Aging is associated with a misregulation of apoptosis in several ways. For example, certain replicating cell types from old mammals have a reduced ability to undergo apoptosis [65], whereas other tissues, such as muscle and fat tissue in mammals and flies, show increased apoptosis with age [66–68]. Clusterin is a sHsp-related protein whose expression is regulated by HSF and up-regulated during aging. Like cellular senescence, apoptosis also functions as an anti-cancer mechanism: Clusterin, Hsp27 and Hsp70 are anti-apoptotic, induced in response to anti-cancer therapies, and implicated in the development and progression of cancers [69]. Elevated levels of Hsps and increased HSF expression are required for tumorigenesis [70], perhaps by inhibiting specific apoptotic regulatory proteins such as p53.
In summary, both cellular senescence and apoptosis are anti-cancer mechanisms that are also involved in normal tissue maintenance. The Hsps are involved in the regulation of these processes, and the abnormal expression of Hsps might be involved in the disruption of these processes during aging.
Areas to stress in the future
Our understanding of the molecular pathways that affect aging and lifespan is advancing rapidly and increasingly implicates the Hsps as key mediators. The ability of Hsps to ameliorate proteotoxicity [71] and to regulate key aging pathways such as senescence, apoptosis and cancer indicates that Hsps are promising targets for future interventions in aging-related diseases. The Hsps also show great promise as the long sought-after biomarkers of aging. An interesting question is whether an ideal biomarker of aging should itself affect the trait it is supposed to forecast, as do the Hsps. Indeed the direct role for Hsps in modulating aging-related pathways might be why Hsp levels can function so well as predictive biomarkers of lifespan. Strikingly, recent experiments in C. elegans have shown a systemic (non-cell-autonomous) control of the heat shock response and Hsp expression that is regulated by signals from the nervous system [72]. It is tempting to speculate that this systemic control of Hsp expression might relate to the systemic control of lifespan by IIS and DR [6, 73], as well as the recently identified systemic control of cellular senescence [74] and stem cell proliferation [75] in mammals. Important additional questions for the future include determining the mechanisms by which Hsps are up-regulated during normal aging and in cancer cells. In addition, the ability of Hsps to act as a predictive biomarkers of lifespan provides an inroad to study one of the most fundamental questions in aging research - why does one animal live longer than another?
Acknowledgements
The author was supported by a grant from the Department of Health and Human Services, National Institute on Aging (AG11833); Scientific Opportunity Funds from the Genetics of Longevity Consortium, California Pacific Medical Center Research Institute (U19 AG032122); and by pilot project funding from the Southern California Environmental Health Sciences Center, which is funded by a grant from the National Institute of Environmental Health Sciences (5P30ES007048).
Glossary
- Aging
A non-specific term that can mean any change over time [76].
- Senescence
A deteriorative change that causes increased mortality [76].
- Extrinsic stress
An environmental factor that causes a change in a biological system that is potentially injurious [77].
- Intrinsic stress
This can take on several forms, including non-optimal genetic constitution (e.g., inbreeding depression), and specific metabolic challenges such as accumulation of waste products and the generation of reactive metabolites during normal metabolism (e.g., reactive oxygen species/ROS) [3].
- Biomarker (of senescence)
A biological parameter of an organism that either alone or in some multivariate composite will, in the absence of disease, better predict survival and functional capability at a later age than will chronological age [18].
- Hormesis
Stimulatory effects of low doses of chemical or environmental stresses that are known to be toxic at higher doses [42].
- Conditioning hormesis
The ability of low doses of a stress to protect against subsequent higher doses of that stress or other stresses, including conferring increased lifespan [42].
References
- 1.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller FL, et al. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 3.Vermeulen CJ, Loeschcke V. Longevity and the stress response in Drosophila. Exp Gerontol. 2007;42:153–159. doi: 10.1016/j.exger.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soti C, Csermely P. Aging cellular networks: chaperones as major participants. Exp Gerontol. 2007;42:113–119. doi: 10.1016/j.exger.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 6.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Cohen E, et al. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 8.Wang MC, et al. JNK Signaling Confers Tolerance to Oxidative Stress and Extends Lifespan in Drosophila. Dev Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- 9.Oh SW, et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landis GN, Tower J. Superoxide dismutase evolution and life span regulation. Mech Ageing Dev. 2005;126:365–379. doi: 10.1016/j.mad.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Macario AJ, Conway de Macario E. Sick chaperones, cellular stress, and disease. N Engl J Med. 2005;353:1489–1501. doi: 10.1056/NEJMra050111. [DOI] [PubMed] [Google Scholar]
- 12.Morrow G, Tanguay RM. Heat shock proteins and aging in Drosophila melanogaster. Semin Cell Dev Biol. 2003;14:291–299. doi: 10.1016/j.semcdb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 13.Morimoto R. Dynamic remodeling of transcription complexes by molecular chaperones. Cell. 2002;110:281–284. doi: 10.1016/s0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- 14.Finch CE, Tanzi RE. Genetics of aging. Science. 1997;278:407–411. doi: 10.1126/science.278.5337.407. [DOI] [PubMed] [Google Scholar]
- 15.Rea SL, et al. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkwood TB, et al. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech Ageing Dev. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–718. doi: 10.1016/j.mad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Baker GT, 3rd, Sprott RL. Biomarkers of aging. Exp Gerontol. 1988;23:223–239. doi: 10.1016/0531-5565(88)90025-3. [DOI] [PubMed] [Google Scholar]
- 19.Johnson TE. Recent results: biomarkers of aging. Exp Gerontol. 2006;41:1243–1246. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Simm A, et al. Potential biomarkers of ageing. Biol Chem. 2008;389:257–265. doi: 10.1515/BC.2008.034. [DOI] [PubMed] [Google Scholar]
- 21.Butler RN, et al. Biomarkers of aging: from primitive organisms to humans. J Gerontol A Biol Sci Med Sci. 2004;59:B560–B567. doi: 10.1093/gerona/59.6.b560. [DOI] [PubMed] [Google Scholar]
- 22.Wu D, et al. Visualizing hidden heterogeneity in isogenic populations of C. elegans. Exp Gerontol. 2006;41:261–270. doi: 10.1016/j.exger.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 24.Ayyadevara S, et al. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Tower J. Expression of hsp22 and hsp70 transgenes is partially predictive of Drosophila survival under normal and stress conditions. J. Gerontol.: Biol. Sci. 2009 doi: 10.1093/gerona/glp054. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grover D, et al. Simultaneous tracking of fly movement and gene expression using GFP. BMC Biotechnol. 2008;8 doi: 10.1186/1472-6750-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norry FM, Loeschcke V. Heat-induced expression of a molecular chaperone decreases by selecting for long-lived individuals. Exp Gerontol. 2003;38:673–681. doi: 10.1016/s0531-5565(03)00057-3. [DOI] [PubMed] [Google Scholar]
- 28.Christensen K, et al. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norry FM, et al. Knockdown resistance to heat stress and slow recovery from chill coma are genetically associated in a quantitative trait locus region of chromosome 2 in Drosophila melanogaster. Mol Ecol. 2007;16:3274–3284. doi: 10.1111/j.1365-294X.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- 30.Singh R, et al. Genetics of human longevity with emphasis on the relevance of HSP70 as candidate genes. Front Biosci. 2007;12:4504–4513. doi: 10.2741/2405. [DOI] [PubMed] [Google Scholar]
- 31.Terry DF, et al. Serum heat shock protein 70 level as a biomarker of exceptional longevity. Mech Ageing Dev. 2006;127:862–868. doi: 10.1016/j.mad.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindner AB, et al. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erjavec N, et al. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura K, et al. Knockdown of Mitochondrial Heat Shock Protein 70 Promotes Progeria-like Phenotypes in Caenorhabditis elegans. J Biol Chem. 2007;282:5910–5918. doi: 10.1074/jbc.M609025200. [DOI] [PubMed] [Google Scholar]
- 35.Yokoyama K, et al. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 2002;516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang HD, et al. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc Natl Acad Sci U S A. 2004;101:12610–12615. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seong KH, et al. Application of the gene search system to screen for longevity genes in Drosophila. Biogerontology. 2001;2:209–217. doi: 10.1023/a:1011517325711. [DOI] [PubMed] [Google Scholar]
- 38.Morrow G, et al. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. Faseb J. 2004;18:598–599. doi: 10.1096/fj.03-0860fje. [DOI] [PubMed] [Google Scholar]
- 39.Bhole D, et al. Doxycycline-regulated over-expression of hsp22 has negative effects on stress resistance and life span in adult Drosophila melanogaster. Mech Ageing Dev. 2004;125:651–663. doi: 10.1016/j.mad.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Kaul SC, et al. Three faces of mortalin: a housekeeper, guardian and killer. Exp Gerontol. 2007;42:263–274. doi: 10.1016/j.exger.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 42.Calabrese EJ, et al. Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose-response framework. Toxicol Appl Pharmacol. 2007;222:122–128. doi: 10.1016/j.taap.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 43.Cypser JR, et al. Hormesis and aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:935–939. doi: 10.1016/j.exger.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gems D, Partridge L. Stress-response hormesis and aging:"that which does not kill us makes us stronger". Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen JG, et al. Sex specific effects of heat induced hormesis in Hsf-deficient Drosophila melanogaster. Exp Gerontol. 2007;42:1123–1129. doi: 10.1016/j.exger.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Galbadage T, Hartman PS. Repeated temperature fluctuation extends the life span of Caenorhabditis elegans in a daf-16-dependent fashion. Mech Ageing Dev. 2008;129:507–514. doi: 10.1016/j.mad.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Curtis C, et al. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lund J, et al. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–1573. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 49.Tsang WY, Lemire BD. The role of mitochondria in the life of the nematode, Caenorhabditis elegans. Biochim Biophys Acta. 2003;1638:91–105. doi: 10.1016/s0925-4439(03)00079-6. [DOI] [PubMed] [Google Scholar]
- 50.Singh V, Aballay A. Heat Shock and Genetic Activation of HSF-1 Enhance Immunity to Bacteria. Cell Cycle. 2006;5:2443–2446. doi: 10.4161/cc.5.21.3434. [DOI] [PubMed] [Google Scholar]
- 51.Garigan D, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brummel T, et al. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A. 2004;101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren C, et al. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melendez A, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 56.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 57.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toth ML, et al. Longevity Pathways Converge on Autophagy Genes to Regulate Life Span in Caenorhabditis elegans. Autophagy. 2008;4 doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 59.Simonsen A, et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 60.Juhasz G, et al. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren C, et al. Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp Gerontol. 2008 doi: 10.1016/j.exger.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherman MY, et al. Molecular chaperones regulate p53 and suppress senescence programs. FEBS Lett. 2007;581:3711–3715. doi: 10.1016/j.febslet.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blagosklonny MV, et al. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc Natl Acad Sci U S A. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leroi AM, et al. What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech Ageing Dev. 2005;126:421–429. doi: 10.1016/j.mad.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 66.Chung L, Ng YC. Age-related alterations in expression of apoptosis regulatory proteins and heat shock proteins in rat skeletal muscle. Biochim Biophys Acta. 2006;1762:103–109. doi: 10.1016/j.bbadis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 67.Zheng J, et al. Differential patterns of apoptosis in response to aging in Drosophila. Proc Natl Acad Sci U S A. 2005;102:12083–12088. doi: 10.1073/pnas.0503374102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marzetti E, Leeuwenburgh C. Skeletal muscle apoptosis, sarcopenia and frailty at old age. Exp Gerontol. 2006;41:1234–1238. doi: 10.1016/j.exger.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 69.Garrido C, et al. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- 70.Dai C, et al. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 72.Prahlad V, et al. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Partridge L. Some highlights of research on aging with invertebrates 2008. Aging Cell. 2008 doi: 10.1111/j.1474-9726.2008.00415.x. [DOI] [PubMed] [Google Scholar]
- 74.Cichowski K, Hahn WC. Unexpected pieces to the senescence puzzle. Cell. 2008;133:958–961. doi: 10.1016/j.cell.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gopinath SD, Rando TA. Stem cell review series: aging of the skeletal muscle stem cell niche. Aging Cell. 2008;7:590–598. doi: 10.1111/j.1474-9726.2008.00399.x. [DOI] [PubMed] [Google Scholar]
- 76.Finch CE. Longevity, Senescence and the Genome. University of Chicago Press; 1990. [Google Scholar]
- 77.Hoffmann AA, Parsons PA. Evolutionary Genetics and Environmental Stress. Oxford University Press; 1991. [Google Scholar]
- 78.Lithgow GJ. Why aging isn't regulated: a lamentation on the use of language in aging literature. Exp Gerontol. 2006;41:890–893. doi: 10.1016/j.exger.2006.06.051. [DOI] [PubMed] [Google Scholar]