Abstract
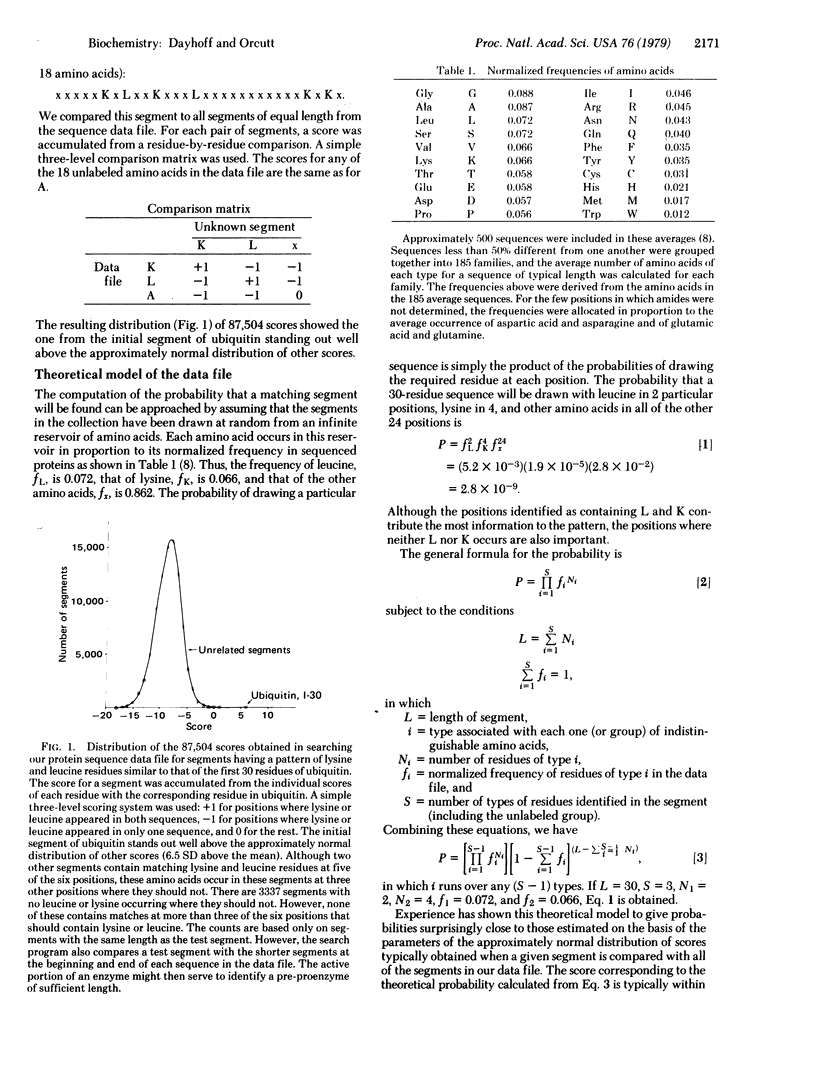
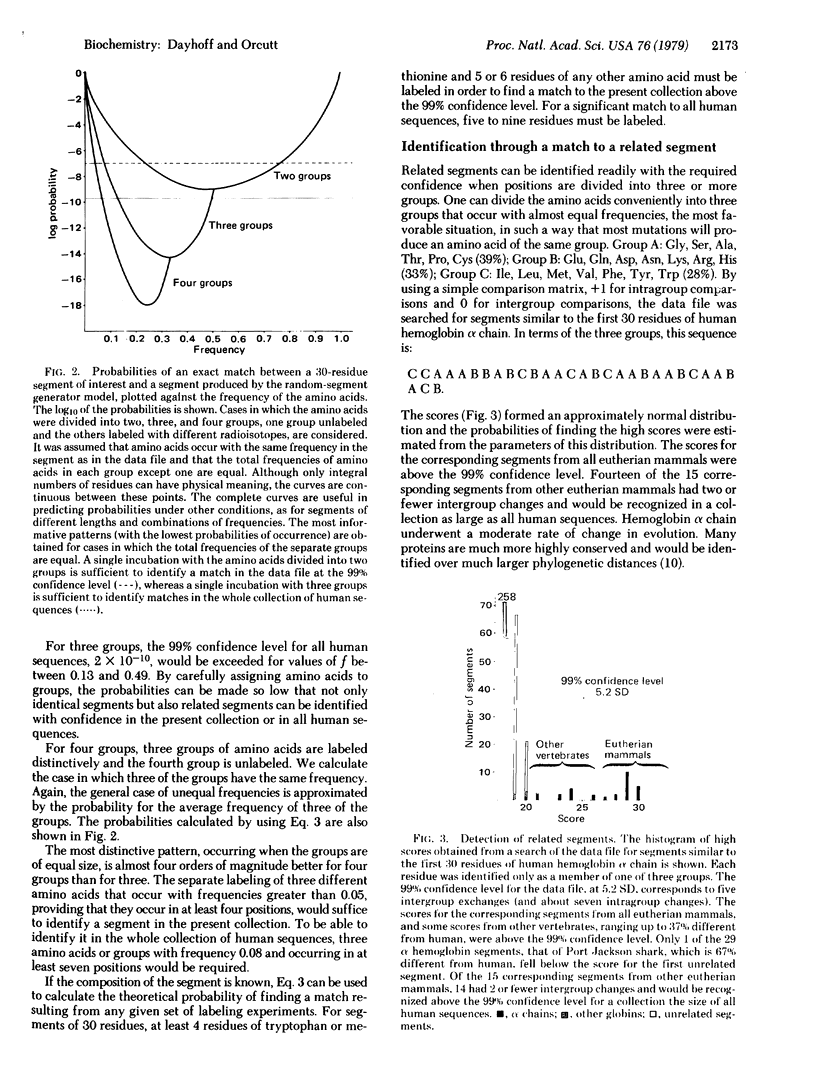
Methods for the identification of a protein segment by using the information from partial sequence analyses are described. If the protein sequence is known, a segment can usually be identified with confidence through comparison with the data file of all known sequences when the identity and position of only seven amino acid residues (not necessarily contiguous) are known. Partial sequences are obtained from extremely sensitive microsequencing procedures. Tissue is incubated with amino acids, one or more of which are distinctively radiolabeled. Proteins of interest are isolated and a sequenator experiment performed to locate the positions of radioactivity in an NH2-terminal segment of approximately 30 residues. We derive and investigate an equation for the probability of finding a unique match to any pattern of radioactivity. From this we suggest a new strategy. In one incubation, several amino acids are labeled with each kind of isotope. The most information is contained in patterns in which approximately equal numbers of positions are occupied by residues distinguished by different labels (including no label). The amino acid composition of the segment will typically not be known in advance. Labeling residues expected to occupy 36% of the positions suffices for a 98% chance of success in uniquely characterizing any human segment. Such a strategy will permit the identification of most proteins from a single tissue incubation. The mathematical discussion is general and applies to any segment from a sequence and to sequences obtained by any method. Improved identification procedures should expedite the accumulation of information on the expression and function of proteins.
Keywords: microsequencing strategy, information in sequences, identifying homologous sequences, radiolabeled amino acid patterns
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hunt L. T., Dayhoff M. O. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem Biophys Res Commun. 1977 Jan 24;74(2):650–655. doi: 10.1016/0006-291x(77)90352-7. [DOI] [PubMed] [Google Scholar]
- Jacobs J. W., Kemper B., Niall H. D., Habener J. F., Potts J. T., Jr Structural analysis of human proparathyroid hormone by a new microsequencing approach. Nature. 1974 May 10;249(453):155–157. doi: 10.1038/249155a0. [DOI] [PubMed] [Google Scholar]
- Lehman W. Hybrid troponin reconstituted from vertebrate and arthropod subunits. Nature. 1975 May 29;255(5507):424–426. doi: 10.1038/255424a0. [DOI] [PubMed] [Google Scholar]
- Schlesinger D. H., Goldstein G., Niall H. D. The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry. 1975 May 20;14(10):2214–2218. doi: 10.1021/bi00681a026. [DOI] [PubMed] [Google Scholar]
- Seidah N. G., Crine P., Benjannet S., Scherrer H., Chrétien M. Isolation and partial characterization of a biosynthetic N-terminal methionyl peptide of bovine pars intermedia: relationship to ubiquitin. Biochem Biophys Res Commun. 1978 Feb 14;80(3):600–608. doi: 10.1016/0006-291x(78)91611-x. [DOI] [PubMed] [Google Scholar]



