Abstract
TTF-1 is an important transcription factor in lung development and lung disease and is essential for lung cell differentiation, specifically surfactant protein (Sftp) expression. The molecular mechanisms that drive the expression and transcriptional control of TTF-1 are not fully understood. In the fetal lung, ErbB4 functions as a transcriptional co-factor and regulates the timely onset of fetal Sftp expression. We speculate that ErbB4 is an upstream regulator of TTF-1 and regulates Sftpb expression via this pathway in alveolar type II cells. Neuregulin-induced ErbB4 and TTF-1 signaling interactions were studied by co-immunoprecipitation and confocal microscopy. Overexpression of ErbB4 and TTF-1 was analyzed in its effect on cell viability, Sftpb expression, TTF-1 expression, and Sftpb and TTF-1 promoter activity. The effect of ErbB4 deletion and ErbB4 nuclear translocation on TTF-1 expression was studied in primary fetal type II epithelial cells, isolated from transgenic HER4heart(−/−) mice. ErbB4 ligand neuregulin induces ErbB4 and TTF-1 co-precipitation and nuclear colocalization. Combined ErbB4 and TTF-1 overexpression inhibits cell viability, while promoting Sftpb expression more than single overexpression of each protein. NRG stimulates TTF-1 expression in ErbB4-overexpressing epithelial cells, while this effect is absent in ErbB4-depleted cells. In primary fetal type II cells, ErbB4 nuclear translocation is critical for its regulation of TTF-1-induced Sftpb upregulation. TTF-1 overexpression did not overcome this important requirement. We conclude that ErbB4 is a critical upstream regulator of TTF-1 in type II epithelial cells and that this interaction is important for Sftpb regulation.
Keywords: lung development, neuregulin, surfactant, type II cell
1. Introduction
Thyroid Transcription Factor (TTF)-1, also known as Nkx2-1, is a homeodomain containing transcription factor and a member of the NK2 homeobox 1 (Nkx2) family, which activates the expression of genes in the thyroid, lung, and brain [1]. TTF-1 is a transcriptional regulator that plays a key role in embryonic lung development and lung cell differentiation [2, 3]. In the lung, TTF-1 regulates the expression of genes coding for the surfactant proteins Sftpa1, Sftpb, and Sftpc, and clara cell secretory protein (CCSP). All these proteins are critical for lung function as surface tension reducing, anti-inflammatory, and immunomodulatory factors [1]. Suppression of TTF-1 translation in a cultured lung explant model results in inhibition of branching morphogenesis [4], while its deletion leads to extensive defects in the ventral region of the forebrain [5] and non-viable pups with a complete lack of lung parenchyma and thyroid tissue [5]. Beside the effects in the developing lung, TTF-1 has a prominent role in epithelial cell regeneration [6, 7] and lung cancer [7]. In lung cancer biology, TTF-1 seems to have a zwitter function, driving oncogenic aspects in some [8-11] and a more favorable prognosis in other lung adenocarcinoma cases [12]. This is paralleled by the observation that ErbB4 a member of the receptor tyrosine kinase family can be either oncogenic or tumor suppressive [13, 14]. Myosin Binding Protein H (MYBPH) has recently been discovered as a TTF-1 downstream target in its inhibition of invasion and metastasis development [15]. The upstream molecular mechanism that controls the expression of TTF-1 and its transcriptional activity is still not fully understood.
ErbB4 is known for its involvement in differentiation processes in the breast, the central nervous system, and the lung [16-21]. ErbB4, similar to TTF-1, is critical for the progression of morphologic and functional late fetal lung development [19, 22]. ErbB4 deletion leads to delayed fetal Sftpb expression [19] and alveolar simplification and a hyperreactive airway system in the adult lung [23]. Recently it has been shown that neuregulin (NRG)-induced ErbB4 activation leads to proteolytic cleavage [24, 25] of the receptor and the 80kDa intracellular domain (4ICD) [26] translocates to the nucleus [27] and interacts with other nuclear receptors and transcription factors. In its promoting effect on Sftpb expression, ErbB4 interacts with Estrogen Receptor (ER)β [28] and the transcription factor Signal Transducer and Activator of Transcription (Stat)5A [29]. There is limited information on the regulation of TTF-1 gene expression by hormones, cytokines and other biological agents [2, 30]. Since both, ErbB4 and TTF-1 are critical in terminal fetal lung development and function we here mechanistically analyze their interactions by hypothesizing that ErbB4 is an upstream regulator of TTF-1.
2. Materials and methods
The mouse lung epithelial cell line (MLE-12) was obtained from American Type Culture Collection (Wesel, Germany); rabbit polyclonal TTF-1 (H-190), rabbit polyclonal ErbB4 (C-18) (suitable for Western Blot), and normal rabbit IgG were obtained from Santa Cruz Biotechnology (Heidelberg, Germany); mouse monoclonal c-ErbB4 antibody (clone HFR-1) (suitable for immunofluorescence) was from Thermo Fisher scientific GmbH (Dreieich, Germany); goat anti-rabbit IgG (HRP-labeled, H+L), goat anti-mouse IgG (HRP-labeled, H+L), and 10 % non–immune goat serum were from Zymed Laboratories Inc. (South San Francisco, CA); Alexa Fluor 488 goat anti-mouse IgG (H+L) and Alexa Fluor 568 anti-rabbit IgG (H+L) were from Molecular Probes (Karlsruhe, Germany); rabbit polyclonal anti-sheep Sftpb was from Chemicon Europe (Schwalbach/Ts, Germany); purified mouse monoclonal anti-phosphotyrosine antibody was from BD Biosciences (Heidelberg, Germany); rabbit polyclonal anti-phosphoserine antibody was from Millipore (Schwalbach/Ts, Germany); mouse monoclonal anti-actin clone AC-40, 4,6-Diamidino-2-phenylindole, dilactate (DAPI), and bovine serum albumine (BSA) were obtained from Sigma (Hamburg, Germany); protein A Sepharose™ CL-4B, Western Blotting Detection Reagents (enhanced chemiluminescence, ECL), and cDNA Kit were from Amersham Biotechnologies (Munich, Germany); Dulbecco’s Modified Eagles Medium containing glutamin (DMEM) was obtained from PAA-Laboratories (Coelbe, Germany). Fetal calf serum was from PAN Biotech GmbH (Aidenbach, Germany); TaqMan Universal PCR Master Mix and ABI PRISM™ Big Dye Terminator Cycle Sequencing Ready Reaction Kit was from Applied Biosystems (Darmstadt, Germany); Total RNA Isolation Reagent (TRIR) was obtained from ABgene (Darmstadt, Germany) and Plasmid Midi Kit from Qiagen (Hilden, Germany). FuGeneR HD Transfection Reagent was from Roche Applied Science (Mannheim, Germany). The Dual-Glo Luciferase Assay System was from Promega (Mannheim, Germany). The forward primers (FP), reverse primers (RP) and probes for Actb, Sftpb and TTF-1 were from Eurogentec (Cologne, Germany). Neuregulin 1β was produced using an expression vector kindly provided by Kermit Carraway III (UC Davis, CA) and purified by Dr. Ann Kane, Phoenix Laboratory (Tufts Medical Center, Boston, MA). NRG was diluted in glutamine containing DMEM and used it in a final concentration of 33 nM.
2.1 Overall experimental approach
The first aspect of our approach was focused on the individual and combined effects of ErbB4 and TTF-1 on cell viability (3.1), Sftpb mRNA and protein expression (3.2) and promoter activity (3.3). This was followed by studies elucidating the interactions of these two proteins by studying their cellular colocalization (3.4), and confirming it in subcellular fractionation (3.5) and co-immunoprecipiation studies (3.6). Next we studied the effect of neuregulin on TTF1 expression (3.7) and promoter activity (3.8). In order to exclude endogenous ErbB4 effects on this interaction we repeated the experiments in primary fetal ErbB4-deleted type II cells. Finally we analyzed whether ErbB4 cleavage and nuclear trafficking were critical for TTF-1-induced Sftpb regulation (3.9) and TTF-1 expression (3.10).
2.2 Plasmids
pEGFP N3 (control), pHER4 (full-length human ErbB4 receptor), pHER4muNLS (human ErbB4 receptor with defective nuclear localization signal), and Sftpb promoter luciferase reporter plasmid were used as previously published [27, 29, 31]. HER4 refers to the human ErbB4 receptor plasmid in the rest of the manuscript, pRC/CMV/Nkx2.1 (TTF-1 expression plasmid) and TTF-1 promoter luciferase reporter plasmid was kindly provided by Dr. Whitsett (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH) [32].
2.3 Transfection experiments
A mouse lung epithelial cell line (MLE-12) was used for most of the transfection experiments. This cell line was established from pulmonary tumors in a mouse transgenic for the SV40 large T antigen under the control of the promoter region of the human Sftpc gene. The cells secrete phospholipids in response to phorbol esters and ATP, but not in response to forskolin [33]. MLE-12 cells are easily transfectable and express Sftpb, TTF-1, and ErbB4. Also, their response to NRG is comparable to fetal type II cells [22]. Transfection protocols were used as previously published [29]. To examine the effects of overexpression of ErbB4 and TTF-1 and to study its effect on Sftpb promoter and TTF-1 promoter activity, MLE-12 cells were transfected with a human ErbB4 construct, [28] a TTF-1 construct, an empty Enhanced Green Fluorescent Protein (EGFP) control construct, and a Sftpb promoter luciferase reporter plasmid [28] or a TTF-1 promoter luciferase reporter plasmid. The amount of DNA was adjusted as suggested by the manufacturer and previously published [28, 29] to keep the amount of transfected DNA and transfection reagent similar in each transfection experiment. To study the importance of the ErbB4 nuclear translocation, cells were transfected with an HER4 mutant, defective in the nuclear localization signal (HER4muNLS) [28]. The cells were transfected for 48 hours using FuGene transfection reagent as previously described [28] in accordance to the manufacturer suggestions.
2.4 MTT-assay
Cell viability was examined using a MTT-assay. In this colorimetric assay the yellow MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, a yellow tetrazole) is reduced to purple formazan in the mitochondria of living, metabolically active cells. The amount of formazan is directly proportional to the cell number [34]. MLE-12 cells were grown in a 96-well plate at a concentration of 2.5 × 103 cells per well. After a 3-hour serum starvation, cells were treated for 24 hours with serum-free DMEM (controls) or NRG (33 nM). After incubating with MTT-solution for 2 hours at 37 °C, the reaction was stopped by isopropanol-HCl. Media was removed and 3 % SDS and isopropanol-HCl were added. Absorption was measured at 570 nm.
2.5 RNA isolation and cDNA synthesis
Cells were plated in 6 well plates until they reached 90 % confluence. After being serum-starved for 3 hours, cells were treated for 24 hours with serum-free DMEM (controls) or NRG (33 nM). Total RNA Isolation Reagent was used for cell lysis. RNA was isolated by guanidinium thiocyanate lysis followed by acid phenol/chloroform extraction [35]. After reversed transcription, 5 μg of total RNA was used for cDNA synthesis in a 15 μl reaction volume containing 1x DTT, 0.2 μg Hexamer Primer, and 5x Bulk Mix for 1 hour. The resulting cDNA was used for real-time amplification reactions.
2.6 Real time polymerase chain reaction (RT-PCR)
The cDNA levels of the Sftpb gene, the TTF-1 gene, and the Actb gene were measured by real-time PCR. Actin was used as a reference gene and internal control to normalize the target gene cDNA level for Sftpb and TTF-1. The linearity of each RT-PCR reaction was confirmed by using serial dilutions of the DNA template (primer efficiency: β-Actin 2.6, Sftpb 2.1, TTF-1 2.1). The 20 μl reaction mixture contained 1 μl of the cDNA template, 10 μl TaqMan universal master mix, 300 nM each of forward primer (FP) and reverse primer (RP), and 200 nM probe (Table 1) [23]. An ABI PRISM 7900 sequence detection system was used for amplification and detection of specific products. The initial denaturation and enzyme activation was done at 95 °C for 10 minutes, followed by 45 cycles at 95 °C for 15 seconds, and 60 °C for 1 minute. The threshold cycle (Ct) was determined for each gene. For relative expression levels, ΔCt values were calculated as differences in the Ct values of the target gene and the reference gene. The difference in the ΔCt values of the treated cells compared to the control cells were calculated as ΔΔCt and presented inversely proportional to the levels of Sftp or TTF-1 mRNA. For each assay, specificity was confirmed by sequencing the PCR products on both strands using capillary electrophoresis with POP-6™-Polymer on an ABI 3100 Genetic Analyzer (Applied Biosystems).
Table 1.
| ß-Actin | |
|---|---|
| Forward Primer (FP): | 5′-AGGTCATCACTATTGGCAACGCA-3′ |
| Reverse Primer (RP): | 5′-CAACGTCACACTTCATGATGGA-3′ |
| Probe: | 5′(FAM)-AGCCTTCCTTCTTGGGTATGGAATCCTGT-(TAMRA)3′ |
|
| |
| Surfactant protein B (Sftpb) | |
| Forward Primer (FP): | 5′-ATGGCCAAGTCGCACCTACT-3′ |
| Reverse Primer (RP): | 5′-CCAGAATTGAGGGCCTTGTG-3′ |
| Probe: | 5′(FAM)-CCCAGGTGCAGCTATCACGTCGG-(TAMRA)3′ |
|
| |
| TTF-1 | |
| Forward Primer (FP): | 5′-CAC Acg ACT CCg TTC TCAgTg T-3′ |
| Reverse Primer (RP): | 5′-CATGCAGGCAAAGCAGTCA-3′ |
| Probe: | 5′(FAM)-TgA CAT CTT gAg TCC CCT g-(TAMRA)3′ |
2.7 Co-immunoprecipitation and Western blotting
MLE-12 cells were serum-starved for 3 hours and stimulated for 2 minutes with NRG (33 nM), treated for 24 hours with NRG (33 nM) or left untreated (controls). Cells were lysed in lyses buffer or co-immunoprecipitation buffer as described before [28, 36]. Lysates were cleared by microcentrifugation for 15 minutes at 4 °C. A Lowry protein assay was used to determine total concentration of protein. For co-immunoprecipitation 300 μg of total protein was incubated for 90 minutes with the specific receptor antibody (anti-ErbB4 C-7, 1:10; anti-TTF-1 H-190, 1:50) at 4 °C. Protein-A-sepharose was added and incubation was continued for another 90 minutes at 4 °C. The beads were collected by microcentrifugation at 14000 rpm for 10 minutes, washed three times with co-immunoprecipitation washing buffer and boiled in Laemmli buffer for 5 minutes at 100 °C. The proteins were separated by 10 % SDS polyacrylamide gel electrophoresis (resolving gel: 10 % (v /v) acrylamide; 400 mM TrisHCl pH 8,8; 0,1 % (v / v) SDS; 0,07 % (v / v) TEMED; 0,05 % (v / v) APS; stacking gel: 4 % (v / v) acrylamide; 125 mM TrisHCl pH 6,8; 0,1 % (v / v) SDS; 0,1 % (v / v) TEMED; 0,05 % (v / v) APS) and transferred to a nitrocellulose membrane. Blots were blocked in 1 % BSA and incubated with antibodies against individual proteins (purified mouse anti-phosphotyrosine antibody, 1:1000; anti-TTF-1 H-190, 1:750; anti-ErbB4 C-18, 1:750; anti-Sftpb, 1:750; or anti-β-Actb, 1:15000) overnight at 4 °C. Secondary antibody (HRP-labeled goat anti-mouse IgG, 1:5000; or goat anti-rabbit IgG, 1:10000) was applied and the proteins were visualized by enhanced chemiluminescence (ECL). Membranes were stripped in strip buffer for 30 minutes at 50 °C, up to three times. An immunoprecipitation with a control rabbit IgG antibody was performed and probed with ErbB4 and TTF-1 antibody to control for the ErbB4 and TTF-1 antibody specificity.
2.8 Luciferase reporter gene assay
Cells were transfected with the Sftpb promoter luciferase reporter plasmid or the TTF-1 promoter luciferase reporter plasmid and the renilla luciferase control vector (pRL-TK) alone or together with the different plasmids as described above. 48 hours after transfection cells were starved for 3 hours and treated with DMEM (control) or NRG (33 nM) for 24 hours. Luciferase assay was done using the Dual-Glo Luciferase assay system. Briefly, luciferase reagent was added to cells in growth media. After 10 minutes incubation the firefly luminescence was measured. Stop and Glo reagent was added to the cells and renilla luciferase was measured after a 10-minute incubation. The ratio of luminescence from the experimental reporter to the luminescence from the control reporter was calculated and normalized to a control well.
2.9 Confocal microscopy
MLE-12 cells were grown on glass cover slips in 24-well plates. After a 3-hour serum starvation, cells were treated with serum-free DMEM (controls) or stimulated for 2 minutes or treated for 24 hours with NRG (33 nM). Cells were washed with PBS, fixed for 20 minutes in 3 % paraformaldehyde, rinsed again with PBS, permeabilized in 0.5 % TritonX-100/ PBS for 5 minutes and blocked for 1 hour with 10 % normal goat serum. After a 30-minute incubation with the primary antibody (ErbB4 clone HFR-1, 1:25; TTF-1 H-190, 1:50), cells were washed with PBS and incubated with a secondary antibody (Alexa Fluor 488, 1:100; Alexa Fluor 568, 1:200) for another 30 minutes. After staining with DAPI (1:100000) for 10 minutes, cells were mounted onto glass slides and examined using a Leica Inverted-2 DM IRB confocal laser scanning microscope connected to a TCS SP2 AOBS scanhead (Leica, Wetzlar, Germany).
2.10 Subcellular fractionation
MLE-12 cells were serum-starved for 3 hours and stimulated for 2 minutes or treated for 24 hours with NRG (33 nM). Cells were scraped in PBS and the pellet was resuspended in Nuclei Buffer (pH 7.9, 10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 1 % Triton X-100, 0.5 mM DTT). After 10-minutes incubation on ice, cells were centrifuged for 10 minutes at 10000g, 4 °C. The supernatant (cytoplasmic fraction) was stored at −80 °C and the pellet was resuspended in high salt buffer (pH 7.9, 10 mM Hepes, 400 mM NaCl, 0.1 mM EDTA, 5 % glycerol, and 0.5 mM DTT) and kept on ice for 20 minutes. The nuclei fraction was cleared by microcentrifugation for 10 minutes at 10000g, 4 °C. The supernatant (nuclear fraction) was stored at −80 °C until further processed. Proteins were separated on a 10 % SDS polyacrylamide gel and further processed as described in the co-immunoprecipitation section.
2.11 Preparation of primary fetal mouse ErbB4-deleted type II epithelial cell cultures
Primary type II cells were freshly isolated from time-dated pregnant transgenic ErbB4 HER4heart(−/−) mice as previously described [29]. These mice have been rescued from their lethal cardiac defects by expressing a human ErbB4 (HER4heart) cDNA under the cardiac-specific α-myosin heavy chain promoter [37]. Due to breeding difficulties of the transgenic mice and a lower transfection efficiency seen in primary fetal type II cells, especially in the experiments where two or three plasmids were used, only experiments focusing on ErbB4 effects on endogenous TTF-1 expression were performed in primary fetal type II cells with an ErbB4 negative background. This eliminated the interference of the plasmid-derived from the endogenous-expressed ErbB4, present in all MLE-12 experiments. The animal research protocol was approved by the institutional IACUC at Hannover Medical School. Pregnant HER4heart (−/−) mice were sacrificed at E17.5 of gestation by CO2 inhalation. The CO2 effect on the fetal lung cells is minimal because the exposure time used for euthanasia of the maternal mice is very short [19, 29, 38, 39]. Fetal lungs were removed, washed in sterile Hank‘s buffered salt solution, minced with a razorblade, and incubated with Collagenase Type II (Worthington Biochemical Corp., Lakewood, NJ) for 2 hours at 37 °C. The reaction was stopped on ice for 30 minutes and the tissue was centrifuged, the pellet resuspended in DMEM, and incubated for another 30 minutes on ice. After a second centrifugation the pellet was resuspended in DNase and trypsin and incubated for 12 minutes at 37 °C. The reaction was stopped by DMEM containing 10 % fetal calf serum (FCS). The cells were filtered, centrifuged, resuspended in DMEM containing 10 % FCS, and plated in culture dishes for 60 minutes at 37 °C (21 % 02 / 5 % C02) to allow differential adherence of lung fibroblasts. For type II cell isolation the supernatant from the first differential adherence was centrifuged, the cell pellet resuspended in DMEM containing 10 % FCS, and plated in culture flasks for 60 minutes at 37 °C for a second differential adherence. Supernatants were removed and centrifuged. Cell pellets were resuspended and plated in DMEM containing 20 % FCS and grown until further use.
2.12 Data analysis
All values were presented as mean ± SEM and the values of the treated groups were presented as relative values normalized for the experimental specific control group unless otherwise stated. The effects of TTF-1 and ErbB4 overexpression on cell viability, TTF-1 and Sftpb expression, and TTF-1 and Sftpb promoter activity were expressed as percentages of controls transfected with EGFP. The effect of NRG was expressed as percentage of EGFP-transfected cells treated with NRG. The effects of the human ErbB4 mutant (HER4muNLS) on TTF-1 and Sftp expression, and TTF-1 and Sftpb promoter activity were expressed as percentages of HER4-transfected cells. To evaluate the results for their statistical significance a two-way ANOVA was used. A post hoc Bonferroni correction for multiple comparisons was used when appropriate. Values of P<0.05 were considered statistically significant.
3. Results
3.1 Co-expression of HER4 and TTF-1 decreases lung epithelial cell viability under control and NRG treatment conditions
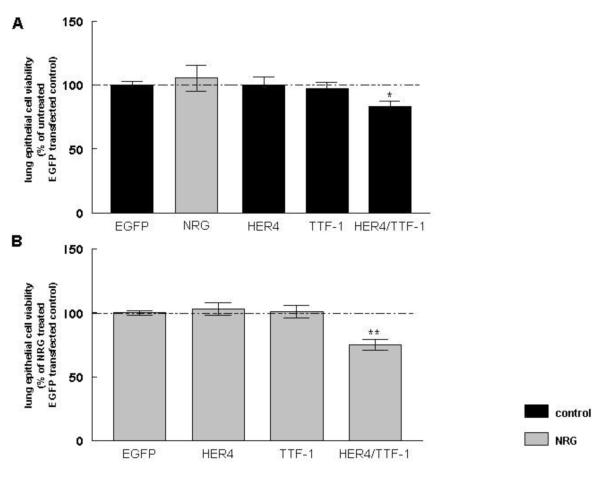
To study the effect of plasmid-derived ErbB4 and TTF-1 on lung epithelial cell viability, untreated and NRG-treated MLE-12 cells were transfected with full-length human ErbB4 (HER4), TTF-1, or a combination of HER4 and TTF-1 (HER4/TTF-1). NRG treatment had no effect on cell viability in EGFP-transfected control cells (105 ± 10 %, n=11, P>0.05). Overexpression of HER4 (100 ± 6 %, n=11, P>0.05) and TTF-1 (97 ± 5 %, n=11, P>0.05) had also no effect, whereas co-expression of HER4 and TTF-1 significantly decreased lung epithelial cell viability to 83 ± 4 % (n=11, P<0.05). All results were compared to untreated (100 ± 3 %, n=11) cells transfected with an EGFP control plasmid (Figure 1A). 24 hours of NRG treatment did not change cell viability in HER4 (103 ± 5 %, n=10, P>0.05) or TTF-1 (101 ± 5 %, n=10, P>0.05) overexpressing cells. NRG did not change the inhibitory effect on cell viability induced by HER4/TTF-1 overexpression (75 ± 4 %, n=10, P<0.001). NRG treatment results were compared to NRG-treated cells transfected, with an EGFP control plasmid (100 ± 2 %, n=10) (Figure 1B).
Figure 1. Co-expression of HER4 and TTF-1 decreases lung epithelial cell viability under control and NRG treatment conditions.
Lung epithelial cell viability was measured in MLE-12 cells, transfected with full-length ErbB4 (HER4), TTF-1 or HER4, and TTF-1 (HER4/TTF-1) under control conditions (black bars) and after 24 hours NRG treatment (grey bars). Results were compared to EGFP -transfected control cells (A) or NRG-treated EGFP-transfected cells (B). *P ≤0.01, **P ≤0.001, n= 10-11.
3.2 Co-expression of HER4 and TTF-1 stimulates Sftpb mRNA and protein expression under control and NRG treatment conditions
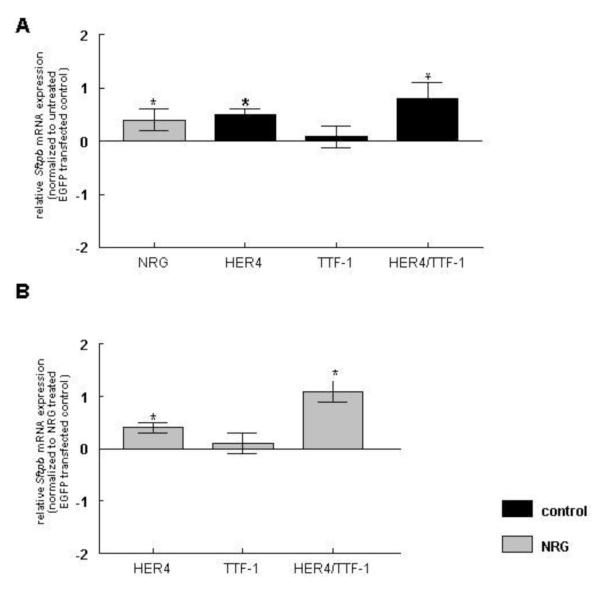
To examine whether plasmid-derived ErbB4 and TTF-1 overexpression had an effect on Sftpb mRNA (Figure 2A, B) and protein expression (Figure 2C, D), untreated and NRG-treated MLE-12 cells were transfected with HER4, TTF-1, and a combination of HER4 and TTF-1 (HER4/TTF-1). NRG treatment had a stimulatory effect on Sftpb mRNA expression in EGFP transfected control cells (0.4 ± 0.2, n=15, P<0.05). Overexpression of human ErbB4 (HER4) significantly increased Sftpb mRNA expression (0.5 ± 0.1, n=10, P<0.01) whereas the overexpression of TTF-1 alone had no effect (0.08 ± 0.2, n=11, P>0.05). The co-expression of both proteins stimulated Sftpb mRNA expression (0.8 ± 0.3, n=9, P<0.01). Results were compared to untreated cells transfected with an EGFP control construct (Figure 2A). NRG treatment did not change the effect of the previously mentioned overexpression on Sftpb mRNA expression, keeping the increase in HER4 (0.4 ± 0.1, n=8, P<0.05) and HER4/TTF-1 (1.1 ± 0.2, n=8, P<0.01) overexpressing cells and did not effect it in TTF-1 overexpessing cells (0.1 ± 0.2 n=9, P>0.05) when compared to NRG-treated cells transfected with an EGFP control construct (Figure 2B).
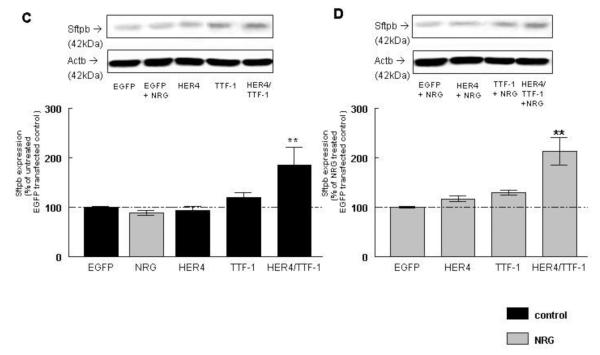
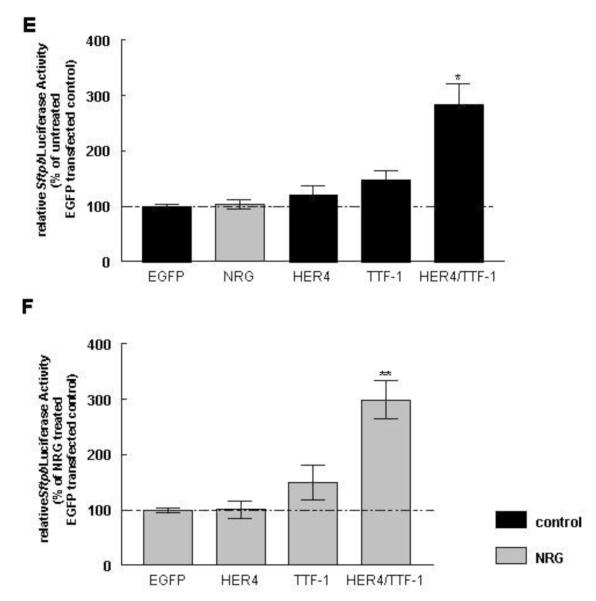
Figure 2. Co-expression of ErbB4 and TTF-1 stimulates Sftpb mRNA, protein expression and promoter activity under control and NRG treatment conditions.
MLE-12 cells were transfected with HER4, TTF-1 or HER4/TTF-1. Sftpb mRNA (A, B) and protein (C, D) expression was measured under control conditions (black bar) and after 24 hours treatment with NRG (grey bars). Results were compared to EGFP-transfected control cells (A, C) or NRG-treated EGFP-transfected cells (B, D). Representative Western Blots are shown on the upper part of Figure 2C and D. Sftpb promoter activity was measured in MLE-12 cells transfected with full-length ErbB4 (HER4), TTF-1 or HER4/TTF-1 under control conditions (black bars) and after NRG treatment (grey bars). Results were compared to EGFP-transfected control cells (E) or NRG-treated EGFP-transfected cells (F). *P ≤0.01, **P ≤0.001, n=8-21
NRG treatment had no additional effect on Sftpb protein expression in EGFP-transfected cells (89 ± 4 %, n=13, P>0.05). HER4 overexpression did not change Sftpb protein expression (94 ± 6 %, n=13, P>0.05), whereas TTF-1 overexpression led to a trend towards an increase (120 ± 9 %, n=13, P>0.05) and the combined expression of HER4 and TTF-1 (185 ± 36 %, n=13, P<0.001) significantly stimulated the Sftpb protein expression, when compared to cells transfected with the EGFP control construct (100 ± 2 %, n=13) (Figure 2C). NRG treatment did not change the Sftpb protein expression in HER4 overexpressing cells (117 ± 6 %, n=13, P>0.05), but led to a trend towards an increase to 130 ± 5 % (n=11, P>0.05) in TTF-1 overexpressing cells. The significant increase in Sftpb protein expression to 213 ± 28 % (n=13, P<0.001) in NRG-treated HER4/TTF-1 overexpressing cells was not statistically different from non NRG-treated transfected cells. Results were compared to EGFP-transfected cells treated with NRG (100 ± 2 %, n=13) (Figure 2D).
3.3 Co-expression of HER4 and TTF-1 induces Sftpb promoter activity under control and NRG treatment conditions
To determine if plasmid-derived ErbB4 and/or TFF-1 expression stimulate Sftpb promoter activity, untreated and NRG-treated MLE-12 cells were transfected with a Sftpb promoter luciferase reporter plasmid. NRG treatment had no effect on luciferase activity in EGFP-transfected cells (103 ± 8 %, n=20, P>0.05). Cotransfection of the Sftpb promoter luciferase reporter with full-length human ErbB4 (HER4) or TTF-1 showed a trend towards an increased luciferase activity in untreated cells (121 ± 16 %, n=18, P>0.05; 147 ± 17 %, n=18, P>0.05, respectively). A combined expression of HER4 and TTF-1 showed a significant increase in luciferase activity to 283 ± 35 % (n=17, P<0.01) when compared to EGFP-transfected untreated cells (100 ± 3 %, n=21) (Figure 2E). NRG did not change these responses in HER4- and in TTF-1-transfected cells (101 ± 16 %, n=17, P>0.05; 149 ± 31 %, n=17, P>0.05, respectively). The combined expression of HER4 and TTF-1 let to a significant increase in luciferase activity to 299 ± 35 % (n=19, P<0.001) in NRG-treated cells, which was similar to the response in non NRG-treated transfected cells. Results were compared to EGFP-transfected, NRG-treated cells (100 ± 4 %, n=20) (Figure 2F).
3.4 NRG induces nuclear translocation of ErbB4 in MLE12 cells
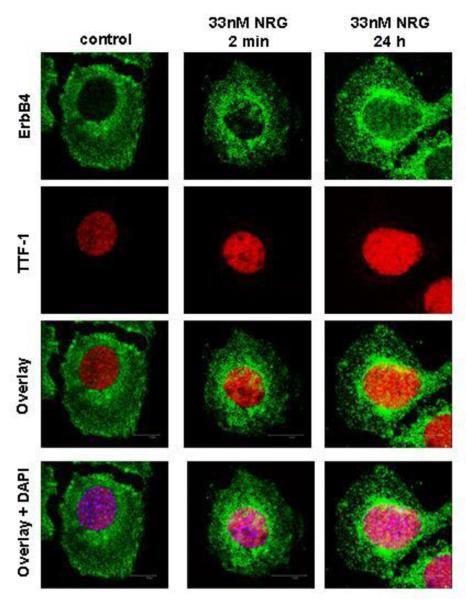
Endogenous TTF-1 localized to the nucleus while endogenous ErbB4 localized to the cell membrane and the cytoplasm in untreated cells only. Short NRG stimulation led to a more pronounced perinuclear and nuclear staining, leading to co-localization of ErbB4 and TTF-1 in this region. This was further enhanced in cells treated for 24 hours with NRG. DAPI staining confirmed nuclear localization of these proteins (Figure 3).
Figure 3. NRG induces nuclear translocation of ErbB4 in MLE-12 cells.
MLE-12 cells were stimulated with NRG for 2 minutes or treated for 24 hours. Confocal microscopy was used to study the cellular localization of ErbB4 (green dye) and TTF-1 (red dye) and the co-localization of both proteins (overlay). DAPI (blue dye) was used to confirm nuclear staining.
3.5 NRG induces ErbB4 cleavage and enrichment of 4ICD in the nuclear fraction
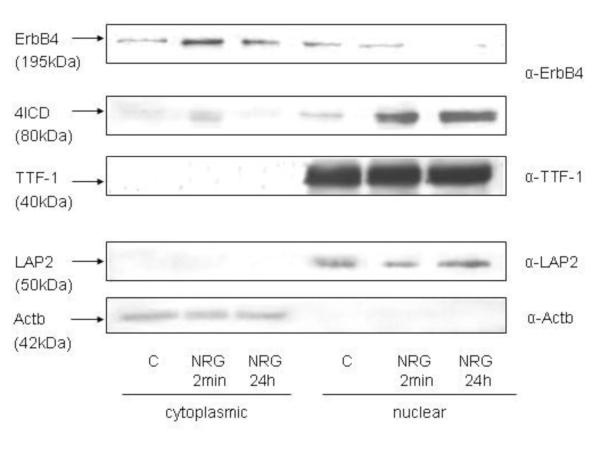
Subcellular fractionation studies isolating the cytoplasmic and nuclear compartments of the cell confirmed the localization of endogenous ErbB4 and endogenous TTF-1 in NRG-stimulated (2 minutes) and NRG–treated (24 hours) MLE-12 cells seen by confocal imaging. Actb (42 kDa) and LAP2 (53 kDa) antibody probing was used to confirm the purity of the cytoplasmic and nuclear fraction [40, 41]. The full-length ErbB4 receptor (195 kDa) was found in the cytoplasmic fraction and in the nuclear fraction in control cells and to a small amount in the nuclear fraction after a 24-hour NRG treatment. Two minutes of NRG stimulation and more pronounced a 24-hour treatment with NRG led to enrichment of ErbB4’s 80 kDa intracellular domain 4ICD in the nucleus. The transcription factor TTF-1 localized to the nucleus independent of NRG treatment (Figure 4).
Figure 4. NRG induces ErbB4 cleavage and enrichment of 4ICD in the nuclear fraction.
Cell fractionation was done in MLE-12 cells under control conditions (C) and after 2 minutes (NRG 2 min) or 24 hours (NRG 24 h) NRG treatment. Blots showing the cytoplasmic and the nuclear fraction were probed with ErbB4, TTF-1, LAP2, and β-Actin (Actb) antibodies.
3.6 ErbB4 and TTF-1 co-precipitate each other
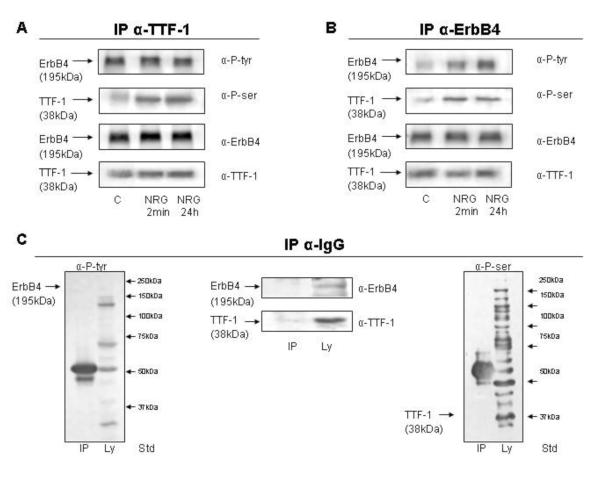
We further confirmed the interaction of endogenous ErbB4 and endogenous TTF-1 by co-immunoprecipitation analyses of both proteins. Both co-precipitated each other under all conditions tested. NRG stimulation induced tyrosine phosphorylation of ErbB4 and serine phosporylation of TTF-1, indicating the tight interactions of these two proteins (Figure 5A and 5B). Immunoprecipitations with a control IgG antibody did not show any nonspecific antibody binding (Figure 5C).
Figure 5. ErbB4 and TTF-1 co-immunoprecipitate each other.
TTF-1 co-immunoprecipitation (Co-IP) (A) and ErbB4 Co-IP (B) were done in MLE-12 cells under control conditions and after 2 minutes (NRG 2 min) or 24 hours (NRG 24 h) NRG treatment. An IgG IP (C) was done to confirm antibody specificity. Blots were probed with anti-phosphotyrosine (α-P-tyr), anti-phosphoserine (α-P-ser), anti-ErbB4, and anti-TTF-1 antibody as stated on the right side next to the blots.
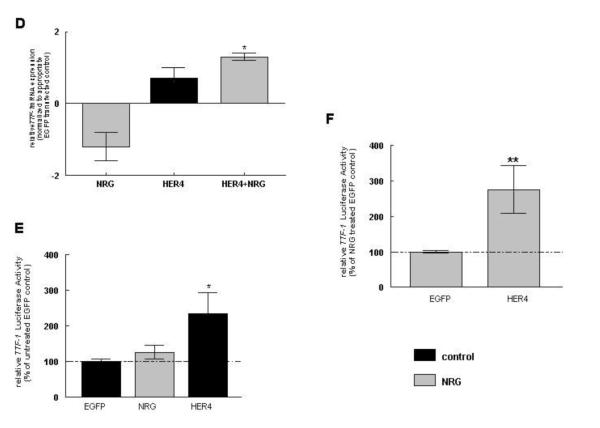
3.7 NRG stimulates TTF-1 mRNA and protein expression in HER4 expressing cells
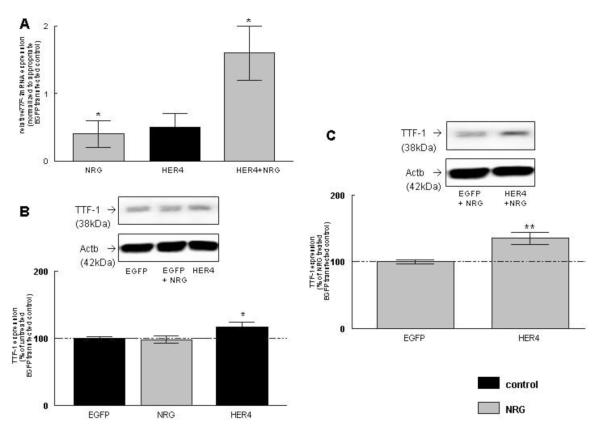
NRG significantly induced TTF-1 mRNA expression in EGFP-transfected control MLE-12 cells (0.4 ± 0.2, n=15, P<0.05). Plasmid-derived HER4 expression showed a trend towards an increase in its expression (0.5 ± 0.2, n=10, P>0.05), but did not reach statistical significance compared to EGFP-transfected control cells. NRG treatment significantly increased TTF-1 mRNA expression in HER4-overexpressing cells (1.6 ± 0.4, n=9, P<0.01) compared to NRG-treated EGFP-expressing control cells (Figure 6A).
Figure 6. NRG stimulates TTF-1 mRNA, protein expression, and promoter activity in ErbB4-overexpressing cells.
TTF-1 mRNA (A) and protein (B, C) expression was measured in MLE-12 cells transfected with HER4 under control conditions (black bar) and after 24 hours of treatment with NRG (grey bars). Results were compared to EGFP-transfected control cells or NRG-treated EGFP-transfected cells. Representative Western Blots are shown above the densitometric graphs (B, C). Fetal HER4heart (−/−) type II epithelial cells were transfected with HER4. TTF-1 mRNA expression was measured under control conditions (black bars) and after 24 hours of treatment with NRG (grey bars). Results were compared to EGFP-transfected control cells or NRG-treated EGFP-transfected cells (D). TTF-1 promoter activity was measured in MLE-12 cells transfected with a control plasmid (EGFP) or full-length ErbB4 (HER4 under control conditions (black bars) and after NRG treatment (grey bars). Results were compared to EGFP-transfected control cells (E) or NRG-treated EGFP-transfected cells (F). *P ≤0.01, **P ≤0.001, n=9-19
NRG treatment did not changed TTF-1 protein expression (98 ± 5 %, n=19, P>0.05) in EGFP-transfected control cells, whereas HER4 overexpression significantly increased the TTF-1 expression to 117 ± 7 % (n=10, P<0.01), when compared to untreated cells transfected with an EGFP control construct (100 ± 2 %, n=19) (Figure 6B). NRG treatment significantly increased TTF-1 protein expression in HER4 overexpressing cells (135 ± 9 %, n=10, P<0.001), when compared to NRG-treated cells transfected with an EGFP control construct (100 ± 3 %, n=19), but this increase was not different from the response in non NRG-treated transfected cells (Figure 6C).
To separate the plasmid-derived from the endogenous ErbB4 effects, we confirmed the effect of NRG stimulation and ErbB4 presence on TTF-1 mRNA expression in primary ErbB4-deleted fetal mouse type II cells. Fetal cells were freshly isolated from HER4heart (−/−) lungs transfected with a full-length ErbB4 or a GFP control vector and treated with NRG. NRG treatment showed a trend towards a decrease in endogenous TTF-1 mRNA expression in ErbB4-deleted EGFP-transfected control cells (−1.2 ± 0.4, n=4, P>0.05), whereas re-expression of the human ErbB4 (HER4) showed a trend towards an increase in TTF-1 mRNA expression in untreated cells (0.7 ± 0.3, n=5, P>0.05), compared to cells transfected with an EGFP control construct. NRG treatment significantly stimulated the TTF-1 mRNA expression in HER4 re-expressing primary type II cells (1.3 ± 0.1, n=3, P<0.01) when compared to NRG-treated type II cells transfected with an EGFP control construct (Figure 6D).
3.8 HER4 induces TTF-1 promoter activity
To determine if expression of plasmid-derived HER4 stimulates the TTF-1 promoter activity, untreated and NRG-treated MLE-12 cells were transfected with a TTF-1 promoter luciferase reporter plasmid. NRG treatment had no significant effect on TTF-1 promoter luciferase activity (126 ± 20 %, n=8, P>0.05) in EGFP-transfected cells, while transfection with full-length human ErbB4 (HER4) increased luciferase activity in untreated cells (234 ± 59 %, n=7, P<0.01) when compared to untreated (100 ± 6 %, n=8) cells transfected with the EGFP control construct (Figure 6E). In NRG-treated HER4 overexpressing cells the luciferase activity was increase (275 ± 68 %, n=6, P<0.001), when compared to NRG-treated control cells transfected with EGFP (100 ± 4 %, n=8) (Figure 6F). This increase was not different from the response in non NRG-treated transfected cells.
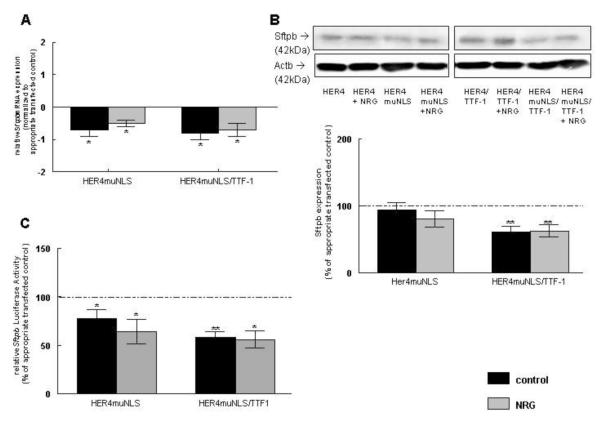
3.9 Nuclear translocation of HER4 is required for the ErbB4/TTF-1-induced Sftpb expression
To examine whether the ErbB4 nuclear translocation is required for ErbB4 effects on Sftpb mRNA (Figure 7A), protein expression (Figure 7B), or Sftpb promoter activity (Figure 7C), untreated and NRG-treated MLE-12 cells were transfected with a plasmid-derived HER4 construct, which has a mutated nuclear localization sequence (HER4muNLS). Prevention of HER4 nuclear entry prevented the ErbB4-induced increase of Sftpb mRNA expression (−0.7 ± 0.2, n=8, P<0.01). Neither NRG treatment (−0.5 ± 0.1, n=8, P<0.01, respectively) nor additional TTF-1 overexpression in untreated and NRG-treated cells did rescue or overcome this inhibition (−0.8 ± 0.2, n=9, P<0.01; −0.7 ± 0.2, n=6, P<0.05, respectively) (Figure 7A). All results were compared to untreated or NRG-treated cells transfected with the full-length ErbB4 receptor construct (HER4) or to untreated or NRG-treated HER4/TTF-1-transfected cells, respectively.
Figure 7. Nuclear translocation of ErbB4 is required for the ErbB4/TTF-1-induced Sftpb expression.
MLE-12 cells were transfected with HER4, HER4/TTF-1, HER4muNLS, or HER4muNLS/TTF-1. Sftpb mRNA (A), protein (B) expression and Sftpb promoter activity (C) was measured under control conditions (black bars) and after 24 hours treatment with NRG (grey bars). Results were compared to untreated or NRG-treated cells transfected with HER4 or HER4/TTF-1. *P ≤0.01 **P ≤0.001, n=5-9 Representative Western Blot is shown above the results of the densitometric readings (B).
In contrast to the effect on gene expression, expression of a plasmid-derived HER4muNLS mutant resulted in minimal change of Sftpb protein expression towards a decrease in untreated (89 ± 10 %, n=7, P>0.05) and in NRG-treated cells (81 ± 10 %, n=7, P>0.05) compared to untreated or NRG-treated HER4-transfected cells (100 %, n=7; 100 %, n=7, respectively). Additional TTF-1 expression somewhat potentiated the decrease in Sftpb protein expression making it statistical significant in untreated (58 ± 7 %, n=7, P<0.001) and NRG-treated cells (59 ± 8 %, n=7, P<0.01). This underlines the importance of ErbB4 nuclear translocation in ErbB4/TTF-1 signaling on Sftpb expression. The values were compared to untreated or NRG-treated HER4/TTF-1-transfected cells (100 %, n=7; 100, n=7, respectively) (Figure 7B).
The inability of HER4 to translocate to the nucleus when expressing plasmid-derived HER4muNLS, decreased Sftpb promoter activity significantly to 78 ± 9 % (n=7, P<0.01) in untreated cells. NRG treatment did not rescue this inhibition (64 ± 13 %, n=5, P<0.01). Results were compared to untreated (100 ± 2 %, n=7) or NRG-treated (100 ± 6 %, n=7) HER4-transfected cells. Additional overexpression of TTF-1 did not significantly change this inhibitory effect, leading to a decrease in Sftpb promoter activity in untreated (58 ± 6 % n=7, P<0.0001) and NRG-treated cells (56 ± 9 %, n=6, P<0.01). Transfected cells were compared to untreated (100 ± 5 %, n=7) or NRG-treated (100 ± 7 %, n=7) HER4/TTF-1-transfected cells (Figure 7C).
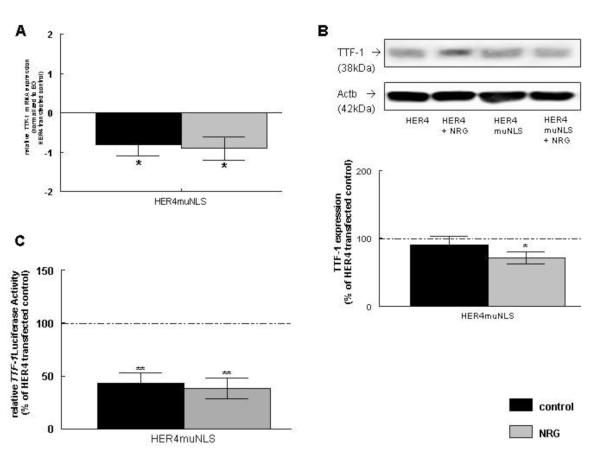
3.10 Nuclear translocation of HER4 is required for TTF-1 expression
To study the effect of ErbB4 nuclear translocation on TTF-1 mRNA (Figure 8A), protein expression (Figure 8B), and TTF-1 promoter activity (Figure 8C), untreated and NRG-treated MLE-12 cells were transfected with plasmid-derived HER4muNLS. Inability of HER4 to localize to the nucleus inhibited TTF-1 mRNA expression in untreated cells (−0.8 ± 0.3, n=8, P<0.05). NRG treatment did not overcome this inhibitory effect (−0.9 ± 0.3., n=10, P<0.01). Results were compared to untreated or NRG-treated cells transfected with the full-length human ErbB4 (HER4) construct (Figure 8A; compare to results in Figure 6A).
Figure 8. Nuclear translocation of ErbB4 is required for TTF-1 expression.
MLE-12 cells were transfected with HER4, or HER4muNLS. TTF-1 mRNA (A), protein (B) expression, and TTF-1 promoter activity (C) was measured under control conditions (black bars) and after 24 hours of treatment with NRG (grey bars). Results were compared to untreated or NRG-treated cells transfected with HER4. *P ≤0.01 **P ≤0.001, n=7-10 Representative Western Blots are shown above the results of the densitometric readings (B).
Inability of HER4 to translocate to the nucleus did not change TTF-1 protein expression when compared to HER4-transfected cells (81 ± 12 %, n=7, P>0.05), but significantly inhibited NRG-induced upregulation of TTF-1 in HER4-transfected cells (76 ± 8 %, n=7, P<0.05). All effects were compared to untreated or NRG-treated cells transfected with the full-length HER4 construct (100, n=7; 100, n=7, respectively) (Figure 8B; compared to results in Figure 6B and C).
Expression of plasmid-derived HER4 mutant also inhibited TTF-1 luciferase activity to 43 ± 10 % (n=8, P<0.0001) in untreated and to 38 ± 10 % (n=8, P<0.001) in NRG-treated cells, showing that there is no additional effect added by NRG treatment. All results were compared to untreated (100 ± 5 %, n=8) or NRG-treated cells (100 ± 8 %, n=9) transfected with full-length HER4 (Figure 8C; compared to results in Figure 6E and F).
4. Discussion
Transcription factors play a central role in regulating expression of tissue-specific genes to control the function, homeostasis, and differentiation of these tissues. TTF-1 is an important transcriptional regulator that plays a key role in cell differentiation [2]. Its function in fetal lung development is illustrated by studies of TTF-1 null mice that lack lung parenchyma [5]. TTF-1 regulates Sftpb promoter activity [42] and its deficiency results in an inhibition of branching morphogenesis [4] and early postnatal death [43]. In the lung, it has been shown that poly (ADP-ribose) polymerases (PARP-2 and PARP-1) enhance TTF-1-mediated activation of the Sftpb gene [44], but there is limited information on the regulation of TTF-1 gene expression by hormones, cytokines, and other biological agents like growth factors [2]. Here we present data for ErbB4 being a potential upstream regulator of TTF-1.
Co-expression of human ErbB4 and TTF-1 results in a decrease in lung epithelial cell proliferation. ErbB4 is known to be mostly involved in differentiation processes, and not primary in cell proliferation like other ErbB receptors [45]. This result is in alignment with the known function of TTF-1 being essential for the morphogenesis and differentiation of the thyroid, the lung, and ventral forebrain [2]. This effect of TTF-1 may relate to the mechanisms in developing maturing cells, where the control of growth and differentiation are reciprocally linked [46].
Surfactant synthesis is a crucial component of fetal lung development in preparation for birth [47]. Deficiency of pulmonary surfactants causes respiratory distress syndrome (RDS) among preterm infants [48]. The growth factor neuregulin (NRG) is required for the initiation of fetal surfactant synthesis [22]. Down-regulation of ErbB4, the signaling receptor for NRG, leads to inhibition of surfactant synthesis in primary fetal rat type II epithelial cells in vitro [36], while in vivo ErbB4 deletion leads to a clinical phenotype of a hyperreactive airway system and a histologic phenotype of alveolar simplification, chronic inflammation, and significant Sftpd downregulation in the postnatal lung [23]. While it would be very interesting to study the interactions of ErbB4 and Sftpd in the adult lung, we here focused on Sftpb, ErbB4, and TTF-1 interactions in the developing fetal lung, since fetal ErbB4 deletion results in delayed fetal lung development, especially delayed expression of Sftpb [19]. Also, Sftpb is the most critical surfactant protein in reducing surface tension in the lung [49-51]. NRG stimulates Sftpb expression by initiation of ErbB4 activation and ErbB4 processing, resulting in its chaperone function for transcription factors like Stat5A [29]. Here we expand on ErbB4 signaling clues, showing that ErbB4 functions as an upstream regulator of TTF-1, one of the most critical transcription factors in lung development and disease [2]. We speculate that in addition to a TTF-1 upregulation through ErbB4, both proteins form a functionally important complex to upregulate Sftpb expression by directly binding to the promoter region of this gene. The region of the Sftpb promoter from bp −111 to - 73 is a known complex binding site for TTF-1 [49, 52]. An upstream enhancer located in the 5′-flanking region of the human Sftpb gene (−439 to −331 base pair) has been found to bind to TTF-1 [53].
We here confirmed that NRG stimulates the interactions of ErbB4 and TTF-1 and that NRG-induced cleavage of ErbB4 results in a translocation of 4ICD to the nucleus, previously published for other cell types [13, 24], which seemed to be a critical process for Sftpb expression. We have previously described that NRG is able to induce 4ICD to function as a nuclear shuttle for the transcription factor Stat5A. This function requires the presence of the nuclear localization sequence (NLS) of 4ICD, [29] while estrogen is able to induce nuclear localization of 4ICD independent of its NLS [28]. This suggests that the estrogen receptor might function as an independent nuclear shuttle mechanism for 4ICD [28]. Here we show that NRG-induced nuclear interactions of ErbB4 and TTF-1 also require 4ICD NLS for its effect on Sftpb expression (Figure 9). Most of the nuclear processes regulated by 4ICD are still unknown. The interactions of these proteins were confirmed by the fact that ErbB4 and TTF-1 co-precipitate each other. Despite the fact that these interactions are baseline interactions and independent of NRG treatment, NRG stimulates tyrosine phosphorylation of ErbB4 and serine phosphorylation of TTF-1. This underlines the potential impact of the strong interactions of those two signaling pathways.
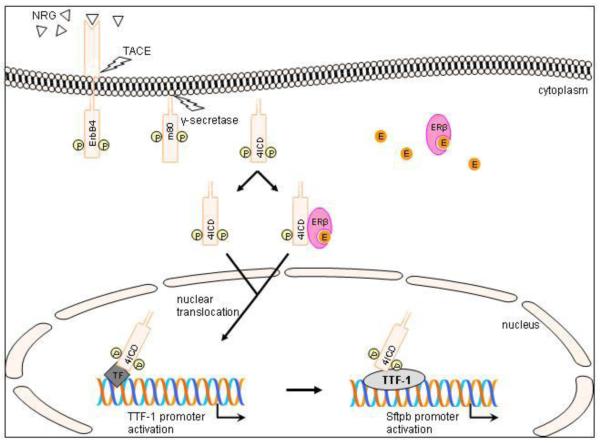
Figure 9. Schematic model of ErbB4 signaling in type II epithelial cells proposed by the authors.
Neuregulin (NRG)-binding leads to activation and proteolytic cleavage of the phosphorylated (P) ErbB4 receptor [24, 25]. The remaining membrane bound part (m80) is further processed by γ-secretase. The resulting 80kDa ErbB4 intracellular domain (4ICD) translocates to the nucleus [26] and enters via its nuclear localization sequence (NLS) [13, 24] or NLS independently using an estrogen (E) activated estrogen receptor β (ERβ) as a nuclear shuttle [28]. In our proposed model, 4ICD leads to TTF-1 promoter activation by binding via an additional transcription factor (TF) or linker to the promoter. This newly formed complex than binds and activates the Sftpb promoter to stimulate Sftpb expression.
Phosphorylation of TTF-1 is required for tissue-specific gene regulation [1]. Here we show that NRG-induced TTF-1 phosphorylation induces Sftpb gene expression in lung epithelial cells. NRG also stimulates TTF-1 mRNA and protein expression more potently in lung epithelial cells which overexpress ErbB4, suggesting that ErbB4 receptor is synergistically enhancing NRG effects on TTF-1 expression. This observation was confirmed by experiments in primary fetal type II cells isolated from HER4heart (−/−) lungs, which do not express ErbB4, emphasizing that TTF-1 expression requires the expression of full-length ErbB4.
There is some evidence that Hoxb3 is a transcriptional regulator of the TTF-1 gene in early embryogenesis [54] and that TTF-1 mRNA accumulation can be regulated by a posttranscriptional, Ras-sensitive mechanism [55]. Our studies demonstrated that ErbB4 leads to TTF-1 promoter activation, suggesting in its upstream regulatory control. Since nuclear translocation of ErbB4 is required for TTF-1 effects on Sftpb expression, it might be speculated that ErbB4 cleavage-induced signaling is most prominent in its effect on TTF-1 transcriptional control. This is in alignment with known regulatory effects of ErbB4 cleavage and 4ICD nuclear translocation on other transcription factors in other organ systems [18, 29, 56]. It remains questionable if ErbB4 directly or indirectly interacts with the TTF-1 promoter. There might be other transcription factors involved, which form a complex with TTF-1 and ErbB4. Since ErbB receptors lack any known structural signatures required for DNA association [57], it is most likely that ErbB4 does not directly interact with genomic DNA, but functions as a transcriptional co-regulator.
In addition to the known function of TTF-1 [8-11] and ErbB4 [13, 14] in lung cancer biology, mutations in TTF-1 and ErbB receptors as well as their interactions can also lead to lung disease. For example, EGFR mutations were significantly more common in tumors expressing TTF-1 [58]. TTF-1 haploinsufficiency is a rare cause of neonatal or infantile respiratory failure, often associated with congenital hypothyroidism and neurologic symptoms recognized as the brain-lung-thyroid syndrome [59].
In summary, ErbB4 regulates TTF-1 expression and the interactions of both proteins stimulate Sftpb expression in type II epithelial cells. NRG induces the co-localization of ErbB4 and TTF-1 and intensifies the stimulatory effect of ErbB4 on TTF1 expression, but does not add additional effects on Sftpb expression and Sftpb promoter activity. NRG also does not overcome the inhibitory effects when ErbB4 cleavage and nuclear translocation is prevented. Viewing our data in the light of known involvements of TTF-1, Sftpb, and ErbB receptors in lung disease, our results impact the knowledge of type II cell developmental biology to create potential approaches to prevent cell injury. Since this is an in vitro approach, further in vivo studies are needed, where manipulating ErbB4 signaling can prove its positive enhancement of TTF-1 effects on the fetal surfactant system and its usefulness in the prevention or treatment of lung disease.
Highlights.
> We examined that ErbB4 and TTF-1 interact with each other. > Interactions of both proteins stimulate Sftpb expression. > NRG stimulates TTF-1 expression. > ErbB4 nuclear translocation is critical for regulation of TTF-1.
Acknowledgements
We thank Dr. M. Gassmann and C. Birchmeier for providing the HER4heart mouse line, Dr. Ballard for supplying the Sftpb promoter luciferase reporter plasmid and Dr. Whitsett for supplying the TTF-1 promoter luciferase reporter plasmid.
Funding: The studies were funded by the German Research Foundation (DFG) Da 378/3-1, 378/3-2 and the National Heart, Lung, and Blood Institute NIH HL-85648.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors contributions: Katja Zscheppang-experimental design, data generation and analysis, manuscript preparation; Ulrike Giese-data generation and analysis; Stefan Hoenzke-data generation and analysis; Christiane Dammann-experimental design, manuscript preparation
The authors have no conflict of interest.
References
- [1].DeFelice M, Silberschmidt D, DiLauro R, Xu Y, Wert SE, Weaver TE, Bachurski CJ, Clark JC, Whitsett JA. TTF-1 phosphorylation is required for peripheral lung morphogenesis, perinatal survival, and tissue-specific gene expression. J Biol Chem. 2003;278:35574–35583. doi: 10.1074/jbc.M304885200. [DOI] [PubMed] [Google Scholar]
- [2].Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin Sci (Lond) 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- [3].Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- [4].Minoo P, Hamdan H, Bu D, Warburton D, Stepanik P, deLemos R. TTF-1 regulates lung epithelial morphogenesis. Dev Biol. 1995;172:694–698. doi: 10.1006/dbio.1995.8080. [DOI] [PubMed] [Google Scholar]
- [5].Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- [6].Pogach MS, Cao Y, Millien G, Ramirez MI, Williams MC. Key developmental regulators change during hyperoxia-induced injury and recovery in adult mouse lung. J Cell Biochem. 2007;100:1415–1429. doi: 10.1002/jcb.21142. [DOI] [PubMed] [Google Scholar]
- [7].White CW, Greene KE, Allen CB, Shannon JM. Elevated expression of surfactant proteins in newborn rats during adaptation to hyperoxia. Am J Respir Cell Mol Biol. 2001;25:51–59. doi: 10.1165/ajrcmb.25.1.4296. [DOI] [PubMed] [Google Scholar]
- [8].Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, Mitsudomi T, Sekido Y, Tanimoto M, Yatabe Y, Takahashi T. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- [11].Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Anagnostou VK, Syrigos KN, Bepler G, Homer RJ, Rimm DL. Thyroid transcription factor 1 is an independent prognostic factor for patients with stage I lung adenocarcinoma. J Clin Oncol. 2009;27:271–278. doi: 10.1200/JCO.2008.17.0043. [DOI] [PubMed] [Google Scholar]
- [13].Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res. 2003;284:66–77. doi: 10.1016/s0014-4827(02)00100-3. [DOI] [PubMed] [Google Scholar]
- [14].Junttila TT, Sundvall M, Lundin M, Lundin J, Tanner M, Harkonen P, Joensuu H, Isola J, Elenius K. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 2005;65:1384–1393. doi: 10.1158/0008-5472.CAN-04-3150. [DOI] [PubMed] [Google Scholar]
- [15].Hosono Y, Yamaguchi T, Mizutani E, Yanagisawa K, Arima C, Tomida S, Shimada Y, Hiraoka M, Kato S, Yokoi K, Suzuki M, Takahashi T. MYBPH, a transcriptional target of TTF-1, inhibits ROCK1, and reduces cell motility and metastasis. Embo J. 2011;31:481–493. doi: 10.1038/emboj.2011.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klien R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- [17].Birchmeier C. ErbB receptors and the development of the nervous system. Exp Cell Res. 2009;315:611–618. doi: 10.1016/j.yexcr.2008.10.035. [DOI] [PubMed] [Google Scholar]
- [18].Jones FE. HER4 intracellular domain (4ICD) activity in the developing mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:247–258. doi: 10.1007/s10911-008-9076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu W, Purevdorj E, Zscheppang K, von Mayersbach D, Behrens J, Brinkhaus MJ, Nielsen HC, Schmiedl A, Dammann CE. ErbB4 regulates the timely progression of late fetal lung development. Biochim Biophys Acta. 2010;1803:832–839. doi: 10.1016/j.bbamcr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- [21].Prevot V, Lomniczi A, Corfas G, Ojeda SR. erbB-1 and erbB-4 receptors act in concert to facilitate female sexual development and mature reproductive function. Endocrinology. 2005;146:1465–1472. doi: 10.1210/en.2004-1146. [DOI] [PubMed] [Google Scholar]
- [22].Dammann CE, Nielsen HC, Carraway KL. Role of Neuregulin1{beta} in the Developing Lung. Am J Respir Crit Care Med. 2003;167:1711–1716. doi: 10.1164/rccm.200205-468OC. [DOI] [PubMed] [Google Scholar]
- [23].Purevdorj E, Zscheppang K, Hoymann HG, Braun A, von Mayersbach D, Brinkhaus MJ, Schmiedl A, Dammann CE. ErbB4 deletion leads to changes in lung function and structure similar to bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L516–522. doi: 10.1152/ajplung.00423.2007. [DOI] [PubMed] [Google Scholar]
- [24].Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, Mei L, Kim TW. Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J Biol Chem. 2002;277:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- [25].Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- [26].Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000;275:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- [27].Williams CC, Allison JG, Vidal GA, Burow ME, Beckman BS, Marrero L, Jones FE. The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 2004;167:469–478. doi: 10.1083/jcb.200403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zscheppang K, Konrad M, Zischka M, Huhn V, Dammann CE. Estrogen-induced upregulation of Sftpb requires transcriptional control of neuregulin receptor ErbB4 in mouse lung type II epithelial cells. Biochim Biophys Acta. 2011;1813:1717–1727. doi: 10.1016/j.bbamcr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zscheppang K, Dork T, Schmiedl A, Jones FE, Dammann CE. Neuregulin receptor ErbB4 functions as a transcriptional cofactor for the expression of surfactant protein B in the fetal lung. Am J Respir Cell Mol Biol. 2011;45:761–767. doi: 10.1165/rcmb.2010-0179OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Das A, Acharya S, Gottipati KR, McKnight JB, Chandru H, Alcorn JL, Boggaram V. Thyroid transcription factor-1 (TTF-1) gene: identification of ZBP-89, Sp1, and TTF-1 sites in the promoter and regulation by TNF-alpha in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2011;301:L427–440. doi: 10.1152/ajplung.00090.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J Biol Chem. 2002;277:38399–38408. doi: 10.1074/jbc.M203188200. [DOI] [PubMed] [Google Scholar]
- [32].Zhou B, Zhong Q, Minoo P, Li C, Ann DK, Frenkel B, Morrisey EE, Crandall ED, Borok Z. Foxp2 inhibits Nkx2.1-mediated transcription of SP-C via interactions with the Nkx2.1 homeodomain. Am J Respir Cell Mol Biol. 2008;38:750–758. doi: 10.1165/rcmb.2007-0350OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA. Production of immortilized distal respiratory epithelial cell lines from surfactant protein C/ simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA. 1993;90:11029–11032. doi: 10.1073/pnas.90.23.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- [35].Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- [36].Zscheppang K, Liu W, Volpe MV, Nielsen HC, Dammann CE. ErbB4 regulates fetal surfactant phospholipid synthesis in primary fetal rat type II cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L429–435. doi: 10.1152/ajplung.00451.2006. [DOI] [PubMed] [Google Scholar]
- [37].Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. PNAS. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Abolhassani M, Guais A, Chaumet-Riffaud P, Sasco AJ, Schwartz L. Carbon dioxide inhalation causes pulmonary inflammation. Am Physiol Lung Cell Mol Physiol. 2009;269:L657–L665. doi: 10.1152/ajplung.90460.2008. [DOI] [PubMed] [Google Scholar]
- [39].Hoeing K, Zscheppang K, Mujahid S, Murray S, Volpe MV, Dammann CE, Nielsen HC. Presenilin-1 processing of ErbB4 in fetal type II cells is necessary for control of fetal lung maturation. Biochim Biophys Acta. 2011;1813:480–491. doi: 10.1016/j.bbamcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bueter W, Dammann O, Zscheppang K, Korenbaum E, Dammann CE. ErbB receptors in fetal endothelium-A potential linkage point for inflammation-associated neonatal disorders. Cytokine. 2007 doi: 10.1016/j.cyto.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Margana RK, Boggaram V. Functional analysis of surfactant protein B (SP-B) promoter. Sp1, Sp3, TTF-1, and HNF-3alpha transcription factors are necessary for lung cell-specific activation of SP-B gene transcription. J Biol Chem. 1997;272:3083–3090. doi: 10.1074/jbc.272.5.3083. [DOI] [PubMed] [Google Scholar]
- [43].Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- [44].Maeda Y, Hunter TC, Loudy DE, Dave V, Schreiber V, Whitsett JA. PARP-2 interacts with TTF-1 and regulates expression of surfactant protein-B. J Biol Chem. 2006;281:9600–9606. doi: 10.1074/jbc.M510435200. [DOI] [PubMed] [Google Scholar]
- [45].Muraoka-Cook RS, Feng SM, Strunk KE, Earp HS., 3rd ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J Mammary Gland Biol Neoplasia. 2008;13:235–246. doi: 10.1007/s10911-008-9080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nielsen HC, Torday JS. Coordination of growth and differentiation in the fetal lung. Exp Lung Res. 1990;188:89–96. doi: 10.1016/0014-4827(90)90281-e. [DOI] [PubMed] [Google Scholar]
- [47].Langston C, Kida K, Reed M. Human lung growth in late gestation and in the neonate. Am Rev Respir Dis. 1984;129:607–613. [PubMed] [Google Scholar]
- [48].Avery ME, Mead J. Surface properties in relation to atelectasis and hyaline membrane disease. Am J Dis Child. 1959;97:517–523. doi: 10.1001/archpedi.1959.02070010519001. [DOI] [PubMed] [Google Scholar]
- [49].Pryhuber GS. Regulation and function of pulmonary surfactant protein B. Mol Genet Metab. 1998;64:217–228. doi: 10.1006/mgme.1998.2722. [DOI] [PubMed] [Google Scholar]
- [50].Weaver TE, Whitsett JA. Function and regulation of expression of pulmonary surfactant-associated proteins. Biochem J. 1991;273(Pt 2):249–264. doi: 10.1042/bj2730249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Whitsett JA, Glasser SW. Regulation of surfactant protein gene transcription. Biochim Biophys Acta. 1998;1408:303–311. doi: 10.1016/s0925-4439(98)00076-3. [DOI] [PubMed] [Google Scholar]
- [52].Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promotor is a traget for thyroid transcription factor 1 and hepatocyte nulcear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yan C, Sever Z, Whitsett JA. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J Biol Chem. 1995;270:24852–24857. doi: 10.1074/jbc.270.42.24852. [DOI] [PubMed] [Google Scholar]
- [54].Guazzi S, Lonigro R, Pintonello L, Boncinelli E, Di Lauro R, Mavilio F. The thyroid transcription factor-1 gene is a candidate target for regulation by Hox proteins. Embo J. 1994;13:3339–3347. doi: 10.1002/j.1460-2075.1994.tb06636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lonigro R, De Felice M, Biffali E, Macchia PE, Damante G, Asteria C, Di Lauro R. Expression of thyroid transcription factor 1 gene can be regulated at the transcriptional and posttranscriptional levels. Cell Growth Differ. 1996;7:251–261. [PubMed] [Google Scholar]
- [56].Linggi B, Carpenter G. ErbB-4 s80 intracellular domain abrogates ETO2-dependent transcriptional repression. J Biol Chem. 2006;281:25373–25380. doi: 10.1074/jbc.M603998200. [DOI] [PubMed] [Google Scholar]
- [57].Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15:6484–6489. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sun PL, Seol H, Lee HJ, Yoo SB, Kim H, Xu X, Jheon S, Lee CT, Lee JS, Chung JH. High incidence of EGFR mutations in Korean men smokers with no intratumoral heterogeneity of lung adenocarcinomas: correlation with histologic subtypes, EGFR/TTF-1 expressions, and clinical features. J Thorac Oncol. 2012;7:323–330. doi: 10.1097/JTO.0b013e3182381515. [DOI] [PubMed] [Google Scholar]
- [59].Guillot L, Carre A, Szinnai G, Castanet M, Tron E, Jaubert F, Broutin I, Counil F, Feldmann D, Clement A, Polak M, Epaud R. NKX2-1 mutations leading to surfactant protein promoter dysregulation cause interstitial lung disease in “Brain-Lung-Thyroid Syndrome”. Hum Mutat. 2010;31:E1146–1162. doi: 10.1002/humu.21183. [DOI] [PubMed] [Google Scholar]