Abstract
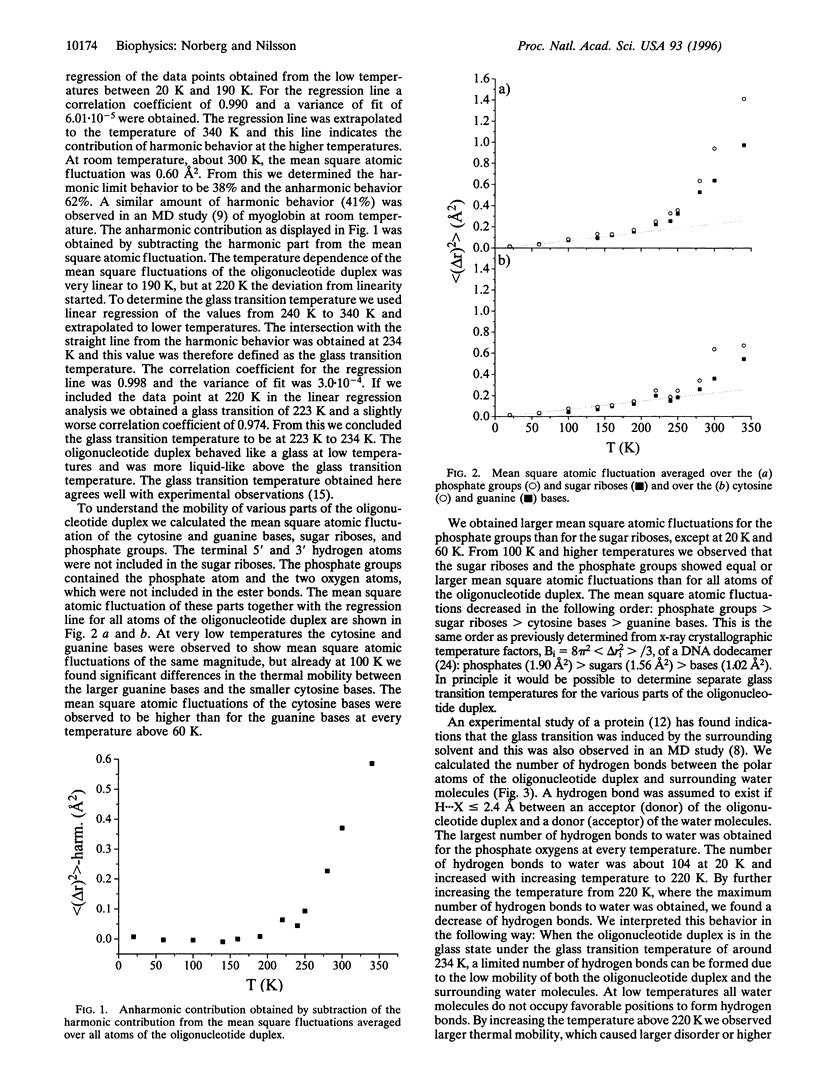
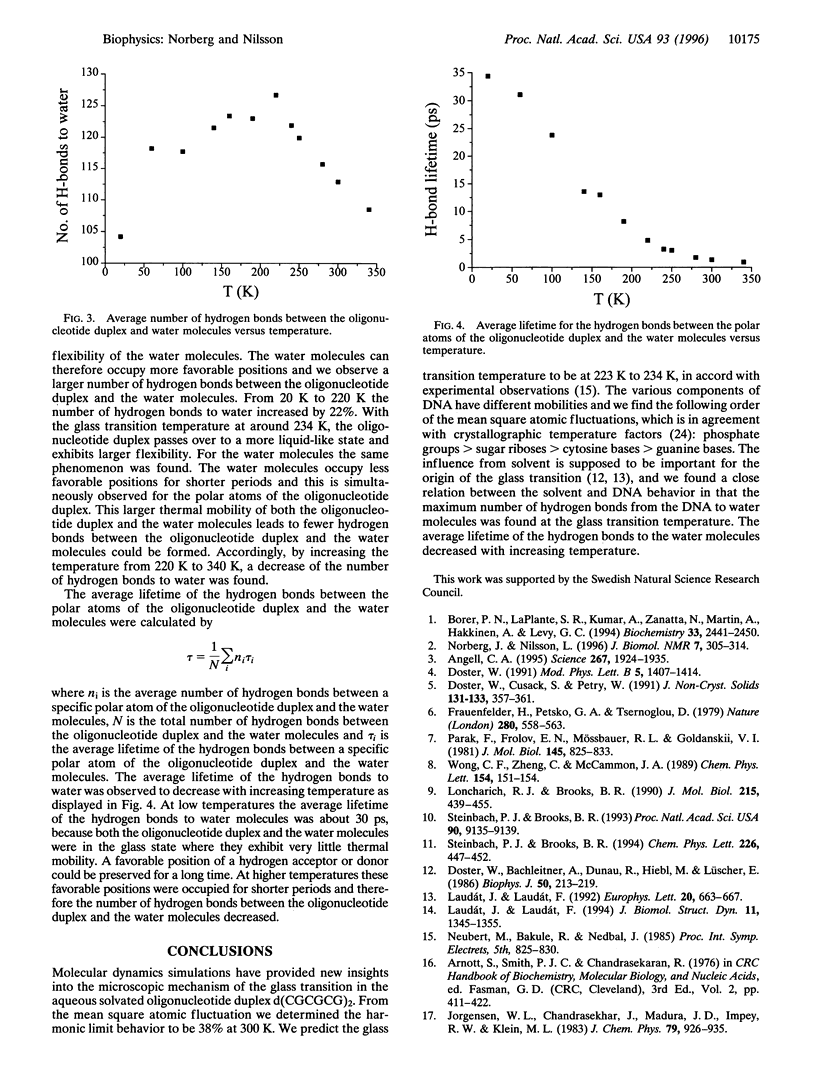
Molecular dynamics simulations of the oligonucleotide duplex d(CGCGCG)2 in aqueous solution are used to investigate the glass transition phenomenon. The simulations were performed at temperatures in the 20 K to 340 K range. The mean square atomic fluctuations showed that the behavior of the oligonucleotide duplex was harmonic at low temperatures. A glass transition temperature at 223 K to 234 K was inferred for the oligonucleotide duplex, which is in agreement with experimental observations. The largest number of hydrogen bounds between the polar atoms of the oligonucleotide duplex and the water molecules was obtained at the glass transition temperature. With increasing temperature we observed a decrease in the average lifetime of the hydrogen bonds to water molecules.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angell C. A. Formation of glasses from liquids and biopolymers. Science. 1995 Mar 31;267(5206):1924–1935. doi: 10.1126/science.267.5206.1924. [DOI] [PubMed] [Google Scholar]
- Borer P. N., LaPlante S. R., Kumar A., Zanatta N., Martin A., Hakkinen A., Levy G. C. 13C-NMR relaxation in three DNA oligonucleotide duplexes: model-free analysis of internal and overall motion. Biochemistry. 1994 Mar 8;33(9):2441–2450. doi: 10.1021/bi00175a012. [DOI] [PubMed] [Google Scholar]
- Doster W., Bachleitner A., Dunau R., Hiebl M., Lüscher E. Thermal properties of water in myoglobin crystals and solutions at subzero temperatures. Biophys J. 1986 Aug;50(2):213–219. doi: 10.1016/S0006-3495(86)83455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Laudát J., Laudát F. Thermally stimulated relaxations in DNA. J Biomol Struct Dyn. 1994 Jun;11(6):1345–1355. doi: 10.1080/07391102.1994.10508072. [DOI] [PubMed] [Google Scholar]
- Loncharich R. J., Brooks B. R. Temperature dependence of dynamics of hydrated myoglobin. Comparison of force field calculations with neutron scattering data. J Mol Biol. 1990 Oct 5;215(3):439–455. doi: 10.1016/s0022-2836(05)80363-8. [DOI] [PubMed] [Google Scholar]
- Norberg J., Nilsson L. Internal mobility of the oligonucleotide duplexes d(TCGCG)2 and d(CGCGCG)2 in aqueous solution from molecular dynamics simulations. J Biomol NMR. 1996 Jun;7(4):305–314. doi: 10.1007/BF00200432. [DOI] [PubMed] [Google Scholar]
- Parak F., Frolov E. N., Mössbauer R. L., Goldanskii V. I. Dynamics of metmyoglobin crystals investigated by nuclear gamma resonance absorption. J Mol Biol. 1981 Feb 5;145(4):825–833. doi: 10.1016/0022-2836(81)90317-x. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Brooks B. R. Protein hydration elucidated by molecular dynamics simulation. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9135–9139. doi: 10.1073/pnas.90.19.9135. [DOI] [PMC free article] [PubMed] [Google Scholar]



