Summary
ComEC is a putative channel protein for DNA uptake in Bacillus subtilis and other genetically transformable bacteria. Membrane topology studies suggest a model of ComEC as a multispanning membrane protein with seven transmembrane segments (TMSs), and possibly with one laterally inserted amphipathic helix. We show that ComEC contains an intramolecular disulphide bond in its N-terminal extracellular loop (between the residues C131 and C172), which is required for the stability of the protein, and is probably introduced by BdbDC, a pair of competence-induced oxidoreductase proteins. By in vitro cross-linking using native cysteine residues we show that ComEC forms an oligomer. The oligomerization surface includes a transmembrane segment, TMS-G, near the cytoplasmic C-terminus of ComEC.
Introduction
Bacillus subtilis can take up exogenous DNA from the environment in a process known as transformation. Competence for transformation depends on the expression of several proteins involved in DNA binding, processing and internalization. These proteins are encoded in five known operons: comE (Inamine and Dubnau, 1995), comF (Londoño-Vallejo and Dubnau, 1993), comG (Albano et al., 1989; Albano and Dubnau, 1989), comC (Mohan et al., 1989) and nucA-nin (van Sinderen et al., 1995; Provvedi et al., 2001), which are upregulated by the transcriptional activator ComK, upon entry into stationary phase.
The comEC locus was identified in a genetic screen for competence mutants and was shown to be absolutely required for DNA uptake but not for DNA binding to the competent cell (Hahn et al., 1987). comEC is the third and last gene in the comE operon, whose transcription is driven from the major promoter in front of the first open reading frame (ORF), comEA, and in addition from a minor promoter in front of the second ORF, comEB (Hahn et al., 1993). lacZ fusion analysis (Inamine and Dubnau, 1995) and transcriptional profiling using microarrays (Berka et al., 2002; Hamoen et al., 2002; Ogura et al., 2002) showed that comE transcription is strongly upregu-lated during competence development, characteristic of competence operons.
ComEA is a DNA-binding protein that serves as a receptor, binding non-specifically to environmental DNA (Provvedi and Dubnau, 1999), but also playing an essential role in transport (Inamine and Dubnau, 1995). In addition to ComEA, all seven ComG proteins encoded in the comG operon are required for DNA binding (Chung and Dubnau, 1998). Because of their apparent inability to bind DNA directly, it was proposed that they modify the cell wall to permit the access of DNA to the receptor ComEA (Provvedi and Dubnau, 1999). Four of the ComG proteins (GC, GD, GE and GG) resemble prepilin proteins and contain an N-terminal hydrophobic sequence preceded by a cleavage site for processing by the competence-induced protease ComC (Chung and Dubnau, 1995; Chung et al., 1998). These proteins can be recovered from the cell wall fraction after processing, and it has been suggested that the ComG proteins form a pilus-like structure that allows DNA to cross the cell wall.
Recently, the bicistronic operon bdbDC was shown to be essential for transformation in B. subtilis and to be under competence regulation (Meima et al., 2002). BdbD and C belong to a family of oxidoreductases involved in disulphide bond formation. In Escherichia coli such redox proteins are located in the membrane or in the periplasm and their active sites face the periplasm (Raina and Missiakas, 1997). ComGC contains an intramolecular disul-phide bond and deletions of either bdbD or C destabilize ComGC (Meima et al., 2002), presumably because correct folding of ComGC requires its oxidation.
The hydrophobic character of ComEC, its large size (776 residues), its membrane localization shown in this report, and its absolute requirement for DNA uptake but not for DNA binding to the cell, suggest that ComEC might be a channel component of the uptake machinery. Because of its toxicity in E. coli (Inamine and Dubnau, 1995), no direct biochemical evidence has been acquired to support this hypothesis. As a first step toward the elucidation of ComEC function, we have studied its topology using phoA and lacZ fusions, and we propose a model in which ComEC crosses the membrane seven times. An intramolecular disulphide bond, introduced by BdbDC, stabilizes the large extracellular N-loop of ComEC, revealing the importance of BdbDC oxidoreductases for the biogenesis of a competence protein other than ComGC. In vitro cross-linking of native cysteines suggests that ComEC exists as an oligomer.
Results
Computer models of ComEC topology
We used 10 hydropathy analysis programs (see Experimental procedures) to predict the membrane topology of ComEC. Seven transmembrane segments (TMSs) were predicted in common by all 10 programs, although the range of such predicted segments was from 12 (HMMTOP) to nine (MEMSAT). All the programs agreed in identifying a large hydrophilic loop, near the N-terminus (referred to hereafter as the N-loop) and a hydrophilic C-terminal region. The programs did not predict consistent orientations of the commonly predicted TMSs.
Predictions of membrane topology can often be enhanced if orthologous proteins are compared. Seven ComEC orthologues from competent species (Streptococcus pneumoniae, Thermus thermophilus, Acinetobacter sp., Neisseria meningitidis, Neisseria gonorrhoeae, Pseudomonas stutzeri and Haemophilus influenzae) were analysed by MEMSAT, HMMTOP, TMHMMM, TOPPRED and TMPRED (see Experimental procedures). Six putative TMSs near the centre of the protein were most commonly predicted. However, the predicted topologies of these seven ComEC orthologues differed with respect to the numbers, identities and orientations of their TMSs, and no consensus topology emerged.
Topology as determined by phoA and lacZ fusions
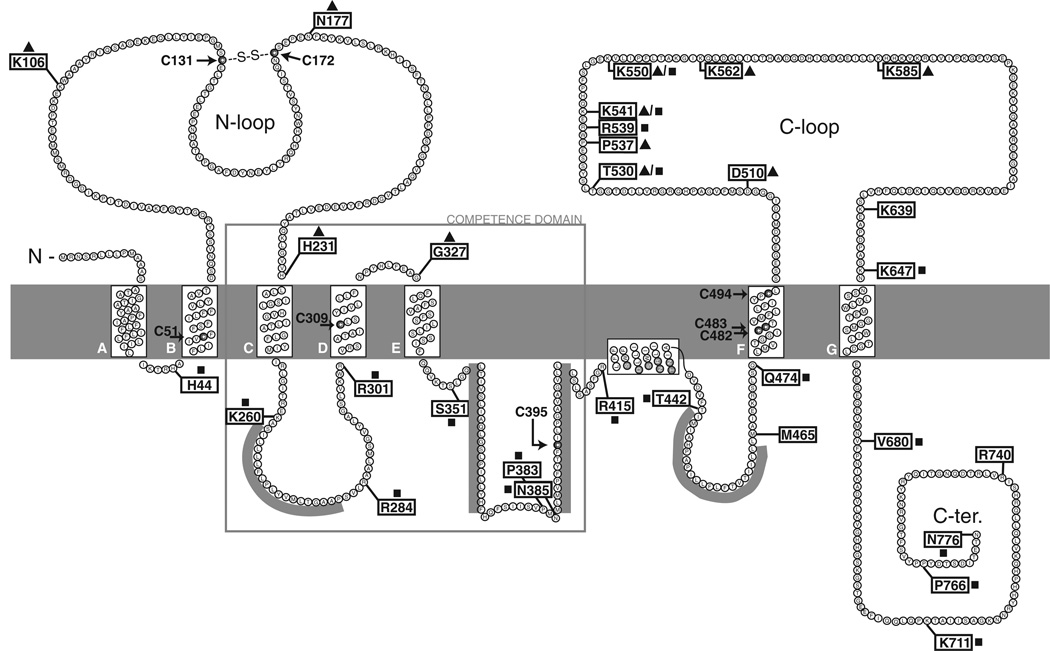
These uncertainties prompted us to examine the topology of ComEC experimentally, using the phoA and lacZ reporter systems (Manoil, 1991). This method is based on the principle that the enzymatic activities of the fusion proteins reveal the cellular locations of the fusion sites. Alkaline phosphatase (PhoA) is folded and assembled into functional dimers only after it is exported across the membrane. Fusions to b-galactosidase (LacZ) are used in a complementary fashion to indicate the cytoplasmic localization of fusion sites. C-terminal truncation fusions of comEC to phoA and lacZ were expressed from the native comEC locus in B. subtilis. Normalized activities, corrected for background, are listed in Table 1. Positions of the fusion sites are indicated in Fig. 1.
Table 1.
β-galactosidase (LacZ) and alkaline phosphatase (PhoA) activities of ComEC fusions measured in competent cultures.
| Fusion site | LacZ activitya | PhoA activitya |
|---|---|---|
| H44 | 76% ± 20% | 1% ± 1% |
| K106 | 0% ± 1% | 100% ± 31% |
| N177 | 1% ± 1% | 33% ± 3% |
| H231 | –b | 25% ± 6% |
| K260 | 43% ± 8% | 0% ± 0% |
| R284 | 17% ± 4% | 4% ± 3% |
| R301 | 100% ± 12% | 1% ± 1% |
| G327 | 1% ± 1% | 29% ± 7% |
| S351 | 46% ± 13% | 1% ± 1% |
| P383 | 18% ± 1% | 4% ± 4% |
| N385 | 15% ± 4% | 1% ± 2% |
| R415 | 24% ± 7% | 2% ± 2% |
| T442 | 21% ± 6% | 1% ± 1% |
| M465 | 8% ± 4% | 1% ± 1% |
| Q474 | 23% ± 6% | 1% ± 1% |
| D510 | 6% ± 2% | 41% ± 21% |
| T530 | 19% ± 4% | 28% ± 10% |
| P537 | 6% ± 1% | 25% ± 3% |
| R539 | 10% ± 2% | 0% ± 0% |
| K541 | 12% ± 3% | 20% ± 3% |
| K550 | 10% ± 2% | 16% ± 3% |
| K562 | 5% ± 2% | 10% ± 5% |
| K585 | 2% ± 1% | 13% ± 6% |
| K639 | 8% ± 2% | 5% ± 1% |
| K647 | 14% ± 4% | 8% ± 2% |
| V680 | 23% ± 1% | 9% ± 4% |
| K711 | 82% ± 54% | 3% ± 3% |
| R740 | 0% ± 0% | 2% ± 4% |
| P766 | 61% ± 7% | 2% ± 2% |
| N776 | 33% ± 15% | 6% ± 4% |
Activities, measured as described in Experimental procedures, were corrected for the background and normalized to the highest activity (16.4 µmol mg−1 protein for LacZ and 1.1 µmol mg−1 for PhoA respectively). These results represent the average of three independent measurements with standard deviations shown. Positive activities, arbitrarily defined as >10%, are shown in bold.
A LacZ fusion to H231 was not constructed.
Fig. 1.
Proposed topology of ComEC. The positions of PhoA and LacZ fusions are indicated by flags. Black triangles next to the flags indicate high PhoA activity and black squares high LacZ activity. Grey bars represent predicted hydrophobic segments that do not cross the membrane according to the fusion data. Arrows point toward eight cysteine residues. The disulphide bond between externally located cysteines is indicated. The competence domain is boxed.
Based on the activities of 59 constructed fusions we propose a model for the topology of ComEC (Fig. 1). This model predicts seven transmembrane segments (TMSs), which we named A–G, as well as three large hydrophilic domains; the N-terminal loop and the C-terminal loop face the extracellular environment, while the C-terminus is in the cytoplasm.
Overpredicted TMSs
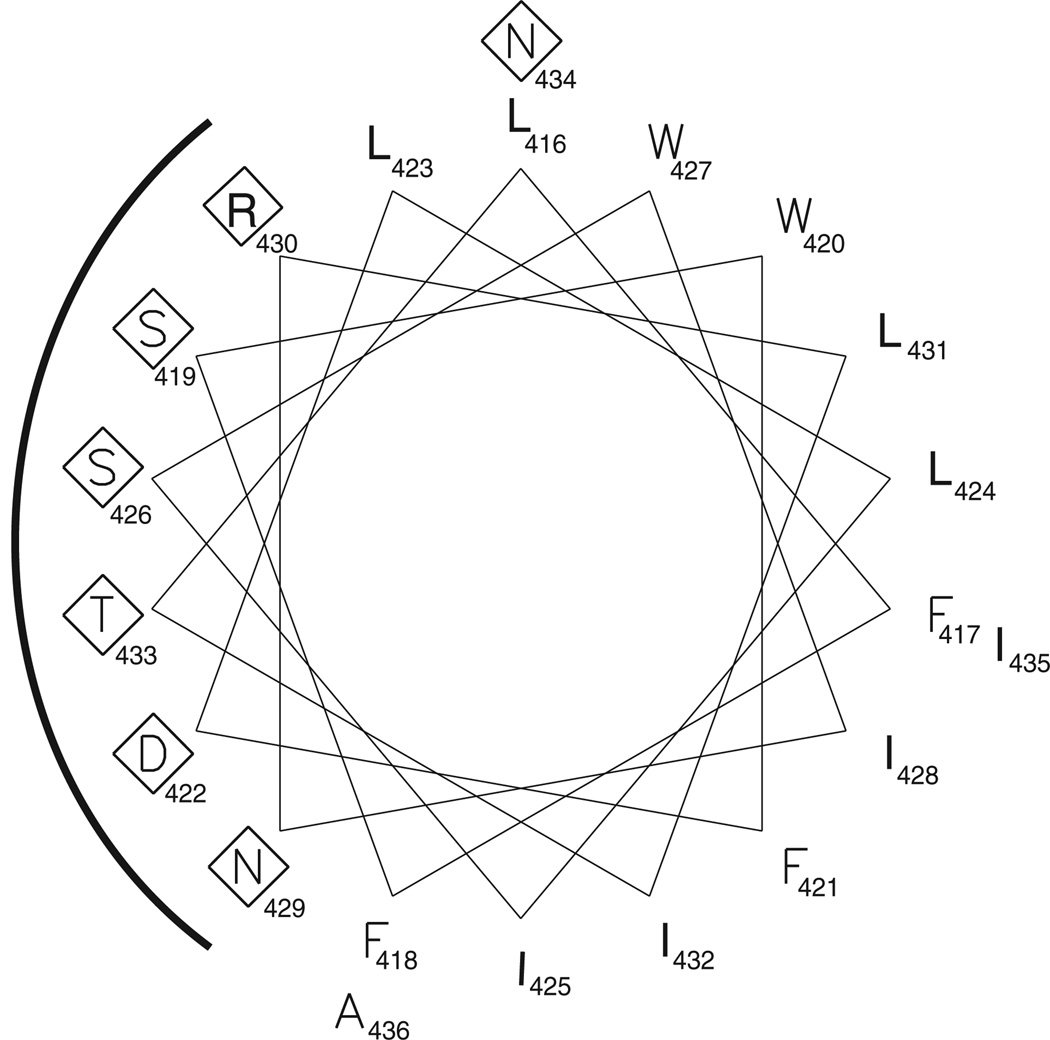
The major discrepancy between the computer predictions and our model is in the number of TMSs. Five hydrophobic segments predicted by most of the computer programs to span the membrane were not confirmed as TMSs by the phoA and lacZ assays. Four of these overpredicted helices are located C-terminally to TMS-E and one between TMS-C and -D. It is possible that one or more of these overpredicted membrane helices are in fact membrane-associated, but do not cross the membrane, a characteristic of amphipathic helices. Hydropho-bic side chains of amphipathic helices are sometimes buried in the lipid bilayer with their hydrophilic side chains exposed to the aqueous phase. As a result these helices are arranged parallel to the membrane surface. We therefore scanned the overpredicted membrane helices for amphipathicity. Only one displayed amphipathic properties and we propose that it forms a membrane-inserted amphipathic a-helix as depicted in Fig. 1. Several arguments support this proposal. First, residues L416 to A436 are predicted to be a-helical by the secondary structure prediction program, PSIPRED (McGuffin et al., 2000). Second, a helical wheel projection of these residues (Fig. 2) reveals the presence of distinct hydrophobic and hydrophilic surfaces. Third, arginine residue 430 provides a positive charge, potentially available for interaction with phospholipid head groups. These features are commonly observed in amphipathic membrane-associated helices (Segrest et al., 1990).
Fig. 2.
Helical wheel projection of residues L416–A436. Polar and charged residues are in diamonds. The hydrophilic side of the helix is indicated by a black line.
All 10 computer programs predict a TMS approximately between residues 260 and 280, which in Fig. 1 are located in the cytoplasm. This segment is flanked on both sides by one or more positively charged residues. According to the ‘positive-inside’ rule, loops with net positive charges are targeted to the cytoplasm (von Heijne, 1992) and the predicted TMS might therefore assume a cytoplasmic location because both surrounding loops have a strong preference for the cytoplasm. Gafvelin and von Heijne, 1994) altered the charge of loops to generate opposing signals and showed that topologically ‘frustrated’ molecules with incompatible orientations, adopt a ‘leave-one-helix-out’ topology. We propose that in ComEC the frustration between flanking regions surrounding the hydrophobic region spanning residues 260–280 is relieved by retaining these residues in the cytoplasm.
Orientation of the N-terminus
We could not unambiguously determine the orientation of the N-terminus based on the fusion data alone. The first available fusion confirmed the cytoplasmic orientation of the fusion site H44. If computer modelling correctly predicts TMS-A, the N-terminus must be outside. However, if TMS-A is not inserted in the membrane, the N-terminus is in the cytosol. In fact, all 10 topology algorithms predict the existence of TMS-A, favouring the first hypothesis. We have isolated membranes by sucrose flotation and shown that TMS-A is indeed membrane-associated, as the β-galactosidase activity of the LacZ fusion at position H44 co-purified with the membranes (data not shown), providing strong support for the outside orientation of the N-terminus.
Location of the C-terminal domain
Fusions following the last TMS, TMS-G, invariably show very high LacZ activities indicating that the C-terminal part of ComEC from E670 to N776 is cytoplasmic. In addition to exhibiting high LacZ activities, fusions P766 and N776 fully support DNA uptake (not shown). This observation strongly supports the location of the extreme C-terminus of ComEC in the cytosol, as functional proteins most likely retain the native topology of ComEC.
Orientation of the C-loop
phoA and lacZ fusions show somewhat contradicting results in the region of the C-terminal loop. Four fusions show strong PhoA activity (D510, P537, K562 and K585), three fusions have high PhoA but also high LacZ (T530, K541, K550), while LacZ is slightly higher than PhoA for fusion R539 and possibly for K647. Since PhoA data is more reliable than LacZ (van Geest and Lolkema, 2000) it is likely that this loop is extracellular, consistent with the C-terminus of the protein being cytoplasmic.
ComEC contains intramolecular disulphide bonds
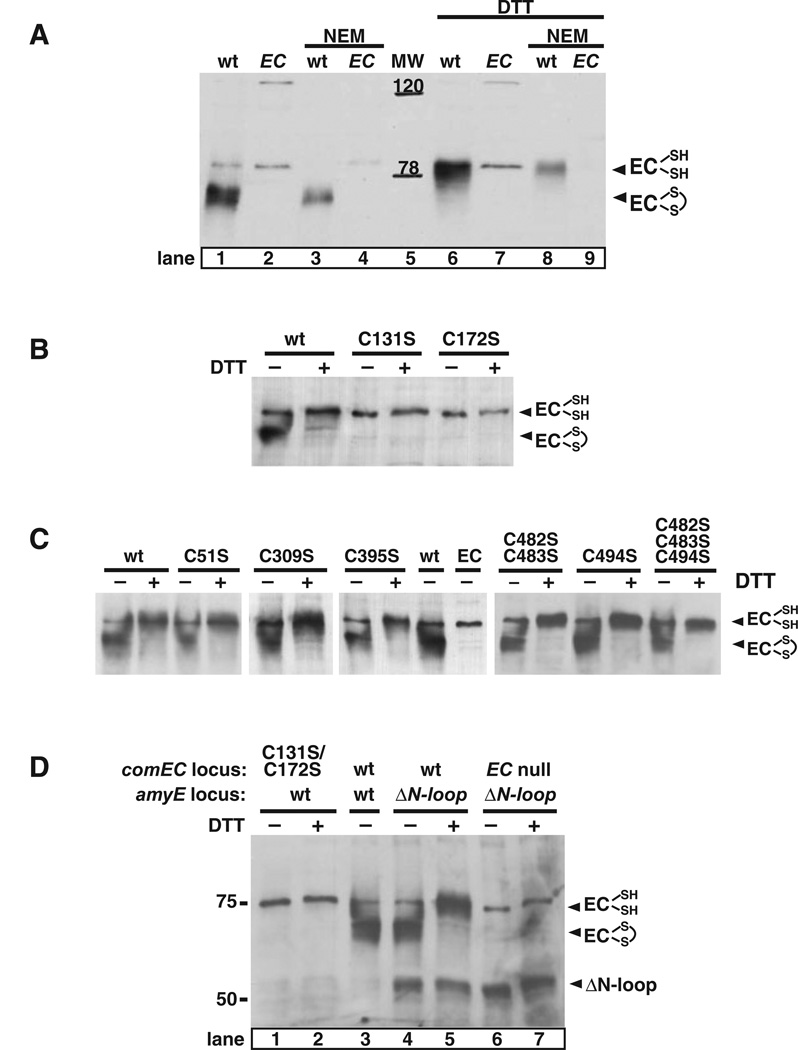
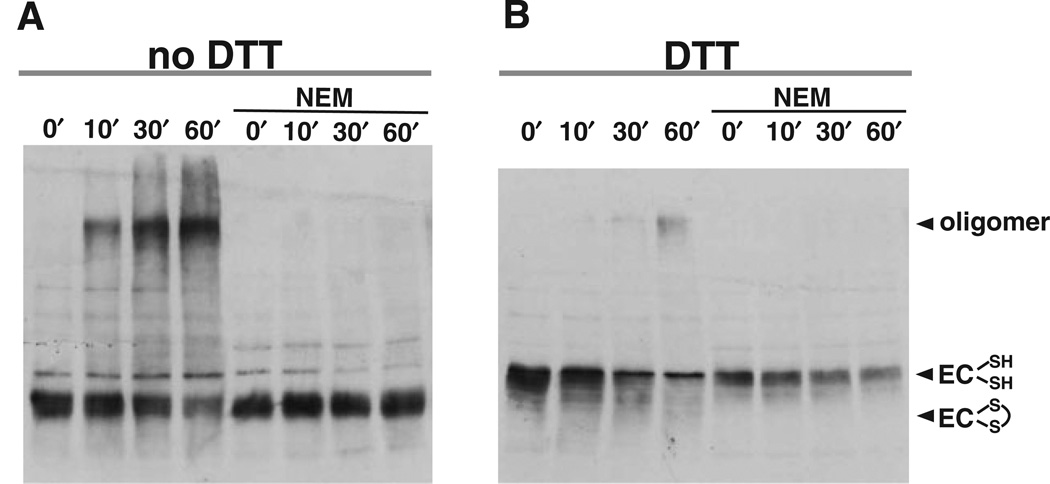
Depending on conditions, ComEC migrates at two positions on SDS-polyacrylamide gels, as shown on the Western blot in Fig. 3A (compare lanes 1 and 6). In the absence of an added reducing agent, ComEC adopts a fast-migrating conformation, while upon addition of dithio-threitol (DTT) it migrates more slowly. These results strongly suggest that ComEC possesses an intramolecular disulphide bond, and is more compact when oxidized.
Fig. 3.
An intramolecular disulphide bond in ComEC is required for stability. Membranes were prepared from competent B. subtilis cultures by flotation (see Experimental procedures), and membrane proteins (10 µg per lane) were resolved by 7% SDS-PAGE, blotted onto nitrocellulose and ComEC was detected using affinity purified anti-comEC antibodies. Samples were loaded in the presence or absence of 100 mM DTT as indicated.
A. To block in vitro disulphide bond formation, NEM (0.1 mM) was added to competent wild-type (wt, BD2528) and comEC (EC, BD2993) cells before harvesting. Positions of the molecular weight standards are indicated.
B. Stability of ComEC mutant proteins with cysteine to serine point mutations: C131S (BD3386), C172S (BD3387).
C. Stability of ComEC mutant proteins with cysteine to serine point mutations: C51S (BD3411), C309 (BD3403), C395 (BD3404), C482S/C483S (BD3495), C494 (BD3496) and C482S/C483S/C494 (BD3497).
D. Disulphide bond formation in ComEC mutant proteins missing both cysteine residues of the N-loop or the entire N-loop: C131S/C172S (BD3487), ΔN-loop (BD3478), EC null ΔN-loop (BD3480). The presence of wild-type copies of genes is indicated by ‘wt’.
To determine whether this disulphide bond is formed after lysis, N-ethyl-maleimide (NEM), was added prior to lysis to block free SH- groups and prevent in vitro disul-phide bond formation. As shown in Fig. 3A, ComEC from NEM-treated cells retains the characteristic migration shift upon addition of DTT (compare lanes 1, 3, 6 and 8), indicating that the disulphide bond exists in the cell.
For reasons we do not understand, the ComEC Western blot signals, as well as the signals of cross-reacting bands were weaker when membranes were prepared in the presence of NEM. ComEC in its reduced form overlaps with a cross-reacting band, which is also present in the extracts from the comEC mutant (Fig. 3A, lanes 2 and 7). To unambiguously distinguish the ComEC signal, samples were routinely loaded with and without DTT.
Two extracellular cysteines form a single disulphide bond
We next determined which of the eight cysteines in ComEC participated in disulphide bond formation. Two of these cysteines (C131 and C172), located in the extracellular N-loop of ComEC were favoured, because according to our model (Fig. 1), only these two would be exposed to an oxidizing environment in vivo. The remaining six cys-teines are either in the plane of the membrane (C51, C309, C482, C483, C494) or in the cytoplasm (C395), shielded from the oxidizing environment.
Each of the eight cysteine residues was replaced by a serine and inserted in the wild-type comEC locus of B. subtilis by Campbell-like recombination. The C131S and C172S mutant proteins were unstable and undetectable by Western blotting (Fig. 3B); only the cross-reacting band was visible. None of the other six cysteine replacements led to detectable instability of ComEC or decreased trans-formability (Fig. 3C, Table 2).
Table 2.
Transformability of comEC mutant strains.
| Strain | Mutation | Relative transformation frequencya |
|---|---|---|
| BD3390 | None | 1.00 |
| BD3410 | C51S | 0.90 |
| BD3386 | C131S | 0.04 |
| BD3387 | C172S | 0.08 |
| BD3487 | C131S/C172S | 0.05 |
| BD3388 | C309S | 1.02 |
| BD3389 | C395S | 0.98 |
| BD3491 | C482S/C483S | 0.80 |
| BD3492 | C494S | 1.00 |
| BD3493 | C482S/C483S/C494S | 1.50 |
| Deletion of the N-loop in comEC | ||
| BD3474 | amyE::DN-loop | <10−6 |
| BD3479 | amyE::ΔN-loop, comEC | <10−6 |
Transforming DNA was incubated with individual strains for 40– 50 min before plating for leucine prototrophy. Data for the cysteine point mutations were normalized to the value for BD3390, which carries the plasmid used to construct the mutant strains.
Although not detectable in the Western blot, some residual C131S and C172S mutant protein must be present, because the transformation frequency was decreased only 10- to 20-fold (Table 2). In contrast, deletion of comEC results in a complete loss of transformability, a decrease of about 107-fold (Inamine and Dubnau, 1995). The instability of only the C131S and C172 mutant proteins, strongly suggests that these two cysteines form the intramolecular disulphide bond. Additional proof is provided below. Our results also demonstrate that the disul-phide bond was not absolutely required for the function of ComEC, as residual transformation was observed in the two cysteine mutants, while deletion of comEC completely abolishes transformability.
To explain the instability of the C131S and C172S mutant proteins two hypothesis were considered: (i) ComEC lacking the intramolecular disulphide bond is often misfolded and therefore degraded and (ii) a surveillance mechanism recognizes and degrades extracellular proteins with free thiols. The second hypothesis predicts that a double mutant protein C131S/C172S would be stable. This double mutant was constructed and shown to be unstable (Fig. 3D, lanes 1 and 2), supporting the first hypothesis. We conclude that formation of the disulphide bond favours the correct folding of the N-loop and that the misfolded protein is degraded.
To confirm this conclusion and to verify that the disul-phide bond is in the N-loop, we deleted this part of the protein. An in-frame deletion of comEC was constructed removing 155 residues from the protein, including both cysteines, and leaving 12 residues as a linker to preserve the overall topology of ComEC. comEC missing the N-loop codons (ΔN-loop) was expressed from an ectopic locus (amyE) under the control of a competence promoter, PcomG, in wild-type cells and also in a comEC null background. The ΔN-loop protein was detected in a Western blot at approximately the same level as the wild-type ComEC and migrated at 55 KDa as predicted (Fig. 3D, lanes 4–7). The ΔN-loop construct was not able to complement the comEC null mutant for transformation nor did it exhibit a dominant-negative effect when coexpressed with the wild-type copy (not shown). The fact that the ΔN-loop protein was stable implied that a misfolded N-loop, lacking the disulphide bond was a signal for degradation of ComEC. Also the ΔN-loop construct unambiguously proved that N-loop cysteines form the intramolecular dis-ulphide bond, because the migration rate of this deletion protein did not respond to the addition of DTT. The location of this in vivo disulphide bond in the N-loop provides strong support for the extracytoplasmic location predicted for this domain by our model.
The competence-induced oxidoreductase pair, BdbDC, is required for the stabilization of ComEC
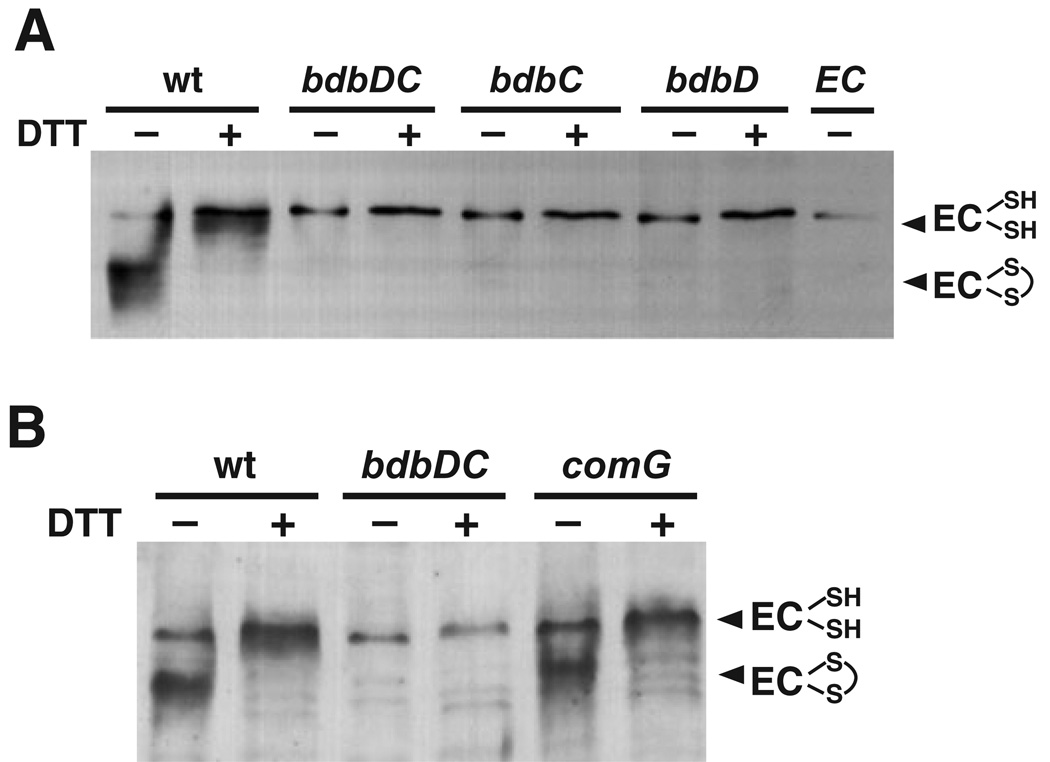
Two thiol-disulphide-oxidoreductase proteins, BdbD and BdbC, are required for transformation (Meima et al., 2002). The operon encoding these two proteins, bdbDC, was shown to be upregulated during competence (Berka et al., 2002; Meima et al., 2002). The essential competence protein ComGC contains two cysteines that form an intramolecular disulphide bond, and ComGC is destabilized in both bdbD and bdbC mutants (Meima et al., 2002). As ComEC also contains an intramolecular disul-phide bond we suspected that formation of this bond was dependent on the competence-specific thiol oxidoreductase system. Membranes were prepared from double (bdbDC) and single (bdbD, bdbC) deletion strains and resolved on SDS-PAGE in the absence or presence of DTT. ComEC was unstable in all three of these strains (Fig. 4A).
Fig. 4.
The BdbDC oxidoreductase pair is required for the stability of ComEC. Western blots for ComEC are shown. (A) wt (BD2528), bdbDC (BD3002), bdbC (BD2999), bdbD (BD3355), EC (BD2993), and (B) wt (BD2528), bdbDC (BD3002) and comG (BD2780).
As the BdbDC proteins are required for stabilization of ComGC, we considered the possibility that the effect of the bdbDC mutants on ComEC stability was indirect and mediated by destabilization of ComGC. Figure 4B shows that, ComEC is stable in a comG null mutant, which is missing all seven ComG proteins including ComGC. Thus, BdbDC is not required for ComEC stability indirectly through stabilization of ComGC or another ComG protein. The BdbDC proteins are not needed to stabilize ComFA or NucA, two other competence proteins that contain cys-teines (not shown).
Oligomerization of ComEC
As channel proteins often function as oligomers, it was of interest to determine whether this was true of ComEC. In the course of experiments with cross-linking reagents we observed a slower-migrating band in ComEC Western blots, even when the cross-linking reagents were omitted. This slowly migrating form of ComEC was observed only when reducing agent (DTT) was omitted from the sample buffer. When membrane vesicles were incubated at 37∞C the amount of the slowly migrating form increased with time (Fig. 5A), and at 60 min up to 75% of ComEC was in this form although the extent of conversion was observed to be somewhat variable. The position of this slowly migrating form indicates that ComEC is part of a higher molecular weight complex. At present, the nature of this complex is not known. Based on its migration in gels, the complex may be a homodimer, a homotrimer or a hetero-oligomer. As expected, when cells were treated with NEM before cell lysis to block free SH- groups, no oligomer formation was observed upon subsequent incubation (Fig. 5A). The addition of DTT to the sample buffer also reversed the formation of the oligomer (Fig. 5B), confirming that the oligomers are held together by disulphide bonds. These data suggest that when membrane vesicles containing ComEC are incubated under oxidizing conditions, an intermolecular disulphide bond forms, indicating that ComEC resides in the membrane as an oligomer, but that disulphide cross-linking is prevented by low redox potential. This in vitro cross-linking provides a convenient tool to detect ComEC oligomers and to identify ComEC residues near its oligomerization surface.
Fig. 5.
In vitro oligomerization of ComEC. Wild-type membranes (BD2528) were prepared using sucrose gradients as described. Membrane protein preparations were incubated at 37°C for 0, 10, 30 or 60 min and loaded on 7% SDS-PAGE in the absence (A) or presence (B) of 100 mM DTT. ComEC was detected using affinity-purified anti-ComEC antibodies. To block in vitro disulphide bond formation, NEM was added to competent cells before harvesting at a final concentration of 0.1 mM.
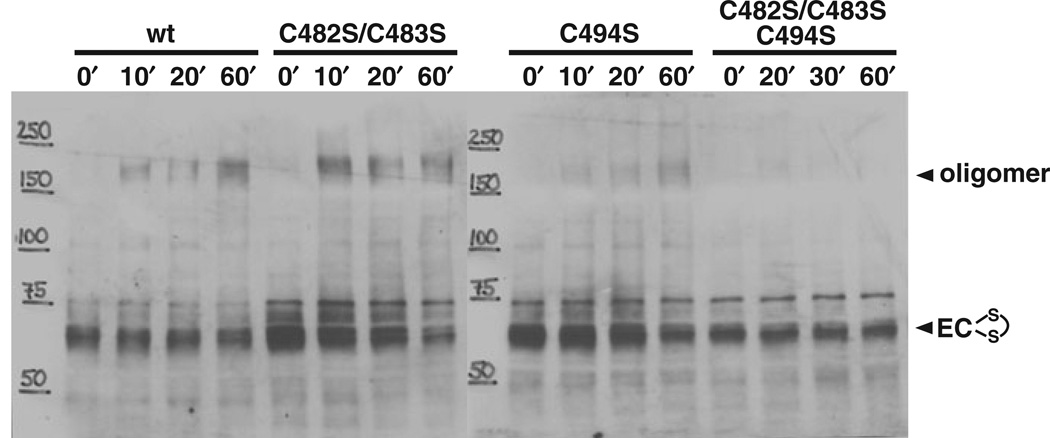
Oligomerization occurs between residues in TMS-F
To identify the residues involved in in vitro cross-linking, we investigated the cysteine replacement mutants described above. Beside mutants lacking the cysteines involved in forming the N-loop intramolecular disulphide bond (C131, C172), all mutants were stably expressed (Fig. 3C) and fully transformable (Table 2). Only replacement of the three cysteines located within TMS-F affected oligomerization, either alone or in combination (Fig. 6). We constructed three mutants, a double mutant strain C482S/C483S, a single mutant strain C494S and a triple mutant strain C482S/C483S/C494S. The C494S protein showed moderately reduced oligomerization, while the C482/483 double mutant had no detectable impact. Oli-gomerization of the triple mutant protein was completely abolished. Clearly, residues in the region between C482 and C494 are located near the oligomerization interface of ComEC. The most parsimonious interpretation of these results is that C494 is directly involved in disulphide cross-linking when the membranes are disrupted and the C-terminal domain is placed under oxidizing conditions, while C482 and C483 are less frequently involved.
Fig. 6.
In vitro oligomerization of ComEC mutant proteins with cysteine to serine point mutations. The oligomerization assay was performed as described in the legend of Fig. 5. wt (BD2528), C482S/C483S (BD3491), C494S (BD3492), C482S/C483S/C494S (BD3493).
Discussion
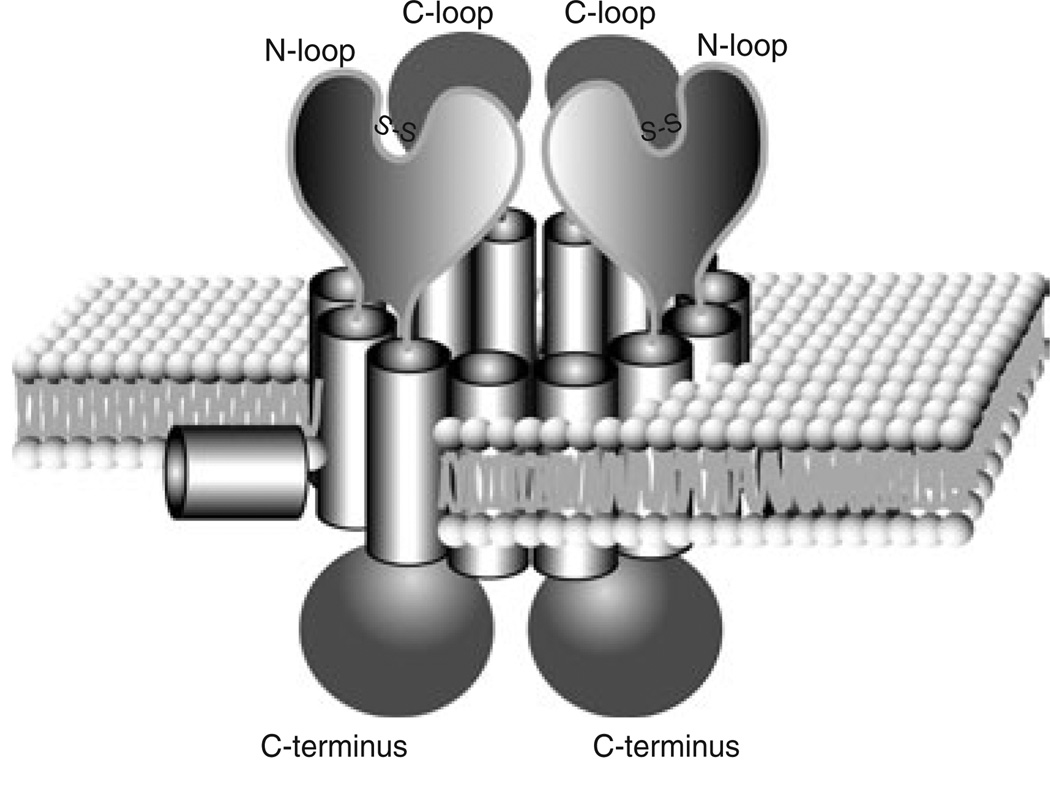
In this report, we have characterized the membrane topology of ComEC, a DNA uptake protein required for genetic competence in B. subtilis. In addition to seven confirmed TMSs in ComEC, other predicted hydrophobic segments might be membrane-associated, as helices that do not cross the membrane completely may remain undetected by the PhoA-LacZ fusion assay. In fact, our model predicts an amphipathic helix, which is proposed to be laterally associated with the cytosolic face of the membrane. Based on in vitro cross-linking with native cysteine residues, we suggest that ComEC non-covalently associates in a higher molecular weight complex, presumably to assemble the DNA-channel. We have also shown that the proper folding of ComEC requires formation of an intramolecular disulphide bond introduced by BdbD and C. A model for the ComEC DNA translocation channel is presented in Fig. 7. In this model, we assume that ComEC is a homodimer. We will now discuss various features of this model.
Fig. 7.
Model for the DNA uptake channel. We propose that two ComEC monomers form a channel in the membrane. Each subunit contains one N-loop, one C-loop, seven TMSs and one laterally inserted amphipathic helix (only one is shown). Subunits oligomerize involving contacts in TMS-F. The external N-loops are stabilized by disulphide bonds.
The competence domain
The competence domain was recently defined in the protein database (Accession number PF03772) as a part of ComEC that is well conserved among competent species. In our model, it includes TMSs C, D and E (Fig. 1). Because of the striking sequence conservation of these TMSs and intervening hydrophobic segments (not shown), we propose that they are important for the function of the channel and might fold to form all or part of an aqueous pore for DNA uptake.
The N-loop
The N-loop is present in all ComEC homologues but its sequence is not conserved beyond the low-GC Gram-positives (not shown). This divergence of the N-loop sequence in otherwise highly conserved proteins suggests that this stretch has diverged to accommodate different needs in Gram-positive and Gram-negative organisms. The uptake process is initiated by the binding of DNA to cell surface receptors. Most likely, receptor proteins contact the channel proteins following the binding of DNA. Perhaps the extracellular N-loop interacts with the DNA receptor ComEA. Gram-negative bacteria have outer membranes and probably an additional receptor protein is needed to bind DNA on the cell surface. At least in N. gonorrhoeae the multiple ComEA orthologues are periplasmic and may act as shuttle proteins transporting DNA from the cell surface to the inner membrane (Chen and Gotschlich, 2001). In B. subtilis ComEA is membrane embedded. Thus, the mechanism of DNA delivery to the channel protein in Gram-positive and -negative bacteria may be different, and this difference may be reflected by divergence in the N-loop sequence. Whatever its role, the N-loop is essential for function in ComEC, as complete deletion of the loop confers a strictly transformation deficient phenotype without affecting the stability of the truncated ComEC (Table 2, Fig. 3D).
Intramolecular disulphide bond
Based on the mobility shift observed upon addition of DTT to samples analysed by SDS-PAGE, we inferred that ComEC contains an intramolecular disulphide bond. Several arguments lead to the conclusion that this disulphide bond is formed between C131 and C172 of the N-loop. Disulphide bonds are usually formed outside the cytoplasm and in fact, based on our topological model, these are the only two extracellularly located cysteines in ComEC. Second, when either one or both of the above cysteines were replaced, ComEC was unstable, as is frequently observed when disulphide bond formation is prevented. Third, when the N-loop containing both cys-teines was deleted, the protein was stable and did not change conformation upon addition of DTT. The C131– C172 disulphide bond is apparently introduced by BdbDC, a competence-induced oxidoreductase protein pair (Meima et al., 2002). Evidence for direct catalysis is lacking, but the degradation of ComEC in bdbDC mutants provides a good indication that this is the case. This conclusion is further supported by the observation that BdbDC does not act on ComEC indirectly through ComFA, NucA or the ComG proteins, as elimination of these proteins does not affect ComEC stability (Fig. 4B and not shown).
Eight cysteines are present in ComEC but only the two that form the intramolecular disulphide bond are needed for function. Alignment of protein sequences from Bacillus halodurans, Bacillus anthracis, Bacillus licheniformis, Bacillus stearothermophilus, Bacillus cereus and Ocean-obacillus iheyensis revealed absolute conservation of the N-loop cysteines, while other cysteines were not conserved. However, the absence of these two cysteine residues from the competent organism S. pneumoniae, which interestingly also lacks BdbDC homologues, argues strongly that the disulphide bond plays a structural role in B. subtilis and is not needed per se for function.
It is of interest that formation of the N-loop disulphide bond appears to require the thiol-disulphide oxidoreduc-tase pair, BdbDC, which is also needed to form an intramolecular disulphide bond in the competence protein, ComGC, and which is under competence control. In both cases, an extracellular domain of these integral membrane competence proteins is oxidized, and disulphide bond formation is required for correct folding and stabilization of the proteins. No reductive pathway responsible for isomerization of incorrectly placed disulphides has been identified in B. subtilis, but these two competence proteins each contain single pairs of cysteine residues in extracellular domains, obviating the need for an isomerization pathway.
Oligomeric structure of ComEC
Native or engineered cysteine residues are widely used as a tool to detect protein–protein interactions in vitro (Khodadad and Weinstein, 1985; Milligan and Koshland, 1988; Lu et al., 1997) and in vivo (Lynch and Koshland, 1991; Hughson et al., 1997). We have used naturally occurring cysteines as cross-linkers to provide evidence for the existence of ComEC oligomers in membrane vesicles. Incubation under oxidizing conditions produced only one high molecular weight product and oligomerization occurred rapidly. These observations suggest that in the cell a ComEC molecule is in close proximity to either another ComEC molecule or to an unknown protein, with cysteine residues in close apposition, able to undergo covalent cross-linking when placed in oxidizing conditions. It is unlikely that the spontaneous cross-linking we have observed is due to non-specific aggregation involving ComEC, because in these experiments we have used membrane vesicles, in which ComEC was maintained in a nearly native environment, and because a unique cross-linked species was detected. If oligomerization were an artefact, we might expect ComEC to become randomly cross-linked to a variety of proteins, which was not observed. If the oligomer we have detected were a ComEC homotrimer, we would expect to also detect an intermediate dimeric form. We favour a model in which ComEC exists as a homodimer (Fig. 7), although other arrangements cannot be ruled out. In our homodimeric model, two molecules of ComEC closely associate to form a channel, a feature commonly observed among ABC transporters. For ABC transporters it has been suggested that five membrane-spanning segments in each of two subunits comprise the minimum for substrate translocation and the orientation of the five transmembrane segments in ComEC, TMS-A to -E, exactly matches that of the proposed minimal unit (N-terminus out) (van der Heide and Poolman, 2002).
Three cysteines within TMS-F were involved in in vitro cross-linking. Helical wheel analysis suggests that C483 and C494 align on a common face of the predicted helix, properly aligned to form disulphide bonds (not shown).
Biogenesis of ComEC
The data described in this report, together with previous work, permit a description of ComEC biogenesis. comEC is a late competence gene under the control of ComK, whose transcription begins upon entry into the stationary phase. ComEC accumulates during about 2 h, after which cells are maximally competent. The newly synthesized protein is inserted in the membrane, most likely via the sec secretion machinery (Asai et al., 1998) as a multiple-spanning membrane protein. Upon export of the N-loop residues, the BdbDC oxidoreductase pair (also under competence control) catalyses the formation of an intramolecular disulphide bond, which aids in the correct folding of the N-loop, thereby stabilizing the protein. Two ComEC monomers associate using contacts, at least some of which are located in membrane-spanning segment F, to form a functional channel with 14 TMSs. Seven helices and an amphipathic helix from each monomer are proposed to contribute to the formation of an aqueous pore for DNA transport (see Fig. 7). The N-loop, the C-loop and cytosolic domains may serve to associate with accessory uptake proteins (e.g. ComEA and ComFA) or to gate the channel.
Experimental procedures
Bacterial strains
All strains in this work are derivatives of B. subtilis 168 (Table 3). B. subtilis cultures were grown under conditions leading to competence in a glucose minimal salt medium (Albano et al., 1987). E. coli XL10 Gold (Stratagene) was used as a host strain for plasmid constructs.
Table 3.
B. subtilis strains used in this study.
| Straina | Genotypeb | Plasmidc | AntibioticR |
|---|---|---|---|
| BD630 | hisA1 leu-8 metB5, wt strain | ||
| BD2528 | pMCS (pUB110::comS) | Kn | |
| BD2780 | comGA::Tn917 (12), pMCS | Em, Kn | |
| BD2993 | comEC::Tn917(518), pMCS | Em, Kn | |
| BD2999 | bdbC::pMutin2, pMCS | Em, Kn | |
| BD3002 | bdbDC::pMutin2, pMCS | Em, Kn | |
| BD3355 | bdbDC::pMutin2amyE::PxylA-bdbC, pMCS | Em, Cm, Kn | |
| phoA and lacZ fusions to comEC in the native locus | |||
| BD3231 | comEC-H44-phoA | pED495 | Cm |
| BD3232 | comEC-H44-lacZ | pED496 | Cm |
| BD2964 | comEC-K106-phoA | pED349 | Cm |
| BD2985 | comEC-K106-lacZ | pED363 | Cm |
| BD3422 | comEC-N177-phoA | pED602 | Cm |
| BD3423 | comEC-N177-lacZ | pED603 | Cm |
| BD2965 | comEC-H231-phoA | PED334 | Cm |
| BD3123 | comEC-K260-phoA | pED415 | Cm |
| BD3124 | comEC-K260-lacZ | pED416 | Cm |
| BD3424 | comEC-R284-phoA | pED604 | Cm |
| BD3425 | comEC-R284-lacZ | pED605 | Cm |
| BD3125 | comEC-R301-phoA | pED417 | Cm |
| BD3126 | comEC-R301-lacZ | pED418 | Cm |
| BD3164 | comEC-G327-phoA | pED447 | Cm |
| BD3171 | comEC-G327-lacZ | pED448 | Cm |
| BD3165 | comEC-S351-phoA | pED449 | Cm |
| BD3172 | comEC-S351-lacZ | pED450 | Cm |
| BD3166 | comEC-N385-phoA | pED451 | Cm |
| BD3173 | comEC-N385-lacZ | pED452 | Cm |
| BD3167 | comEC-R415-phoA | pED453 | Cm |
| BD3174 | comEC-R415-lacZ | pED454 | Cm |
| BD3168 | comEC-T442-phoA | pED455 | Cm |
| BD3175 | comEC-T442-lacZ | pED456 | Cm |
| BD3169 | comEC-Q474-phoA | pED457 | Cm |
| BD3176 | comEC-Q474-lacZ | pED458 | Cm |
| BD2966 | comEC-P537-phoA | pED336 | Cm |
| BD2967 | comEC-P537-lacZ | pED337 | Cm |
| BD3426 | comEC-K550-phoA | pED606 | Cm |
| BD3427 | comEC-K550-lacZ | pED607 | Cm |
| BD3430 | comEC-K639-phoA | pED610 | Cm |
| BD3431 | comEC-K639-lacZ | pED611 | Cm |
| BD3428 | comEC-K647-phoA | pED608 | Cm |
| BD3429 | comEC-K647-lacZ | pED609 | Cm |
| BD3620 | comEC-V680-phoA | PED695 | Cm |
| BD3621 | comEC-V680-lacZ | pED696 | Cm |
| BD3622 | comEC-K711-phoA | pED697 | Cm |
| BD3623 | comEC-K711-lacZ | pED698 | Cm |
| BD3624 | comEC-R740-phoA | pED699 | Cm |
| BD3625 | comEC-R740-lacZ | pED700 | Cm |
| BD3170 | comEC-P766-phoA | pED459 | Cm |
| BD3177 | comEC-P766-lacZ | pED460 | Cm |
| BD3626 | comEC-N776-phoA | pED701 | Cm |
| BD3627 | comEC-N776-lacZ | pED702 | Cm |
| Cysteine to serine point mutations in comEC | |||
| BD3388 | comEC C309S | pED558 | Cm |
| BD3389 | comEC C395S | pED559 | Cm |
| BD3390 | comEC (wt), truncated comEC C172S | pED557 | Cm |
| BD3491 | comEC C482S, C483S | pED567 | Cm |
| BD3492 | comEC C494S | pED568 | Cm |
| BD3493 | comEC C482S, C483S, C494S | pED569 | Cm |
| BD3411 | BD3410, pMCS | Cm, Kn | |
| BD3400 | BD3390, pMCS | Cm, Kn | |
| BD3401 | BD3386, pMCS | Cm, Kn | |
| BD3402 | BD3387, pMCS | Cm, Kn | |
| BD3403 | BD3388, pMCS | Cm, Kn | |
| BD3404 | BD3389, pMCS | Cm, Kn | |
| BD3488 | BD3487, pMCS | Cm, Kn | |
| BD3495 | BD3491, pMCS | Cm, Kn | |
| BD3496 | BD3492, pMCS | Cm, Kn | |
| BD3497 | BD3493, pMCS | Cm, Kn | |
| Deletion of the N-loop in comEC | |||
| BD3474 | amyE::comECΔN-loop | Cm | |
| BD3478 | amyE::comECΔN-loop, pMCS | Cm, Kn | |
| BD3479 | comEC::Tn917(518), amyE::comECΔN-loop | Em, Cm | |
| BD3480 | comEC::Tn917(518), amyE::comECΔN-loop, pMCS | Em, Cm, Kn | |
All strains, except for the lab strains BD630 and BD2528, and strains constructed by Meima et al. (2002), BD2999, BD3002, BD3355, were constructed for this work.
All strains are derivatives of BD630 and are auxotrophic for histidine (hisA1), leucine (leu-8) and methionine (metB5).
Plasmids used in transformation to obtain the B. subtilis strains.
Computer analysis of ComEC
The following programs were used to predict the membrane topology for ComEC: HMMTOP 2.0 (Tusnady and Simon, 1998) (http://www.enzim.hu/hmmtop/index.html); TMHMM 2.0 (Krogh et al., 2001) (http://www.cbs.dtu.dk/services/TMHMM/); MEMSAT 2 (Jones et al., 1994) (http://www.psipred.net.); DAS (Cserzo et al., 1997) (http://www.sbc.su.se/~miklos/DAS/maindas.html); TOPPRED (Claros and von Heijne, 1994) (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html); PRED-TMR 1.0 (Pasquier et al., 1999); Swiss protein (http://us.expasy.org/cgi-bin/sprotsearch-ac?P39695); SOSUI (http://sosui.proteome.bio.tuat.ac.jp/cgi-bin/sosui.cgi?/sosui_submit.html); SPLIT (Juretic et al., 2002) (http://garlic.mefos.hr/split/); TMpred (Hofmann and Stoffel, 1993) (http://www.ch.embnet.org/software/TMPRED_form.html).
Construction of comEC-phoA and comEC-lacZ fusions
Fusions of comEC to either phoA or lacZ were generated by cloning fragments of comEC, amplified by PCR, in frame with the phoA reporter gene of plasmid pUCCMPHOA (Piazza et al., 1999) or with the lacZ reporter gene of pJF751 (Ferrari et al., 1986). The following 5’-primers were used for PCR amplifications (in combination with fusion specific 3’-primers listed in Table 4): EC-FP#0A–BamHI (CGGGATCCTTTG GAGGGTGATGAATGCGTAATTCG), EC-FP#0Z–EcoRI (CGGAATTCTTTGGAGGGTGATGAATGCGTAATTCG), EC-FP#26A–BamHI (CGGGATCCAACGGCTGGAATTACTG), EC-FP#27Z–EcoRI (CGGAATTCAACGGCTGGAATTACTG), EC-FP#132-EcoRI (CGGAATTCTGCTCTTTATATATCCGTGTC), EB-FP#3-EcoRI (CGGAATTCATAGTCAGAGACAAACGC) and EB-FP#12-PvuII (CGCAGCTGATA GTCAGA GACAAACGC). The last two primers start within comEB to provide sufficient fragment length for recombination. Amplified comEC fragments for fusions were cloned into pUCCMPHOA using the underlined BamH I/SalI sites with the exception of the H44-phoA fragment, which was digested with EcoRI(filled-in)/Sal I, and cloned into BamHI(filled-in)/ SalI sites. To obtain lacZ fusions, PCR fragments were cloned into pJF751 using EcoRI/BamHI sites. Exceptions were the K106-lacZ fusion (cut PvuII(filled-in)/BamHI and cloned into AflIII(filled-in)/BamHI sites of pUCCM18) and the K711 -lacZ, R740-lacZ and N776-lacZ fusions, which were digested with MfeI (present in comEC) and BamHI and cloned into EcoRI/ BamHI sites of pJF751. Plasmids carrying the phoA and lacZ reporter fusions to comEC are listed in Table 4. These plasmids were integrated in the native site of B. subtilis strain BD630 by Campbell-like recombination to generate chromo-somally located fusions under native competence control. Fusions that showed no enzymatic activities were confirmed by sequencing. The resulting Bacillus strains are listed in Table 3. Because comEC is the last gene in the operon, downstream genes were not affected by the plasmid integrations.
Table 4.
Primers used for the construction of phoA and lacZ fusions to comEC.
| Fusiona | 5′ primerb | 3′ primer (sequence)c | Plasmidd |
|---|---|---|---|
| H44-phoA | EB-FP#3 | EC-RP#53 (ACTGCGGTCGACAGTGCCTCGTTTTGATTAAA) | pED495 |
| H44-lacZ | EB-FP#3 | EC-RP#54 (CGGGATCCACGTGCCTCGTTTTGATT) | pED496 |
| K106-phoA | EC-FP#0A | EC-RP#9 (ACTGCGGTCGACATTTTTCCTTATCAGGTGTCTC) | pED349 |
| K106-lacZ | EC-FP#12 | EC-RP#12 (CGCGGATCCCATTTTTCCTTATCAGGTGTC) | pED363 |
| N177-phoA | EC-FP#26A | EC-RP#82 (ACTGAGGTCGACAATTTTCAGGTTCGCTGC) | pED602 |
| N177-lacZ | EC-FP#27Z | EC-RP#83 (CGGGATCCAAATTTTCAGGTTCGCTGC) | pED603 |
| H231-phoA | EC-FP#0A | EC-RP#1 (ACTGCGGTCGACGATGGACAACACCAAGC) | PED334 |
| K260-phoA | EC-FP#26A | EC-RP#26 (ACTGCGGTCGACACTTTTCTCTAGTTATACCAAGG) | pED415 |
| K260-lacZ | EC-FP#27Z | EC-RP#27 (CGGGATCCAGCTTTTCTCTAGTTATACCAAGG) | pED416 |
| R284-phoA | EC-FP#26A | EC-RP#84 (ACTGCGGTCGACAGCGTAGCACTGAAGGAG) | pED604 |
| R284-lacZ | EC-FP#27Z | EC-RP#85 (CGGGATCCAAGCGTAGCACTGAAGGAG) | pED605 |
| R301-phoA | EC-FP#26A | EC-RP#28 (ACTGCCGTCGACAACGCCATTTGACAAGAC) | pED417 |
| R301-lacZ | EC-FP#27Z | EC-RP#29 (CGGGATCCTTACGCCATTTGACAAGAC) | pED418 |
| G327-phoA | EC-FP#26A | EC-RP#33 (ACTGCTGTCGACTACCGGCTTCAAAGAGATG) | pED447 |
| G327-lacZ | EC-FP#27Z | EC-RP#34 (CGCGGATCCAAACCGGCTTCAAAGAG) | pED448 |
| S351-phoA | EC-FP#26A | EC-RP#35 (ACTGCGGTCGACTGGAGGTTTTAACCTGCTG) | pED449 |
| S351-lacZ | EC-FP#27Z | EC-RP#36 (CGGGATCCAAGGAGGTTTTAACCTGC) | pED450 |
| P383-phoA | EC-FP#26A | EC-RP#75 (ACTGAGGTCGACACGGTACGCTGATGATAGA) | pED568 |
| P383-lacZ | EC-FP#27Z | EC-RP#76 (CGGGATCCAACGGTACGCTGATGATAGA) | pED569 |
| N385-phoA | EC-FP#26A | EC-RP#37 (ACTGAGGTCGACAATTCATCGGTACGCTGA) | pED451 |
| N385-lacZ | EC-FP#27Z | EC-RP#38 (CGGGATCCAAATTCATCGGTACGCTGA) | pED452 |
| R415-phoA | EC-FP#26A | EC-RP#39 (ACTGCGGTCGACATCTCCCAAACGAAGCG) | pED453 |
| R415-lacZ | EC-FP#27Z | EC-RP#40 (CGCGGATCCAATCTCCCAAACGAAGC) | pED454 |
| T442-phoA | EC-FP#26A | EC-RP#41 (ACTGCGGTCGACTCGTGAACACATCAACAT) | pED455 |
| T442-lacZ | EC-FP#27Z | EC-RP#42 (CGGGATCCATCGTGAACACATCAACA) | pED456 |
| M465-phoA | EC-FP#26A | EC-RP#101 (ACTGCGGTCGACACATAAGCAATAGGATGATCGT) | pED681 |
| M465-lacZ | EC-FP#27Z | EC-RP#102 (CGGGATCCACCATAAGCAATAGGATGATCGT) | pED682 |
| Q474-phoA | EC-FP#26A | EC-RP#43 (ACTGAGGTCGACACTGCGACAAGGAGCGT) | pED457 |
| Q474-lacZ | EC-FP#27Z | EC-RP#44 (CGGGATCCAACTGCGACAAGGAGCG) | pED458 |
| D510-phoA | EC-FP#26A | EC-RP#103 (ACTGCGGTCGACAGTCACCCTGTCCAATATC) | pED683 |
| D510-lacZ | EC-FP#27Z | EC-RP#104 (CGGGATCCACGTCACCCTGTCCAATATC) | pED684 |
| T530-phoA | EC-FP#26A | EC-RP#105 (ACTGCGGTCGACAAGTGCCGCCGGTATCA) | pED685 |
| T530-lacZ | EC-FP#27Z | EC-RP#106 (CGGGATCCAAAGTGCCGCCGGTATCAA) | pED686 |
| P537-phoA | EC-FP#0A | EC-RP#4 (ACTGCGGTCGACAAGGCTCTGACGAGTAAG) | pED336 |
| P537-lacZ | EC-FP#0Z | EC-RP#5 (CGCGGATCCCAAGGCTCTGACGAGTAAG) | pED337 |
| R539-phoA | EC-FP#26A | EC-RP#107 (ACTGAGGTCGACAGCGCCAAGGCTCTGACG) | pED687 |
| R539-lacZ | EC-FP#27Z | EC-RP#108 (CGGGATCCAAGCGCCAAGGCTCTGACG) | pED688 |
| K541-phoA | EC-FP#26A | EC-RP#109 (ACTGCGGTCGACATTTTTCGCGCCAAGGCTC) | pED689 |
| K541-lacZ | EC-FP#27Z | EC-RP#110 (CGGGATCCAATTTTTCGCGCCAAGGCTC) | pED690 |
| K550-phoA | EC-FP#26A | EC-RP#86 (ACTGCGGTCGACACTTTTCCCCCAGTGAAAAC) | pED606 |
| K550-lacZ | EC-FP#27Z | EC-RP#87 (CGCGGATCCACCTTTTCCCCCAGTGAAAAC) | pED607 |
| K562-phoA | EC-FP#26A | EC-RP#111 (ACTGCGGTCGACACTTGATTCCCTTAGCAGTT) | pED691 |
| K562-lacZ | EC-FP#27Z | EC-RP#112 (CGGGATCCAACTTGATTCCCTTAGCAGTT) | pED692 |
| K585-phoA | EC-FP#26A | EC-RP#113 (ACTGCGGTCGACACTTCAGCAGAATCTCCG) | pED693 |
| K585-lacZ | EC-FP#27Z | EC-RP#114 (CGGGATCCAACTTCAGCAGAATCTCCG) | pED694 |
| K639-phoA | EC-FP#26A | EC-RP#90 (ACTGCGGTCGACACGGTGACAGTACATGGAA) | pED610 |
| K639-lacZ | EC-FP#27Z | EC-RP#91 (CGGGATCCAACGGTGACAGTACATGGAA) | pED611 |
| K647-phoA | EC-FP#26A | EC-RP#88 (ACTGAGGTCGACATTTGCTTGCCGGATCAG) | pED608 |
| K647-lacZ | EC-FP#27Z | EC-RP#89 (CGGGATCCAATTTGCTTGCCGGATCAG) | pED609 |
| K680-phoA | EC-FP#26A | EC-RP#115 (ACTGCGGTCGACACACGTTCATCACCTCTTG) | pED695 |
| K680-lacZ | EC-FP#27Z | EC-RP#116 (CGGGATCCAACACGTTCATCACCTCTTG) | pED696 |
| K711-phoA | EC-FP#26A | EC-RP#117 (ACTGCGGTCGACATTTCGGCTGAAGCTGTTG) | pED697 |
| K711-lacZ | EC-FP#27Z | EC-RP#118 (CGGGATCCAATTTCGGCTGAAGCTGTTG) | pED698 |
| R740-phoA | EC-FP#26A | EC-RP#119 (ACTGCGGTCGACAGCGGATAGAATGTCTCTG) | pED699 |
| R740-lacZ | EC-FP#132 | EC-RP#120 (CGGGATCCAAGCGGATAGAATGTCTCTG) | pED700 |
| P766-phoA | EC-FP#26A | EC-RP#45 (ACTGCGGTCGACATGGAGGATAGACAGAAAAG) | pED459 |
| P766-lacZ | EC-FP#27Z | EC-RP#46 (CGGGATCCAATGGAGGATAGACAGAAAAG) | pED460 |
| N776-phoA | EC-FP#26A | EC-RP#121 (ACTGCGGTCGACAGTTCGTCTCTGTTATATCTGA) | pED701 |
| N776-lacZ | EC-FP#132 | EC-RP#122 (CGGGATCCAAGTTCGTCTCTGTTATATCTGA) | pED702 |
In frame phoA or lacZ fusion was made at the indicated amino acid residues in comEC.
See text for the primer sequences.
Restriction sites are underlined.
Plasmids bearing the fusion constructs.
Assay of alkaline phosphatase and β-galactosidase activities
For the alkaline phosphatase assay, a protocol from Piazza et al. (1999) was adopted. The competent cells from 1 ml of culture were resuspended in 110 µl of Buffer A [1 M Tris (pH = 8), 0.1 mM ZnCl2] and incubated with 140 µl of p-nitrophenyl phosphate (1.4 mg ml−1 in Buffer A) for 1 to 2 h at 37°C. Reactions were stopped by the addition of 25 µl 1 M KH2PO4, and after centrifugation, activities were determined by measuring OD420 of the supernatants. The protocol to measure the β-galactosidase activity was similar. Buffer Z (0.1 M NaPO4 pH = 7, 0.001 M MgSO4, 0.1 M β-mercaptoethanol) was used instead of Buffer A. 140µl of o-nitrophenyl-β-D-galactoside (1.4 mg ml−1 in Buffer Z) was added. The reaction was stopped by the addition of 25 µl of 1 M Na+-carbonate. Cells were permeabilized with toluene for 30 min on ice prior to the addition of o-nitrophenyl-β-D-galactoside.
Construction of cysteine to serine point mutations in comEC
comEC missing the first 43 bp was amplified by primers EC-A15-FP#27Z–EcoRI (CGGAATTCAACGGCTGGAATTACTG) and EC-STOP-RP#8-BglII (GAAGATCTTTAGTTCGTCTCTGTTATATCTG) and cloned into pPCR-Script (Invitrogen) to generate plasmid pED523. The integrity of the insert was verified by sequencing. The comEC fragment was excised from pED523 with NcoI and EcoRV and blunt ligated into pUCCM18 predigested with HindIII and BanII to obtain plas-mid pED537. To avoid extensive sequencing of comEC, an additional plasmid containing an internal HindIII-KpnI fragment of comEC was made. To obtain this plasmid (pED530), a HindIII-KpnI fragment of comEC was cut out from pED523 and cloned into pUC19.
PCR mutagenesis was carried out on template plasmids listed in Table 5 using a QuickChange™ site-directed mutagenesis kit (Stratagene). For each of the eight cys-teine to serine substitutions, a complementary set of primers was designed. Codons for serine were chosen so that new restriction sites were introduced into the comEC by mutagenesis (Table 5). Because none of the codons created a new restriction site for the C309S replacement, bases at the third position of the two preceding amino acids were changed to introduce a SauIIIA site. Introduction of the mutations was confirmed by sequencing. For plasmid constructs pED551-pED554, only a small region surrounding the mutation was sequenced and subcloned into a fully sequenced ComEC construct pED537 using the following restriction sites: BseRI-HindIII for pED551 and pED552, HindIII-BanII for pED553 and BanII-BstEII for pED554 resulting in final constructs pED556-59. Finally, plasmids pED570, 556, 557, 558, 559, 600, 657, 658 and 659 (Table 5) were integrated at the native comEC locus of B. subtilis 168 by Campbell-like recombination. Two types of cross-over reaction were expected: (i) recombination before the mutation site leading to mutated comEC and (ii) recombination after the mutation site reconstructing the intact comEC locus with mutation remaining in the downstream truncated copy of comEC. Transformants were screened for the first type of recombination by restriction pattern analysis of PCR products. The resulting mutant strains are listed in Table 3.
Table 5.
Construction of cysteine to serine point mutations in comEC.
| Mutation | Template for PCR mutagenesis |
5′-mutagenic primera | New siteb | Plasmid after PCRc |
| C51S | pED537 | CGAGGCACGCTTTTCTTATTATTGTTAGCTTCTTCTCTTTT ATATTGTTTTTTG |
AluI | pED570 |
| C131S | pED523 | GAACCAGGAATGTCATCTGAGTTGACTGGTACATTGG | DdeI | pED551 (pED556) |
| C172S | pED523 | CTGTCACGTCTATCCAAAACTCGAGCGAACCTGAAAATTTC | XhoI | pED552 (pED557) |
| C131S, C172S | pED556 | CTGTCACGTCTATCCAAAACTCGAGCGAACCTGAAAATTTC | DdeI | pED600 |
| C309S | pED530 | GTCCGCTCTGCAACTGCGATCAGCCTTTCGTACATCGTCC | SauIIIA | pED553 (pED558) |
| C395S | pED537 | GTACCATTTTATACCTTCTCGATTTTGCCGGGAGCTGTAG | TaqI | pED554 (pED559) |
| C482S, C483S | pED537 | GATGGTAACCGGAGGCATTTCGAGCACGGTGATGTTTCTG | TaqI | pED657 |
| C494S | pED537 | CTGCTCTTTATATATCCGAGTCTTAGTTCCGAAGGAGAAG | HnfI | pED658 |
| C482S, C483S, C494S | pED657 | CTGCTCTTTATATATCCGAGTCTTAGTTCCGAAGGAGAAG | TaqI, HinfI | pED659 |
5′-primers and their complements were used for PCR. Mutagenic bases are underlined.
Restriction sites introduced into comEC sequence by PCR mutagenesis.
Template plasmids containing mutations introduced by PCR. In the parentheses are plasmids obtained by further subcloning of the mutants (see Experimental procedures).
Construction of an N-loop deletion (ΔN-loop)
We constructed a comEC deletion that removed 155 residues from the N-terminal loop. A total of 12 residues of the originally 167 residue-N-loop were left as a linker region. The deletion was constructed by amplifying the N-terminal (PCR#93) and the C-terminal (PCR#94) parts of comEC separately and joining them by ligation in the vector pDG67. This vector carries a comG promoter, a ribosome-binding site and homology to amyE for double cross-over recombination. PCR#93 (213 bp) was carried out using primers FP#93-SmaI-EC-ATG (TCCCCCGGGATGCGTAATTCGCGCTTA) and RP#93-HindIII-EC-S71 (ACTGCGAAGCTTAGAGACATTCTGAGAATCTG). Primers for PCR#94 (1700 bp) consisted of 5′EC-ISP (GGTGACAGATTTTACGTGG) and RP#94-ClaI-EC-STOP (CCATCGATTTAGTTCGTCTCTGTTATATCTG). The product of PCR#93 was digested at the underlined SmaI/ HindIII sites and of PCR#94 at ClaI/HindIII. Both digested products were ligated into pDG67 predigested with SmaI/ ClaI. Triple ligation (SmaI-HindIII-ClaI) produced a plasmid pED601, verified by sequencing, which was transformed into the amyE locus of BD630 to produce BD3474. BD3474 was transformed with pMCS to obtain BD3478 and with comEC::Tn917(518) chromosomal DNA to obtain BD3479. BD3479 was transformed with pMCS to obtain BD3480.
Isolation and treatment of membranes
Competent cells, harvested from 200 ml of cultures at T2, were resuspended in 5 ml of P-buffer (100 mM sodium phos-phate pH = 7.5, 5 mM MgCl2, 1 mM EDTA, 10 mM Protease inhibitor cocktail, Roche) and lysed in a French pressure cell. After 30 min digestion with DNase I (10 µg ml−1, Roche) at 4°C, unbroken cells were removed by 5 min centrifugation at low speed. Membranes were then sedimented at 100 000 g for 45 min, washed twice with P-buffer and resuspended in 200 µl of P-buffer for further purification through a sucrose gradient. Membrane suspensions were mixed with 1 ml 90% sucrose (72% final conc.) and injected at the bottom of a 5 ml polycarbonate tube containing 1.5 ml 52% sucrose overlaid with 1 ml 42% sucrose. Sucrose solutions were prepared in P-buffer. The tubes were spun for 18 h at 100 000 g and the floated membranes were collected from the 56%-42% sucrose interface. These membranes were diluted seven times in P-buffer, pelleted at 100 000 g for 90 min and resus pended in P-buffer to a final protein concentration of 2–5 mg ml−1.
Western blot analysis
Membrane protein extracts were mixed with a glycerol sample buffer (with or without 100 mM DTT as indicated) and equilibrated at room temperature for 5 min. Proteins (10 µg per lane) were resolved by SDS-PAGE (7% for ComEC and 15% for ComEA detection) at constant voltage (72 V) and transferred to nitrocellulose membranes for 2 h at 12 V in a semi-dry transfer apparatus (Bio-Rad). Transferred proteins were detected using affinity purified anti-ComEC antibodies (1:200), followed by secondary anti-rabbit antibodies (Zymed #401315, 1:10 000). Secondary antibodies were detected using ECL+ (Amersham).
Production of antibodies against ComEC
A sequence corresponding to the hydrophilic C-terminus of ComEC (residues 519–776) was amplified using primers CtermEC-Up (GAGGCATGCAGCGGGGTCGTGTCTTAAT) and CtermEC-Lo (CAAGGATCCGTTCGTCTCTGTTATATCT GAT). The resulting PCR product was cloned into a his6-tag expression vector pQE70 (Qiagen) using the underlined SphI and BamHI sites (pED309). Production of the resulting fusion protein was induced in E. coli M15 growing in Luria broth containing ampicillin (100 µg ml-1) and kanamycin (50 µg ml−1), by the addition of isopropyl-β-D-thiogalactopyranoside (1 mM) to an exponentially growing culture. Cells, harvested after 1 h induction, were broken in a French pressure cell and the fusion protein was purified on Ni2+-resin (Qiagen) under denaturing conditions as suggested by the manufacturer. The eluted material, which was largely homogeneous as determined by SDS-polyacrylamide gel electrophoresis, was excised from the gel and used for antibody production in rabbits. The immunization protocol was carried out by Research Genetics (Invitrogen). Antibodies from the final bleed were affinity purified on a nitrocellulose membrane containing isolated fusion protein.
Acknowledgements
We thank all the members of our lab for useful discussions and advice. Special thank to Roberta Provvedi for generating the ComEC antibody. We would like to thank an anonymous reviewer for a suggestion that greatly improved the quality of this paper. This work was supported by NIH grant GM43756.
References
- Albano M, Dubnau DA. Cloning and characterization of a cluster of linked Bacillus subtilis late competence mutations. J Bacteriol. 1989;171:5376–5385. doi: 10.1128/jb.171.10.5376-5385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M, Hahn J, Dubnau D. Expression of competence genes in Bacillus subtilis . J Bacteriol. 1987;169:3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M, Breitling R, Dubnau DA. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989;171:5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai K, Fujita M, Kawamura F, Takahashi H, Kobayashi Y, Sadaie Y. Restricted transcription from sigma H or phosphorylated spo0A dependent promoters in the temperature-sensitive secA341 mutant of Bacillus subtilis . Biosci Biotechnol Biochem. 1998;62:1707–1713. doi: 10.1271/bbb.62.1707. [DOI] [PubMed] [Google Scholar]
- Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, et al. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- Chen I, Gotschlich EC. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J Bacteriol. 2001;183:3160–3168. doi: 10.1128/JB.183.10.3160-3168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YS, Dubnau D. ComC is required for the processing and translocation of comGC, a pilin-like competence protein of Bacillus subtilis . Mol Microbiol. 1995;15:543–551. doi: 10.1111/j.1365-2958.1995.tb02267.x. [DOI] [PubMed] [Google Scholar]
- Chung YS, Dubnau D. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis . J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YS, Breidt F, Dubnau D. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis . Mol Microbiol. 1998;29:905–913. doi: 10.1046/j.1365-2958.1998.00989.x. [DOI] [PubMed] [Google Scholar]
- Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofs-son A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- Ferrari E, Howard SMH, Hoch J. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986;166:173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G, von Heijne G. Topological ‘frustration’ in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–412. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Albano M, Dubnau D. Isolation and characterization of Tn917lac-generated competence mutants of Bacillus subtilis . J Bacteriol. 1987;169:3104–3109. doi: 10.1128/jb.169.7.3104-3109.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Inamine G, Kozlov Y, Dubnau D. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake of transforming DNA. Mol Microbiol. 1993;10:99–111. doi: 10.1111/j.1365-2958.1993.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30:5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heide T, Poolman B. ABC transporters: one, two or four extracytoplasmic substrate-binding sites? EMBO Rep. 2002;3:938–943. doi: 10.1093/embo-reports/kvf201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMBASE – A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hughson AG, Lee GF, Hazelbauer GL. Analysis of protein structure in intact cells: crosslinking in vivo between introduced cysteines in the transmembrane domain of a bacterial chemoreceptor. Protein Sci. 1997;6:315–322. doi: 10.1002/pro.5560060206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamine GS, Dubnau D. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J Bacteriol. 1995;177:3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Juretic D, Zoranic L, Zucic D. Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci. 2002;42:620–632. doi: 10.1021/ci010263s. [DOI] [PubMed] [Google Scholar]
- Khodadad JK, Weinstein RS. Band 3 protein of the red cell membrane of the llama: crosslinking and cleavage of the cytoplasmic domain. Biochem Biophys Res Commun. 1985;130:493–499. doi: 10.1016/0006-291x(85)90444-9. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, Dubnau D. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol Microbiol. 1993;9:119–131. doi: 10.1111/j.1365-2958.1993.tb01674.x. [DOI] [PubMed] [Google Scholar]
- Lu HM, Motley ST, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and extracellular protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- Lynch BA, Koshland DE., Jr Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli . Proc Natl Acad Sci USA. 1991;88:10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Manoil C. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Meth Cell Biol. 1991;34:61–75. doi: 10.1016/s0091-679x(08)61676-3. [DOI] [PubMed] [Google Scholar]
- Meima R, Eschevins C, Fillinger S, Bolhuis A, Hamoen LW, Dorenbos R, et al. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J Biol Chem. 2002;277:6994–7001. doi: 10.1074/jbc.M111380200. [DOI] [PubMed] [Google Scholar]
- Milligan DL, Koshland DE., Jr Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- Mohan S, Aghion J, Guillen N, Dubnau D. Molecular cloning and characterization of comC, a late competence gene of Bacillus subtilis . J Bacteriol. 1989;171:6043–6051. doi: 10.1128/jb.171.11.6043-6051.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS, Hamodrakas SJ. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 1999;12:381–385. doi: 10.1093/protein/12.5.381. [DOI] [PubMed] [Google Scholar]
- Piazza F, Tortosa P, Dubnau D. Mutational analysis and membrane topology of ComP, a quorumsensing histidine kinase of Bacillus subtilis controlling competence development. J Bacteriol. 1999;181:4540–4548. doi: 10.1128/jb.181.15.4540-4548.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis . Mol Microbiol. 1999;31:271–280. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- Provvedi R, Chen I, Dubnau D. NucA is required for DNA cleavage during transformation of Bacillus subtilis . Mol Microbiol. 2001;40:634–644. doi: 10.1046/j.1365-2958.2001.02406.x. [DOI] [PubMed] [Google Scholar]
- Raina S, Missiakas D. Making and breaking disulfide bonds. Annu Rev Microbiol. 1997;51:179–202. doi: 10.1146/annurev.micro.51.1.179. [DOI] [PubMed] [Google Scholar]
- Segrest JP, De Loof H, Dohlman JG, Brouillette CG, Anantharamaiah GM. Amphipathic helix motif: classes and properties. Proteins. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- van Sinderen D, Kiewiet R, Venema G. Differential expression of two closely related deoxyribonu-cleases, nucA and nucB.Bacillus subtlis . Mol Microbiol. 1995;15:213–223. doi: 10.1111/j.1365-2958.1995.tb02236.x. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]