Abstract
We have characterized the 200-nucleotide-long insertion found in f1 after segregation of a chimeric phage containing the genomes of f1 and pSC101 [Ohsumi, M., Vovis, G.F. & Zinder, N.D. (1978) Virology 89, 438--449]. The insertion in this novel f1 species, called f1', is derived from pSC101 and has the potential to form an extended base-paired secondary structure, as determined by nucleotide sequence analysis. A five-nucleotide direct repeat, derived from f1 sequences, is present in f1'. The 200 additional nucleotides that are inserted into the DNA sequence coding for the carboxy terminus of f1 gene IV protein have generated a novel carboxy terminus for the f1' gene IV protein. In vitro transcription--translation studies demonstrate that a read-through protein can be expressed, as predicted from the f1' nucleotide sequence results. This 200-nucleotide-long sequence appears to be a transposable element found within pSC101 and is similar in sequence to the inverted repeat found in Tn3. Restriction enzyme analysis of the chimeric phage DNA, coupled with the nucleotide sequencing results, allows us to predict a structure for the genomic organization of this chimera.
Full text
PDF
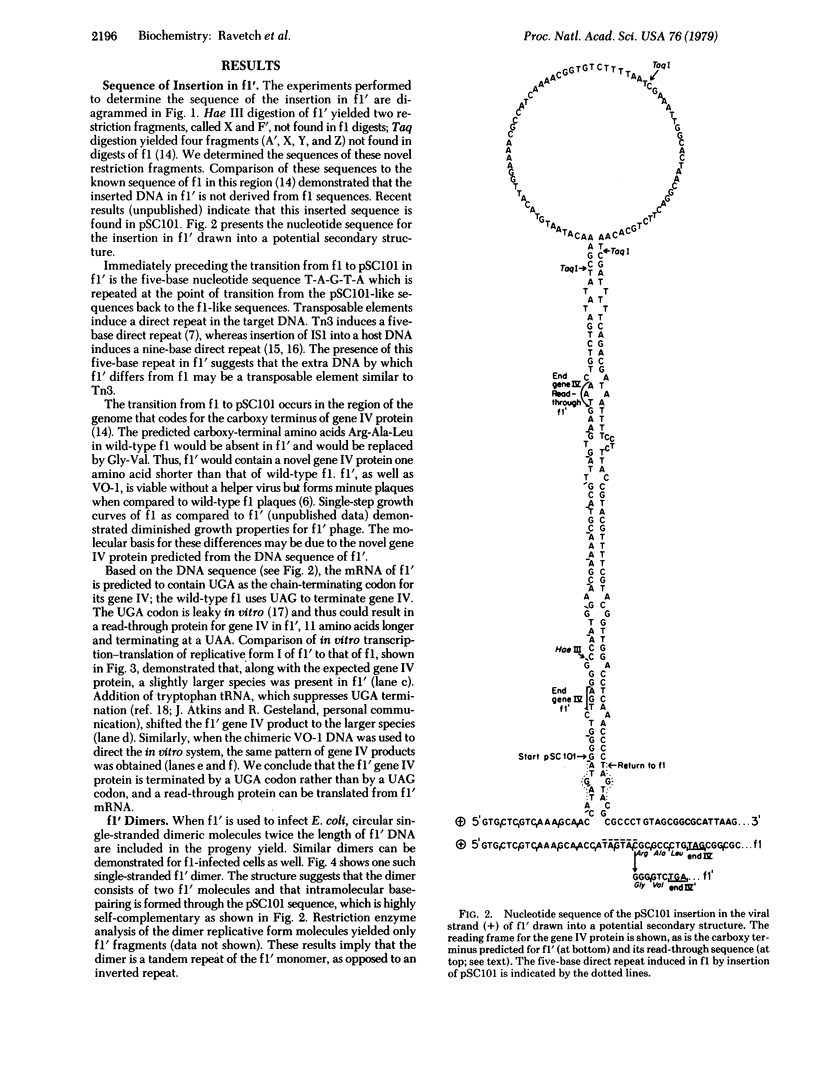

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D., Gold L. Translation of the UGA triplet in vitro by tryptophan transfer RNA's. J Mol Biol. 1971 Jun 14;58(2):459–468. doi: 10.1016/0022-2836(71)90363-9. [DOI] [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Enea V., Zinder N. D. Cleavage map of bacteriophage f1: location of the Escherichia coli B-specific modification sites. J Mol Biol. 1975 Jun 25;95(2):147–165. doi: 10.1016/0022-2836(75)90388-5. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model P., Webster R. E., Zinder N. D. The UGA codon in vitro: chain termination and suppression. J Mol Biol. 1969 Jul 14;43(1):177–190. doi: 10.1016/0022-2836(69)90087-4. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Nomura N., Yamagishi H., Oka A. Isolation and characterization of transducing coliphage fd carrying a kanamycin resistance gene. Gene. 1978 Feb;3(1):39–51. doi: 10.1016/0378-1119(78)90006-9. [DOI] [PubMed] [Google Scholar]
- Ohsumi M., Vovis G. F., Zinder N. D. The isolation and characterization of an in vivo recombinant between the filamentous bacteriophage f1 and the plasmid pSC101. Virology. 1978 Sep;89(2):438–449. doi: 10.1016/0042-6822(78)90186-1. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Horiuchi K., Zinder N. D. DNA sequence analysis of the defective interfering particles of bacteriophage f1. J Mol Biol. 1979 Mar 5;128(3):305–318. doi: 10.1016/0022-2836(79)90090-1. [DOI] [PubMed] [Google Scholar]
- Ravetch J. V., Horiuchi K., Zinder N. D. Nucleotide sequences near the origin of replication of bacteriophage f1. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4219–4222. doi: 10.1073/pnas.74.10.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D. S., Kook K. Insertion of the Tn3 transposon into the genome of the single-stranded DNA phage M13. Gene. 1978 Oct;4(2):109–119. doi: 10.1016/0378-1119(78)90024-0. [DOI] [PubMed] [Google Scholar]