Abstract
Single cell mass cytometry facilitates high-dimensional, quantitative analysis of the effects of bioactive molecules on cell populations at single-cell resolution. Datasets are generated with antibody panels (upwards of 40) in which each antibody is conjugated to a polymer chelated with a stable metal isotope, usually in the Lanthanide series of the periodic table. Isotope labelled antibodies recognize surface markers to delineate cell types and intracellular signaling molecules to provide a measure of the network state—and thereby demarcating multiple cell state functions such as apoptosis, DNA damage and cell cycle. By measuring all these parameters simultaneously, the signaling state of an individual cell can be measured at its network state. This review will cover the basics of mass cytometry as well as outline steps already taken to allow it to stand aside traditional fluorescence based cytometry in the immunologist’s analytical arsenal in their study of immune states during infection.
Keywords: Mass cytometry, flow cytometry, drug discovery, pathogens, animal models
Introduction
Fluorescence-based flow cytometry has proven an invaluable technology for both immunologists and clinicians alike [1]. Importantly, it provides critical biological information at the single-cell level regarding ploidy, immunophenotype, frequency of cell subsets, expression levels of proteins, as well as functional characterization [2-7]. Furthermore, the potential of this technology can be significantly extended by interrogating single cells not only in their basal state but also after their exposure to exogenous stimuli. The latter has given rise to fluorescence-based phospho-flow cytometry, which has enabled determination of the activity of intracellular pathways [8-17]. Interrogated revelation of cellular states is key to the mechanistic understanding of the diseased cell, immune systems perturbed during infection, and to elucidating the positive or negative effects on these pathways wherein cells have been exposed to therapeutic and potential therapeutic agents in vitro or in vivo [14, 18-20].
However, as powerful as fluorescence-based flow cytometry can be, it falls somewhat short of uncovering the well-recognized complexity of the immune system when determined simultaneously with intracellular network states. The primary drawback of traditional fluorescence-based flow cytometry is ironically the same tool that has enabled it to be so useful for nearly three3 decades. In fluorescence-based flow cytometry, the number of markers that can be simultaneously analyzed is inherently limited by fluorescence spectral overlap— that measurement beyond three fluorophores becomes more complex with increasing parameters added, involving corrections for spectral overlap as well as experimental appreciation for the cellular autofluorescence of certain cell types [21, 22]. Even with such corrections & understanding, the practical multiplexing limit of flow cytometry is about ten markers wherein significant training or effort is involved in designing panels with multiple markers. Investigation of intracellular pathways under such restrictions is unwieldy (if not impossible), since the bulk of the parameters for a fluorescence based analysis will be assigned to surface markers that call out specific cell types, leaving only a few channel to measure phosphorylation states or levels of intracellular proteins. So what do we go from here?
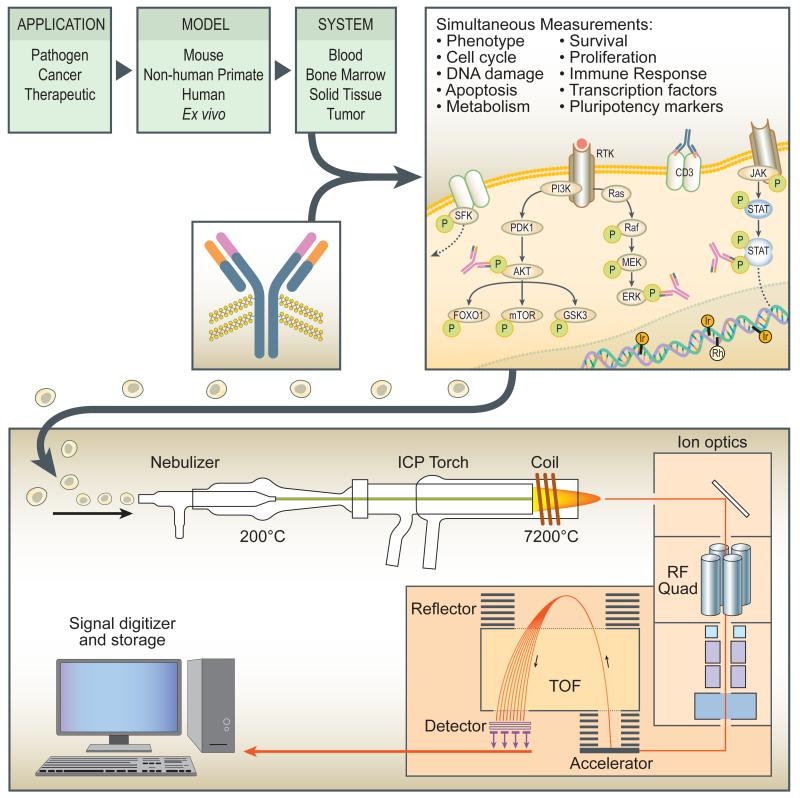
A new generation of single-cell analysis technology called mass cytometry overcomes most of these limitations (Figure 1). The CyTOF (CYtometry Time Of Flight) is a newly developed single cell mass spectrometry-flow cytometry hybrid instrument that replaces fluorophores with stable elemental mass reporters [23-28]. The increased signal resolution (Figure 1) of this new platform offers, at the least, a 10-fold improvement in the number of simultaneous parameters that can be readily measured. Mass cytometry offers a number of significant advantages compared to fluorescence-based applications. It is quantitatively accurate with detection at the single cell level being linear across 4 orders of magnitude where unlike fluorescence the difference in sensitivity between the best and worst reporters is only ~2-fold (1). Further advances such as increased numbers of deployable isotopes, novel nano-crystal configurations, and computational tools promise to extend cytometry well into the “omics” arena and eventually provide system-wide views of immune function in healthy and diseased patients in infectious disease and other inflammatory states that lead to morbidity and mortality.
Figure 1. Mass cytometry enables high-dimensional analysis of diseases and therapies.
Diseases including cancers and infections perturb cellular signaling. Mass cytometry provides a readout for up to 52 simultaneous measurements, both of those disease-induced perturbations and importantly of counter-perturbations induced by candidate therapeutics. Furthermore, the simultaneous inclusion of cell phenotype and cell cycle provide a more detailed picture than possible before. After cells are stained with antibodies and other metallic assay reagents, they are introduced into the mass cytometer as a stream of single cells, then atomized and ionized as they pass through the inductively coupled plasma (ICP) torch. Low-mass elements (including carbon and nitrogen) are filtered out by a radio frequency quadrupole before entering the time-of-flight (TOF) detector. A high-speed, online analysis system produces data equivalent to that of traditional fluorescence-based cytometry.
Basic Concepts of Mass Cytometry
To address the limitation of traditional fluorescence based cytometry, namely the simultaneously measured parameter ceiling, Scott Tanner and colleagues at the University of Toronto embarked upon a remarkable adaptation of inductively coupled plasma mass spectrometry (ICP-MS). ICP-MS is routinely used in the mining, metallurgy and semi-conductor industries and is the method of choice for measuring the elemental composition of materials since it can detect the contamination of, for example, blood with lead and drinking water with arsenic, beryllium or heavy metals. In ICP-MS, samples are atomized and ionized in plasma at temperatures approximating that of the surface of the sun (7500K). The mass spectrometer can then resolve and quantify elemental components, based on mass-to-charge ratio (m/z), with a level of sensitivity of parts per quadrillion. Key features of traditional ICP-MS instrumentation that make it a prime technology for adaptation to cytometry include the absence of interference between elemental masses and a linear dynamic range of greater than 108.
Tanner and colleagues realized that incorporating such attributes into flow cytometry might dramatically increase the number of parameters that could be measured per single cell. They reasoned that, rather than being conjugated to fluorophores, antibodies could be conjugated to stable metal isotopes, such as lanthanides, that are absent or at low abundance in biological systems, and then adapt ICP-MS instrumentation for their detection [23, 28-33]. This was the foundational concept upon which the mass cytometer was developed.
By tagging each antibody with a unique lanthanide isotope, the readout from each isotope can be correlated with a particular antibody, which in turn can be correlated to levels of antigen associated with an individual cell. Thus, the number of simultaneously measurable parameters per cell is now only limited by the number of stable isotopes suitable for conjugating to antibody reagents. For this to be accomplished, two fundamental technical challenges needed to be overcome. One was to develop reagents to tag antibodies with stable metal isotopes. Another was to adapt the ICP mass spectrometer to simultaneously detect multiple isotope tags, in the form of an ion cloud, associated with a single cell event.
In many respects the workflow or pipeline of mass cytometry is analogous to traditional flow cytometry (see Figure 1). By taking advantage of a long history of fluorescence based innovations, it has been possible to “recreate” the utility of many reagents with isotope labelled tags. This and extension specific to mass cytometry will be outlined below.
Attaching Metal-Chelating Polymers to Antibodies
For the attachment of multiple atoms of a given isotope to a selected antibody, acrylic acid polymers were synthesized with a uniform number polymer units and functionalized with multiple copies of a chelator, such as 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid or diethylene triamine pentaacetic acid (DTPA), compatible with the chemistry of trivalent metal lanthanide ions [34]. The resultant chelated lanthanide has a Kd of 10−16 and is therefore nearly impervious to losses or exchanges between other chelated metals within an antibody panel. A terminal maleimide group on the polymer permits its conjugation to selectively reduced disulfide groups in the hinge region of the immunoglobulin heavy chain.
Typically two to four polymers bind to each antibody, with each polymer chain capable of carrying up to 30 metal isotopes [32, 34, 35]. The first and current generation of isotope-chelating polymers can bear up to 120 lanthanide ions per antibody molecule [34]. That, combined with the level of ion transmission at 1 in 10,000, means the lower limit of detection for any given cellular parameter is about 300-1000 target protein copies (comparable to many fluorophores by traditional fluorescence). Unlike photomultiplier tubes in photon-driven cytometry which can show non-linear sensitivity across the dynamic range, the sensitivity of the ICP-MS readout is linearly proportional to the number of elemental isotopes conjugated to each antibody. Nevertheless, the sensitivity of lanthanide-conjugated antibodies is currently about two-fold lower than the brightest fluorophores such as phycoerythrin.
As will be discussed below, the lanthanides dominate as the group of metals compatible with current polymer tag chemistries. However, in order to expand the number of simultaneously measureable parameters, new chelation chemistries are under development with the aim of including the noble metals [36]. Their atomic size falls within the range suitable for mass cytometry and could thus significantly increase the panel of metal-tagged antibodies available for single-cell analysis.
Assigning Stable Metal Isotopes to Measurements of Cellular Parameters
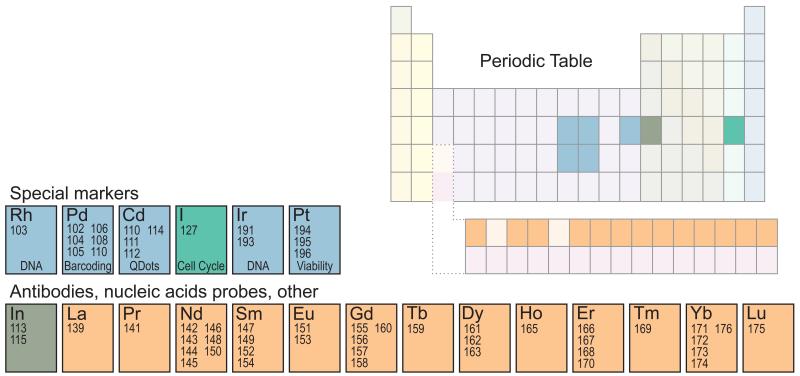
At present, a total of 37 purified, stable lanthanide isotopes are available and compatible with the metal chelating polymer chemistry (Figure 2). Of those, 27 are available at enrichment purities above 97 percent; the rest are available at enrichment purities above 92 percent. Care must be taken when assigning antibodies to isotopes to ensure that impurities in other channels do not result in false-positives during analysis. Additionally, certain metals are prone to oxidation, which results in signal in other channels. For example, measurements of gadolinium 157 are prone to interference from +16 oxidation of praseodymium 141. In addition to the lanthanides, indium, a post-transition metal, has two isotopes that are compatible with the chelating chemistry, but their sensitivity is low and thus only suitable for detection of highly abundant proteins, such as CD45 expressed on leukocytes. One additional parameter can be measured with quantum dots (Q-dots) using the cadmium as a major constituent of Q-dots.
Figure 2. A large number of metals are available for a variety of measurements.
The lanthanides provide 37 stable isotopes for measuring antigen-bound antibodies, MHC tetramers and nucleic acid probes with high sensitivity. Indium provides two additional low-sensitivity channels for highly expressed markers, and quantum dots provide one additional channel (cadmium). Rhodium and iridium, as DNA intercalators, register a cell event. Platinum, in the form of cisplatin:sulfur complexes, is used as a viability marker. Iodine, as iodo-deoxyuridine, is used for cell cycle measurements. Six palladium isotopes allow for mass tag “barcoding” and multiplexed measurement of samples.
Given the absence of light scatter properties to record a cell event, measurement of rhodium or iridium DNA intercalators, as well as a cell event-induced ion cloud duration measurement (“cell length”) can be used to demarcate cells in terms of their DNA content and approximate size, respectively [27, 32]. A variety of other measurements (viability, cell cycle and multiplexing bar-code reagents) are discussed below, bringing the total number of quantifiable parameters for a single cell to 52. It might be possible someday to place a standard forward and side scatter device prior to the ICP-MS plasma, but there remain problems associating the scatter information to the post-plasma cell events. This is currently due to the fact that while every cell that passes a light detector is measured, not every cell makes the passage through the plasma as a uniform ion cloud. Thus, deciding which forward/side scatter event relates to which cloud of ions will require, we believe some orthogonal determination of which cell via light scatter can be assigned to which set of ion events in the mass spectrometer.
Adapting ICP-MS to Measuring Single-Cell Ion Clouds
It was necessary to adapt the ICP-MS to retain the temporal information of a multi-ion cloud derived from a single cell [23, 29, 32]. As with conventional ICP-MS, liquid samples, but now containing a cell suspension, are nebulized into single-cell droplets, rapidly dried in a heated spray chamber and then delivered into the central channel of the 7500K argon plasma where they are vaporized, atomized and ionized to create clouds of ions that correspond to the cells. The ion cloud derived from a single cell has a measurement span of approximately 200 to 300 microseconds. Each data acquisition cycle for an ion cloud representing a cell is achieved over approximately a 200-microsecond time frame. Currently this limits the throughput of mass cytometry to 1000 cells per second when cells are passed by Poisson random input.
Of fundamental importance is the observation that fluorescence and mass cytometry yield very comparable results when analyzed by traditional 2D flow plots, histograms and heat-maps [24, 37]. Yet there are clear differences between the two platforms. Although the deep dive into a single cell at the level of greater than 35 parameters provides an unprecedented level of detail unavailable by fluorescence, the latter platform efficiently determines measures of, for example, cellular calcium and reactive oxygen species, for which as yet there are no mass cytometry equivalents. However, over the past year a number of new reagents have been created for mass cytometry that can now be incorporated into the “tool kit” as discussed below.
Data Normalization with Bead Standards
As with any quantitative technology, there is a stringent requirement for internal and external reagent standards for data normalization. In the case of mass cytometry, variation in instrument performance can be caused by factors such as instrument drift and build-up of cellular debris on the sample introduction components. In order to correct for these factors, polystyrene beads infused with precise amounts of several lanthanide isotopes are acquired simultaneously with samples for normalization of data over time. A sliding window correction derived from the bead signature is now routinely applied to the raw mass cytometry data before any further analysis takes place [38]. The software for its implementation is available at www.cytobank.org/nolanlab.
Increasing Throughput and Decreasing Variability by Mass Tag Barcoding
Amine-reactive fluorescent dyes, such as Pacific Blue, Alexa Fluor 488, Alexa Fluor 700 and Alexa Fluor 750 each attached to N-hydroxysuccinimidyl (NHS) ester, can be used in different combinations for fluorescence “barcoding” of separate samples (primary cells or cell lines) that are subsequently pooled, stained in a single tube with a fluorescently-tagged antibody panel and analyzed simultaneously on a flow cytometer. Data are then deconvoluted according to the fluorescent barcode signatures of the component samples [39]. There are three significant advantages to sample barcoding: i) all samples are stained in the same tube with the same antibody mix, eliminating cell-to-antibody ratio-dependent effects on staining, ii) reduced antibody consumption and iii) increased sample throughput.
These principles can also apply to barcoding reagents available for mass cytometry (mass cell barcoding (MCB)). In a recent study, metal barcode reagents were prepared by chelating lanthanides with a bifunctional macrocyclic compound, maleimido-mono-amide-DOTA (mDOTA), which labels cells by covalent attachment to intracellular thiol groups. Samples were labeled with a unique binary combination of seven mDOTA-lanthanide reagents, multiplexed and deconvoluted to accurately recover samples with given barcodes. This foundational study lends support to the use of metal barcoding reagents in mass cytometry [40]. Unlike fluorescence-based cytometric analysis, a fluidics purge lasting several minutes is required between sample introductions on the mass cytometer. The throughput gained by barcoding is thus particularly significant. As a refinement of the barcoding reagents used in the published study, the mDOTA-lanthanides have now been replaced with six palladium isotopes. These have masses (102 to 110) below that of the smallest lanthanide and therefore do not occupy channels that can otherwise be used for metal-tagged antibodies (E. Zunder, G. Behbehani, R. Finck, C. Thom, G.P. Nolan manuscript in preparation).
Cell Viability Determinations
A large set of fluorescence-based reagents exists with which to measure cell viability. They operate on the principle that a compromised cell with a damaged plasma membrane permits reagent entry into the cytoplasm, whereas a healthy cell with an intact plasma membrane does not. Specifically, reagents are available that: i) intercalate non-covalently into DNA (for example 7-Aminoactinomycin D (7-AAD) or propidium iodide) [41, 42], ii) covalently attach to DNA (TdT dUTP nick end labeling (TUNEL) [43] iii) covalently attach to proteins (Invitrogen Fixable LIVE/DEAD1) [44] iv) monitor alterations in mitochondrial membrane potential (differences in fluorescence of the monomeric and aggregate forms of 5,5′,6,6′-tetrachloro-1,1′,3,3′ tetraethylbenzimidazolylcarbocyanine iodide (JC-1)) [45]. Similarly, a variety of reagents is available for determination of cell viability by mass cytometry and operates under the same principles as fluorescent reagents, albeit with a different readout. These include rhodium and iridium-containing metal-intercalators [27] and an amine-reactive chelator, 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid mono (N-hydroxysuccinimide ester) (DOTA-NHS-ester) [24]. Recently, a protocol was described for using cisplatin to determine cell viability [46]. Although well known as a chemotherapeutic agent because of its ability to form extremely stable DNA-platinum adducts, cisplatin has an alternative activity in which it reacts on a much more rapid timescale (minutes as opposed to days) with protein thiols, forming covalent platinum: sulfur bonds. Furthermore, in an independent study, acrylamide polymers bearing platinum or palladium recognized non-viable cells [36]. Platinum has six stable isotopes, of which three are dominant (194, 195 and 196 Daltons) and well separated from the lanthanides, making cisplatin an ideal reagent for determinations of cellular viability by mass cytometry.
Measuring the Cell Cycle by Mass Cytometry
No biological evaluation would be complete without including measurements of cell cycle phase. An abundance of fluorophore-based reagents have been used for decades to stage cells based on their DNA content using traditional flow cytometry. Included in the list are supra-vital stains in the Hoechst group, such as 4′, 6-diamidino-2-phenylindole (DAPI) that bind to A-T-rich regions within the minor groove of DNA, or membrane-impermeable reagents such as propidium iodide, bromodeoxyuridine [3, 47]. These fluorescent stains can all be used with a limited number of antibodies that characterize a specific cell cycle phase.
In a recent study, Behbehani et al. designed a panel of metal-chelated antibodies with which to perform a comprehensive analysis of the cell cycle progression machinery [37]. The panel included antibodies against cyclins, phosphorylated retinoblastoma (Rb), phosphorylated Cdk1, and phosphorylated histone H3 and Ki67 to denote cells in cycle covering the G1, G2 and M phases. Of its many roles, retinoblastoma is pivotal for cell cycle progression, with a complex mechanism defining its role in G1 to S progression [48, 49]. However, the antibody used here was against an epitope encompassing residues pS807 and pS811, which are substrates for cyclin C/Cdk3 and are necessary for quiescent cells to enter into cycle [50]. To identify cells in S phase, multiple studies have used halopyrimidines (bromo-, iodo- and chloro-deoxyuridine) which become incorporated into newly synthesized DNA. Antibodies recognizing these groups necessarily require a DNA denaturation step to gain access to the modified site [51, 52]. However, iododexoyuridine, without an accompanying antibody, can be measured directly by mass cytometry: the incorporated iodine in the newly synthesized DNA has an atomic mass of 127, which falls within the requisite range for mass cytometry. Its inclusion in the panel gave a direct and clear measure of the percentage of S-phase cells which was also beneficial in increasing the resolution between the G1 and G2 phases.
Using a variety of cancer cell lines, cycling T lymphocytes and primary human bone marrow, Behbehani et al. identified all cell cycle phases with a core cell cycle panel: p-Rb (pS807/S811), IdU, cyclin B and p-Histone H3 (pS28). Importantly, the cell cycle phases were validated in side-by-side measurements by fluorescence flow cytometry [37]. The importance of this study lies in the ability to now measure many other biological parameters within the context of the cell cycle. For example, signaling, DNA damage, and metabolic pathways can now be examined at defined phases in the cell cycle (G.K Behbehani, W.J. Fantl, G.P. Nolan, S. Lowe, P. Mallik unpublished). In the area of infectious disease, vaccinia, influenza and hepatitis C virus infections are all known to alter cell cycle progression [53-56]. Conversely the host cell cycle affects the replication of viruses such as Ebola virus, in which case the virus depends on actively proliferating host cells for replication itself [57]. This core marker set would be equally significant in the development of therapeutic agents, many of whose activities are known to be influenced by cell cycle state [58-62].
Measurements of Cytokines: Regulators of Immune Cell Subsets and Beyond
Traditional flow cytometry has greatly benefited from single cell measures of cytokine activity. The cytokine superfamily includes interleukins, chemokines, colony-stimulating factors, interferons, as well as the transforming growth factor and tumor necrosis factor families, all with a large array of diverse biological functions. They have well-described functions in innate immunity (inflammation, chemotaxis, allergy, macrophage and NK cell activation) as well as in adaptive immunity (cellular and humoral) [63-70]. However, cytokines are now known to be produced by and mediate their effects on cells other than immune cells and have been implicated in the pathologies of, for example, cancer, stroke and pulmonary arterial hypertension [71-73].
Thus, given their far-reaching effects in multiple tissues, defining cellular phenotypes based on their cytokine expression is another essential parameter to include in the mass cytometry toolkit. Fluorescence-based flow cytometry protocols have measured cytokine production in a variety of T cell subsets [74-77]. Recently, mass cytometry applied to CD8+ T cells after stimulation with PMA/ionomycin has remarkably revealed there to be about 200 distinguishable subtypes based on the combinatorial diversity of the nine functional attributes, with even greater diversity revealed when taking into account different expression patterns of surface markers [78]. In the same study, a large range of distinct patterns of functional diversity were seen when examining CD8+ T cell subsets responsive to different viral peptide-major histocompatibility complex (MHC) complexes [78]. This study has set the stage for measuring the functional diversity of both immune and non-immune cell subsets.
Major Histocompatibility Class-Peptide Tetramers Conjugated to Metal Chelating Polymers
Antigen-specific T cell subsets are generated when their T cell receptors (TCR) interact with pathogen-derived peptide-major histocompatibility complexes expressed on antigen presenting cells [79]. At a given moment, there will be numerous T cell subsets throughout the body with different antigen specificities. However, their low frequency and low affinity interaction of their receptor with peptide-bound MHC precluded a detailed characterization of their properties. To circumvent this problem, Altman and Davis constructed a peptide-MHC tetramer in which four identical biotinylated MHC-peptide molecules were complexed with streptavidin conjugated to a fluorophore, resulting in increased avidity [80]. The peptide-MHC complex will only bind to the specific T-cells that respond to that peptide. The tetramer can then be detected by flow cytometry via fluorescent label [80, 81]. Recently, the fluorophore on streptavidin was replaced by a metal chelating polymer allowing a multidimensional analysis to be performed by mass cytometry with a panel of cytokine antibodies as described above. In this way it was possible to identify 56 to 106 combinations of functional attributes for several viral-specific T cell subsets, revealing a far more complex view of the cytokine network that seen before [78].
Building Panels of Thirty-Something Antibodies for Deep Proteomic Profiling
One undisputed advantage of single-cell mass cytometry is the ability to measure multiple parameters on a single-cell basis, without the need to compare smaller panels or computationally join data files from separate smaller antibody panels [5, 7]. As mentioned above, most isotopes are assigned to antibodies, as they are at the crux of the mass cytometry “tool kit.” As with any antibody, the conditions for their use must be optimized. The key steps are conjugation to the metal-containing polymer and performing the appropriate titrations to measure signal to noise ratios (also referred to as “stain index”).
Current antibody panels are able to deeply profile both phenotypic and intracellular signaling attributes of single cells. Panels can be designed with about 40 antibodies focused completely on surface markers to delineate cellular hierarchy, or a combination of surface markers and intracellular signaling molecules. The latter are focused on the activation states of intracellular signaling pathways. This approach has provided new information about established cell types, as well as previously unidentified cell types revealed by new combinations of surface markers. In addition to providing an increased understanding of the immune system (see discussion below), mass cytometry can also provide new information about solid tumors. Using appropriate protocols to dissociate tumors [82, 83] into their constituent single cells, new cellular hierarchies have been revealed in ovarian cancer (J. Stewart, B. Neel, B. Bodenmiller, W. Fantl and G. Nolan unpublished). This increased level of detail regarding signaling potential at the single-cell level, regardless of the tissue of derivation, provides a new backdrop for drug discovery, discussed below.
Deep Proteomic Profiling of the Human Immune System
The first deep proteomic study evaluated signaling responses in specific immune cell subsets within the hematopoietic continuum [24]. Two panels each comprising 31 metal polymer-conjugated antibodies were assembled. The first “phenotypic panel” was designed to measure surface molecules expressed on immune cell subsets, and successfully identified known and distinct immune cell subsets, including B, T, NK-cells and monocytes. However, transitional cells were also seen, not previously captured in prior studies but that are consistent with an immune continuum rather than abrupt conversions to distinct differentiation stages. The second panel maintained 13 surface marker antibodies from the first panel but had an additional 18 intracellular signaling molecules representing the activation status for a number of pathways. Human bone marrow was treated with twenty extracellular modulators including growth factors, cytokines, chemokines and three small molecule kinase inhibitors (dasatinib, Jak1 inhibitor and the U0126 Mek inhibitor). Thus, Ras/Raf/Erk, NF-kB, p38/MAPKAPK2, STATs 1, 3 and 5, CREB and BCR signaling were all included and measured simultaneously [24]. Using spanning tree progression analysis of density-normalized events (SPADE, discussed in more depth below) signaling responses were seen within tight population boundaries as well as across multiple immune cell types. This systems-level view is the first in a series of studies to generate a human immune reference map as a resource for therapeutic and vaccine studies (Unpublished Z. Bjornson, G. Fragiadakis, M. Spitzer, M. Davis, G.P. Nolan).
This foundational study also demonstrated a paradigm for multi-dimensional analysis of complex primary tissues, namely establishing a phenotypic hierarchy using surface markers and then selecting a subset of surface marker antibodies to combine with antibodies that measure activated intracellular signaling molecules. Since then, many additional antibody panels have been optimized to interrogate a broad variety of cellular functions including: EGFR signaling, epithelial-mesenchymal transition, the Wnt pathway, apoptosis, survival, proliferation, DNA damage response, cell cycle, metabolism, embryonic stem cells and induced pluripotent stem cells. As will be discussed below, the value of this technology platform is in its ability to measure multiple cellular functions, which will be invaluable for drug development programs. However, it is first necessary to have the tools to analyze high dimensional data.
Analyzing High-Dimensional Mass Cytometry Data
Although mass and fluorescence-based flow cytometry use entirely different instrumentation, the data from both platforms provide equivalent information [24, 37]). However, there are several notable differences in the data. In fluorescence cytometry, significant background signals arise from spectral overlap and auto-fluorescence, the natural fluorescence of cellular structures. In mass cytometry, because “auto-mass” does not exist, there is minimal background and consequently less spread around zero. Nonetheless, in both cases, a transformation such as the inverse hyperbolic sine function is typically applied to compress values around zero, resulting in a more coherent negative population (one that lacks a marker of interest). However, the standard transformation used with mass cytometry data does not compress the data as strongly as standard transformations for fluorescence data, resulting in data that is truer to the measured signal (Figure S2 in [24]).
We and others have adapted a variety of algorithms to the analysis of high-dimensional, single-cell mass cytometry data. One obvious choice for processing large datasets is automated clustering, the grouping of similar cells, which has been applied extensively to microarray data [84, 85]. One of the first algorithms developed to analyze mass cytometry data was spanning tree progression analysis of density-normalized events (SPADE) [24, 86, 87]. It uses hierarchical, agglomerative clustering after performing density-dependent downsampling in an effort to preserve rare cell types that would otherwise be drowned out by far more frequent cell types. The resulting clusters can be placed into a minimum-spanning tree [24], or into a more highly connected graph with a force-directed layout (E. Zunder and G.P. Nolan, manuscript in preparation).
Automatic determination of known, biologically relevant clusters is still a difficult problem in flow cytometry data analysis because it is difficult to determine the edge of where one cell population begins in the other ends. This is an especially difficult problem when one considers transitions and progressions – were cells dynamically exist along a framework of cell states. On one end, under-segmentation results in clusters containing multiple cell types whereas on the other end, over-segmentation needlessly divides homogenous cell types. In a new approach, over-segmentation of cell subsets and their subsequent merging by affinity propagation resulted in larger, biologically relevant cell subpopulations (Frey, Dueck et al. 2007; Chen, Clutter et al., in preparation). This technique works especially well for analyzing continuous progressions of cells, such as the cell cycle, where manual demarcation of cell subsets is difficult.
Summary and Conclusions
Reiterating a central theme of this essay we have discussed how mass cytometry can be applied to generate single-cell network reference maps for humans and model organisms such as non-human primate and murine immune systems. While it is clear that a far greater understanding of animal models than presently exists is urgently needed, they can never act as perfect surrogates for humans. Although mass cytometry, as it stands with panels of 45 parameters in routine use, provides a level of detail about protein function not previously possible, further improvements are needed. These include increased sensitivity of antibody tags, changes in the instrumentation to increase sample flow rate and reduce sample loss as well as new computational tools. Regardless, multi-dimensional, single-cell mass cytometry is currently positioned to have dramatic consequences on drug development and therapeutic programs for multiple indications ranging from pathogen infection, vaccine development, cancer and potentially will be extendable to other inflammatory conditions such heart failure, stroke, diabetes and obesity, and surgical trauma.
Research Highlights.
40 quantitative protein or mRNA measures per cell enabled by mass cytometry
Isotope based cell labeling replaces fluorophores
Deep functional cell profiling and functional network states
Acknowledgements
The authors wish to thank Drs Scott Tanner, Olga Ornatsky, Dmitry Bandura, Mitch Winnik and Mark Nitz for their critical reading of this manuscript. This work was supported by the Rachford and Carlota A. Harris Endowed Chair to GPN as well as NIH grants PN2EY018228, 0158 G KB065, U19 AI057229, P01 CA034233-22A1, HHSN272200700038C, 1R01CA130826, RFA CA 09-011, RFA CA 09-009 and CIRM grant USC DR1-01477 to GPN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: G.P.N. has personal financial interest in the company DVS Sciences, the manufacturers of the instrument and reagents described in this manuscript.
References
Select recent references of special (*) or outstanding (**) interest, with an annotation that gives a brief description of the major findings and the importance of the study
- 1.Chattopadhyay PK, Roederer M. Cytometry: today’s technology and tomorrow’s horizons. Methods. 2012;57(3):251–8. doi: 10.1016/j.ymeth.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sigal A, et al. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat Protoc. 2007;2(6):1515–27. doi: 10.1038/nprot.2007.197. [DOI] [PubMed] [Google Scholar]
- 3.Darzynkiewicz Z. Critical aspects in analysis of cellular DNA content. Curr Protoc Cytom. 2011 doi: 10.1002/0471142956.cy0702s56. Chapter 7: p. Unit 7 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobberger JW, et al. A new biomarker for mitotic cells. Cytometry A. 2008;73(1):5–15. doi: 10.1002/cyto.a.20501. [DOI] [PubMed] [Google Scholar]
- 5.Biancotto A, et al. High dimensional flow cytometry for comprehensive leukocyte immunophenotyping (CLIP) in translational research. J Immunol Methods. 2011;363(2):245–61. doi: 10.1016/j.jim.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biancotto A, et al. OMIP-004: in-depth characterization of human T regulatory cells. Cytometry A. 2012;81(1):15–6. doi: 10.1002/cyto.a.21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Lochem EG, et al. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin Cytom. 2004;60(1):1–13. doi: 10.1002/cyto.b.20008. [DOI] [PubMed] [Google Scholar]
- 8.Krutzik PO, et al. Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol. 2004;110(3):206–21. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- * 9.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–28. doi: 10.1016/j.cell.2004.06.028. This report demonstrates that perturbation responses in individual cancer cells can be related leukemia patient clinical outcomes (Irish et al., Cell 2004). An advantage of this single cell approach was that signaling could be characterized in rare populations of cancer cells and contrasted with the bulk cancer cell population. This study revealed and characterized the cell signaling heterogeneity of AML and showed that signaling in individual cancer cells can be closely linked to the clinical behavior of the disease.
- 10.Irish JM, et al. Kinetics of B cell receptor signaling in human B cell subsets mapped by phosphospecific flow cytometry. J Immunol. 2006;177(3):1581–9. doi: 10.4049/jimmunol.177.3.1581. [DOI] [PubMed] [Google Scholar]
- 11.Irish JM, et al. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006;108(9):3135–42. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 12.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6(2):146–55. doi: 10.1038/nrc1804. In this paper we described the advantages of a single cell, high dimensional flow cytometry in translational cancer research. This review defines much of the new language in the field (e.g. ‘signaling nodes’ and ‘signaling profiles’) and highlights key challenges in cancer research that can now be addressed using ‘single-cell proteomics’ approaches like mass cytometry.
- * 13.Palazzo AL, et al. Association of reactive oxygen species-mediated signal transduction with in vitro apoptosis sensitivity in chronic lymphocytic leukemia B cells. PLoS ONE. 2011;6(10):e24592. doi: 10.1371/journal.pone.0024592. Multi-parametric flow cytometry of primary chronic B-cell leukemia samples identified distinct cell subpopulations, within and between samples. B-Cell receptor network proteins of these subpopulations showed varying intracellular signaling responses to ex vivo treatment with hydrogen peroxide, a reactive oxygen species which acts as an intracellular second messenger. A link was seen between the magnitudes of these responses and their apoptotic proficiency after ex vivo fludarabine exposure. Such single cell analysis has the potential to monitor the therapeutic benefit of this standard-of-care drug.
- * 14.Rosen DB, et al. Distinct patterns of DNA damage response and apoptosis correlate with Jak/Stat and PI3kinase response profiles in human acute myelogenous leukemia. PLoS ONE. 2010;5(8):e12405. doi: 10.1371/journal.pone.0012405. Multi-parameter flow cytometry of primary acute myeloid leukemia samples revealed multiple distinct cell subpopulations identifiable by their surface marker expression, cytokine and growth factor-mediated signaling pathway responses, as well as their response to DNA-damaging agents. The activation states of the JAK/STAT and PI3 kinase pathways were strongly associated with ex vivo and in vivo responsiveness to DNA damaging agents.
- 15.Hotson AN, et al. The T cell STAT signaling network is reprogrammed within hours of bacteremia via secondary signals. J Immunol. 2009;182(12):7558–68. doi: 10.4049/jimmunol.0803666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Gorman WE, et al. The initial phase of an immune response functions to activate regulatory T cells. J Immunol. 2009;183(1):332–9. doi: 10.4049/jimmunol.0900691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Gorman WE, et al. Alternate mechanisms of initial pattern recognition drive differential immune responses to related poxviruses. Cell Host Microbe. 2010;8(2):174–85. doi: 10.1016/j.chom.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krutzik PO, et al. High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol. 2008;4(2):132–42. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 19.Kornblau SM, et al. Dynamic single-cell network profiles in acute myelogenous leukemia are associated with patient response to standard induction therapy. Clin Cancer Res. 2010;16(14):3721–33. doi: 10.1158/1078-0432.CCR-10-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong FK, Chow S, Hedley D. Pharmacodynamic monitoring of BAY 43-9006 (Sorafenib) in phase I clinical trials involving solid tumor and AML/MDS patients, using flow cytometry to monitor activation of the ERK pathway in peripheral blood cells. Cytometry B Clin Cytom. 2006;70(3):107–14. doi: 10.1002/cyto.b.20092. [DOI] [PubMed] [Google Scholar]
- 21.Chattopadhyay PK, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12(8):972–7. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 22.Perfetto SP, et al. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat Protoc. 2012;7(12):2067–79. doi: 10.1038/nprot.2012.126. [DOI] [PubMed] [Google Scholar]
- 23.Baranov VI, et al. A sensitive and quantitative element-tagged immunoassay with ICPMS detection. Anal Chem. 2002;74(7):1629–36. doi: 10.1021/ac0110350. [DOI] [PubMed] [Google Scholar]
- ** 24.Bendall SC, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–96. doi: 10.1126/science.1198704. First large scale demonstration of developed reagents and analysis methods for a single-cell mass cytometry and leveraging of the mutual information encoded in these novel, high parameter single-cell datasets. Besides being the first practical demonstration of the technology, it also provided an analytical resource of regulatory signaling information in the human hematopoietic and immune system that continues to be utilized in subsequent investigations in a fashion akin to gene expression and genomic sequence repositories (www.cytobank.org/nolanlab).
- 25.Ornatsky O, et al. Multiple cellular antigen detection by ICP-MS. J Immunol Methods. 2006;308(1-2):68–76. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Ornatsky OI, et al. Development of analytical methods for multiplex bio-assay with inductively coupled plasma mass spectrometry. J Anal At Spectrom. 2008;23(4):463–469. doi: 10.1039/b710510j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ornatsky OI, et al. Study of cell antigens and intracellular DNA by identification of element-containing labels and metallointercalators using inductively coupled plasma mass spectrometry. Anal Chem. 2008;80(7):2539–47. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- 28.Razumienko E, et al. Element-tagged immunoassay with ICP-MS detection: evaluation and comparison to conventional immunoassays. J Immunol Methods. 2008;336(1):56–63. doi: 10.1016/j.jim.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bandura DR, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81(16):6813–22. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- * 30.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotechnol. 2012;30(7):639–47. doi: 10.1038/nbt.2283. One of the most pressing issues facing next-generation single cell analysis platforms is addressing the cancer heterogeneity and how it relates to the overall disease progression and outcome. These approaches vary widely, from imaging and mass spectrometry of expressed epitopes and molecules, to molecular and sequencing analysis of gene expression and genomic content. Bendall and Nolan compare and contrast a number of these approaches, providing real-world applications of how they have been used to decipher complex cellular systems.
- 31.Bendall SC, et al. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33(7):323–32. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ornatsky O, et al. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361(1-2):1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Tanner SD, et al. Flow cytometer with mass spectrometer detection for massively multiplexed single-cell biomarker assay. Pure Appl Chem. 2008;80(12):2627–2641. [Google Scholar]
- 34.Lou X, et al. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed Engl. 2007;46(32):6111–4. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdelrahman AI, et al. Metal-Containing Polystyrene Beads as Standards for Mass Cytometry. J Anal At Spectrom. 2010;25(3):260–268. doi: 10.1039/b921770c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majonis D, et al. Curious results with palladium- and platinum-carrying polymers in mass cytometry bioassays and an unexpected application as a dead cell stain. Biomacromolecules. 2011;12(11):3997–4010. doi: 10.1021/bm201011t. [DOI] [PubMed] [Google Scholar]
- ** 37.Behbehani GK, et al. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A. 2012;81(7):552–66. doi: 10.1002/cyto.a.22075. A methodology to allow measurement of all phases of the cell cycle by mass cytometry. This methodology (which utilizes the mass cytometer’s ability to directly detect Iodo-deoxyuridine) was extensive compared to standard fluorescent approaches and yields equivalent results across a range of cell lines and primary cell types. More importantly, mass cytometric cell cycle analysis allows for the simultaneous measurement of up to 30-35 additional parameters, permitting the measurement of cell cycle state in complex samples or in conjunctions with multiple other measurements of cellular function. As a proof of principle, the authors simultaneously measured the cell cycle state of 25 different immunophenotypic populations of normal human bone marrow.
- * 38.Finck R, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013 doi: 10.1002/cyto.a.22271. Bead-based normalization of mass cytometry data uses the signal intensities of metal-embedded beads to account for the effects of instrument variation, and thus enables a more accurate interpretation of the biological differences between samples measured on the mass cytometer. The method, which is implemented on freely available software, applies a multiplicative correction derived from slopes fitted between smoothed bead signals and their global averages. This expands the types of analyses available using mass cytometry by allowing comparisons to be made across data acquired over periods of weeks or longer.
- 39.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3(5):361–8. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- ** 40.Bodenmiller B, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30(9):858–67. doi: 10.1038/nbt.2317. This publication describes a mass-tag based, cellular multiplexing approach (MCB) for mass cytometry to increase measurement throughput and reduce experimental variation. MCB was applied to characterize inhibitor impact on human peripheral blood mononuclear cell signaling networks under 96 conditions, allowing to classify inhibitor and cell type selectivity. This study highlights that high-content, high-throughput screening with MCB might be useful for drug discovery, preclinical testing and mechanistic investigation of human disease.
- 41.Moore A, et al. Simultaneous measurement of cell cycle and apoptotic cell death. Methods Cell Biol. 1998;57:265–78. doi: 10.1016/s0091-679x(08)61584-8. [DOI] [PubMed] [Google Scholar]
- 42.Schmid I, et al. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13(2):204–8. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 43.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. The Journal of cell biology. 1992;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perfetto SP, et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313(1-2):199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Cossarizza A, et al. A new method for the cytofluorimetric analysis of mitochondrial membrane potential using the J-aggregate forming lipophilic cation 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1) Biochem Biophys Res Commun. 1993;197(1):40–5. doi: 10.1006/bbrc.1993.2438. [DOI] [PubMed] [Google Scholar]
- * 46.Fienberg HG, et al. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 2012;81(6):467–75. doi: 10.1002/cyto.a.22067. Cisplatin is the first viability dye that is compatible with the harsh fixation, permeabilization and wash steps that are most often used in analysis of intracellular proteins by mass cytometry (Fienberg et al., 2012). Since platinum isotopes are not routinely conjugated to antibodies for mass cytometry, this viability stain does not entail loss of a protein measurement channel. Furthermore, although cisplatin is a known DNA-damaging agent, cisplatin staining does not induce DNA damage or apoptosis if used in the “pulse” application that optimally discriminates viable from non-viable cells.
- 47.Darzynkiewicz Z, Crissman H, Jacobberger JW. Cytometry of the cell cycle: cycling through history. Cytometry A. 2004;58(1):21–32. doi: 10.1002/cyto.a.20003. [DOI] [PubMed] [Google Scholar]
- 48.Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012;26(13):1409–20. doi: 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi C, Sasaki N, Kitajima S. Twists in views on RB functions in cellular signaling, metabolism and stem cells. Cancer Sci. 2012;103(7):1182–8. doi: 10.1111/j.1349-7006.2012.02284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren S, Rollins BJ. Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell. 2004;117(2):239–51. doi: 10.1016/s0092-8674(04)00300-9. [DOI] [PubMed] [Google Scholar]
- 51.Burns KA, Kuan CY. Low doses of bromo- and iododeoxyuridine produce near-saturation labeling of adult proliferative populations in the dentate gyrus. Eur J Neurosci. 2005;21(3):803–7. doi: 10.1111/j.1460-9568.2005.03907.x. [DOI] [PubMed] [Google Scholar]
- 52.Svetlova M, et al. Differential incorporation of halogenated deoxyuridines during UV-induced DNA repair synthesis in human cells. DNA Repair (Amst) 2005;4(3):359–66. doi: 10.1016/j.dnarep.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Wali A, Strayer DS. Infection with vaccinia virus alters regulation of cell cycle progression. DNA Cell Biol. 1999;18(11):837–43. doi: 10.1089/104454999314836. [DOI] [PubMed] [Google Scholar]
- 54.Yoo NK, et al. Vaccinia virus-mediated cell cycle alteration involves inactivation of tumour suppressors associated with Brf1 and TBP. Cell Microbiol. 2008;10(3):583–92. doi: 10.1111/j.1462-5822.2007.01047.x. [DOI] [PubMed] [Google Scholar]
- 55.Jiang W, et al. Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J Virol. 2013 doi: 10.1128/JVI.03176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kannan RP, et al. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol. 2011;85(16):7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kota KP, et al. High content image based analysis identifies cell cycle inhibitors as regulators of Ebola virus infection. Viruses. 2012;4(10):1865–77. doi: 10.3390/v4101865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malumbres M. Cell cycle-based therapies move forward. Cancer Cell. 2012;22(4):419–20. doi: 10.1016/j.ccr.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 59.Dent P, et al. CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Interv. 2011;11(2):133–40. doi: 10.1124/mi.11.2.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang YW, Hunter T, Abraham RT. Turning the replication checkpoint on and off. Cell Cycle. 2006;5(2):125–8. doi: 10.4161/cc.5.2.2308. [DOI] [PubMed] [Google Scholar]
- 61.Malumbres M, Barbacid M. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer. 2001;1(3):222–31. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 62.Ewald B, Sampath D, Plunkett W. H2AX phosphorylation marks gemcitabine-induced stalled replication forks and their collapse upon S-phase checkpoint abrogation. Mol Cancer Ther. 2007;6(4):1239–48. doi: 10.1158/1535-7163.MCT-06-0633. [DOI] [PubMed] [Google Scholar]
- 63.Melo RC, et al. Eosinophil-derived cytokines in health and disease: unraveling novel mechanisms of selective secretion. Allergy. 2013;68(3):274–84. doi: 10.1111/all.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38(1):13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140(6):845–58. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 67.Cox MA, Kahan SM, Zajac AJ. Anti-viral CD8 T cells and the cytokines that they love. Virology. 2013;435(1):157–69. doi: 10.1016/j.virol.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur J Immunol. 2009;39(8):2076–82. doi: 10.1002/eji.200939722. [DOI] [PubMed] [Google Scholar]
- 69.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol. 2011;11(10):645–57. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takata H, Naruto T, Takiguchi M. Functional heterogeneity of human effector CD8+ T cells. Blood. 2012;119(6):1390–8. doi: 10.1182/blood-2011-03-343251. [DOI] [PubMed] [Google Scholar]
- 71.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339(6117):286–91. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shichita T, et al. Novel therapeutic strategies targeting innate immune responses and early inflammation after stroke. J Neurochem. 2012;123(Suppl 2):29–38. doi: 10.1111/j.1471-4159.2012.07941.x. [DOI] [PubMed] [Google Scholar]
- 73.Hassoun PM, et al. Inflammation, growth factors, and pulmonary vascular remodeling. J Am Coll Cardiol. 2009;54(1 Suppl):S10–9. doi: 10.1016/j.jacc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 74.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 75.Donaldson MM, et al. Optimization and qualification of an 8-color intracellular cytokine staining assay for quantifying T cell responses in rhesus macaques for pre-clinical vaccine studies. J Immunol Methods. 2012;386(1-2):10–21. doi: 10.1016/j.jim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamoreaux L, Roederer M, Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 2006;1(3):1507–16. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 77.Lovelace P, Maecker HT. Multiparameter intracellular cytokine staining. Methods Mol Biol. 2011;699:165–78. doi: 10.1007/978-1-61737-950-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ** 78.Newell EW, et al. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–52. doi: 10.1016/j.immuni.2012.01.002. This paper includes a description of the development and use of peptide-MHC tetramer staining in conjunction with mass cytometry to identify and profile antigen-specific T cells with a large number of phenotypic and functional markers. Computational methods were applied that provide a new view of the functional and phenotypic diversity of the CD8+ T cell compartment. This analysis shows that CD8+ T cells from normal human donors display a broad continuum phenotypic profiles with remarkable diversity in their abilities to produce various cytokines.
- 79.Blum JS, Wearsch PA, Cresswell P. Pathways of Antigen Processing. Annu Rev Immunol. 2013 doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–6. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 81.Davis MM, Altman JD, Newell EW. Interrogating the repertoire: broadening the scope of peptide-MHC multimer analysis. Nat Rev Immunol. 2011;11(8):551–8. doi: 10.1038/nri3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panchision DM, et al. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells. 2007;25(6):1560–70. doi: 10.1634/stemcells.2006-0260. [DOI] [PubMed] [Google Scholar]
- 83.Chang Q, Hedley D. Emerging applications of flow cytometry in solid tumor biology. Methods. 2012;57(3):359–67. doi: 10.1016/j.ymeth.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 84.Do JH, Choi DK. Clustering approaches to identifying gene expression patterns from DNA microarray data. Mol Cells. 2008;25(2):279–88. [PubMed] [Google Scholar]
- 85.Nugent R, Meila M. An overview of clustering applied to molecular biology. Methods Mol Biol. 2010;620:369–404. doi: 10.1007/978-1-60761-580-4_12. [DOI] [PubMed] [Google Scholar]
- ** 86.Qiu P, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–91. doi: 10.1038/nbt.1991. SPADE is a cytometry visualization tool that uses a tree-like representation to convey the relatedness of cell phenotypes, including rare cell types. The paper shows how SPADE can be used to identify immune subsets based on non-canonical markers or to compare marker expression under different experimental conditions. SPADE was the first algorithm that was purpose-built for investigating mass cytometry data, and SPADE diagrams have appeared in several subsequent mass cytometry publications.
- 87.Linderman MD, et al. CytoSPADE: high-performance analysis and visualization of high-dimensional cytometry data. Bioinformatics. 2012;28(18):2400–1. doi: 10.1093/bioinformatics/bts425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26(3):303–4. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]