Abstract
Joints are essential for skeletal flexibly and form, yet the process underlying joint morphogenesis is poorly understood. Zebrafish caudal fins are comprised of numerous segmented bony fin rays, where growth occurs by the sequential addition of new segments and new joints. Here, we evaluate joint gene expression during fin regeneration. First, we identify three genes that influence joint formation, evx1, dlx5a, and mmp9. We place these genes in a common molecular pathway by evaluating both their expression patterns along the distal-proximal axis (i.e. where the youngest tissue is always the most distal), and by evaluating changes in gene expression following gene knockdown. Prior studies from our lab indicate that the gap junction protein Cx43 suppresses joint formation. Remarkably, changes in Cx43 activity alter the expression of joint markers. For example, the reduced levels of Cx43 in the sof b123 mutant causes short fin ray segments/premature joints. We also find that the expression of evx1-dlx5a-mmp9 is shifted distally in sof b123, consistent with premature expression of these genes. In contrast, increased Cx43 in the alf dty86 mutant leads to stochastic joint failure and stochastic loss of evx1 expression. Indeed, reducing the level of Cx43 in alf dty86 rescues both the evx1 expression and joint formation. These results suggest that Cx43 influences the pattern of joint formation by influencing the timing of evx1 expression.
Introduction
The precise location of joints provides both flexibility and form to the vertebrate skeleton. We use the zebrafish regenerating fin as a model to study skeletogenesis, including the appropriate formation of fin ray joints. Fin ray joints have been termed “fibrous joints” [1] since the articulation occurs in the bone matrix while the central mesenchyme remains continuous. These joints are distinct from synovial joints which completely articulate previously uninterrupted cartilaginous templates of the endochondral skeleton [2]. The fin grows rapidly during regeneration, fully restoring fin size and pattern. The fin is comprised of multiple bony fin rays or lepidotrichia, and each fin ray is comprised of multiple bony segments separated by fin ray joints (or simply, joints). Each fin ray consists of two hemirays of bone matrix surrounding the mesenchyme, and several layers of epidermal cells surrounding the bone matrix. Actinotrichia are bundles of collagen-like fibers that emanate from the distally located basal epidermal cells and serve as a substrate for osteoblasts to align and secrete bone matrix directly [3]. The mesenchyme medial to the actinotricha includes dividing cells, undifferentiated cells, blood vessels, nerves, and connective tissues [4,5]. The mesenchyme lateral to the actinotrichia includes the bone matrix, osteoblasts, and joint-forming cells [6,7]. Joint-forming cells are a subpopulation of lateral mesenchymal cells that condense on the surface of the uninterrupted bone matrix and form an elongated row of cells at the site of the presumptive joint. These cells later separate into two rows of cuboidal cells that flank a newly established articulation in the bone matrix [7]. Thus, these cells appear responsible for the articulation of the fin ray jointsWe refer to the osteoblasts and joint-forming cells collectively as skeletal precursor cells.
During growth and regeneration, the fin regenerates in the proximal to distal direction where new segments and new joints are continually added to the distal end of the fin ray. Thus, youngest tissue is always located more distally than mature tissue [8]. Following amputation, the regenerate undergoes three main stages: wound healing, blastema formation, and outgrowth [5,9]. The blastema is a specialized compartment comprised of rapidly proliferating cells, and is located in the distal and medial mesenchyme. These cells are the source of new tissue during regeneration. Recent studies show that several cell types in the regenerating fin are lineage restricted, meaning that new cells in the regenerating fin arise from precursor cells of the same cell type [10-13]. These cells undergo de-differentiation, cell proliferation, and re-differentiation, in order to replace lost tissue. This may not represent the only way to replace lost tissue, as others have found that osteoblasts are capable of de novo differentiation during fin regeneration [11]. Osteoblasts and joint-forming cells appear to be derived from a common lineage [13]. To date, little is known about the genes required for differentiation of joint-forming cells, or indeed, the signals required to initiate this process.
The transcription factor Even-skipped 1 (Evx1) belongs to a family of vertebrate eve-related homeobox genes [14]. In zebrafish regenerating fins, the expression of evx1 was observed strongly in the distal-most and youngest joints [1]. Sections of evx1 following in situ hybridization (ISH) showed a strong expression level of evx1 mRNA in the lateral compartment where skeletal precursor cells reside [1]. More recently, evx1 was shown to be required for joint formation since an evx1 mutant fails to produce fin ray joints during regeneration [15]. Our evaluation of two other fin mutants, short fin (sof b123) and another long fin (alf dty86), suggest that the gap junction protein Connexin43 (Cx43) also contributes to joint formation. Both cx43 mRNA and Cx43 protein are expressed throughout the medial mesenchyme, adjacent to the lateral populations of skeletal precursor cells [16]. The sof b123 mutant exhibits reduced levels of cx43 mRNA and protein (without a lesion in the coding sequence) that lead to reduced cell proliferation, short segments (i.e. premature joints) and short fin length [17]. In contrast, the alf dty86 mutant exhibits fin overgrowth and overlong segments on average due to stochastic joint failure [18]. The alf dty86 phenotype is not caused by mutations in cx43 but coincidently has increased levels of cx43 mRNA [7]. We have shown that morpholino-mediated cx43 knockdown in alf dty86 rescues joint formation, suggesting that the higher levels of cx43 in this mutant contributes to the loss of fin ray joints [7]. Thus, reduced cx43 leads to premature joints while increased cx43 leads to joint failure. We interpret these findings to indicate that Cx43 suppresses joint formation, perhaps by communication between the medial cx43-positive mesenchyme and the lateral evx1-positive mesenchyme.
As an initial attempt to understand the events initiating and controlling joint formation, we first wished to define additional molecular players acting downstream of evx1. Here, we describe the addition of two evx1-dependent joint gene markers that also contribute to joint formation: distal-less homeobox-5a (dlx5a) and matrix-metalloproteinase-9 (mmp9). We also exploited the characteristics of low and high Cx43 activity in sof b123 and alf dty86 to address the relationship between the expression of these joint genes and Cx43 activity during joint patterning. We found that the onset of joint gene expression correlates with the level Cx43 activity. These results suggest that Cx43 may regulate joint formation by influencing the timing of evx1 expression.
Materials and Methods
Statement on the ethical treatment of animals
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols used for this manuscript were approved by Lehigh’s Institutional Animal Care and Use Committee (IACUC) (protocol identification 128, approved 11/14/2012). Lehigh University’s Animal Welfare Assurance Number is A-3877-01. All experiments were performed to minimize pain and discomfort.
Fish maintenance
Zebrafish were derived from the C32 strain. Mutant fish used in these studies include sof b123 [19], alf dyt86 [18], homozygous evx1-/- mutant fish, and heterozygous carriers [15]. All fish were raised and cared for at constant temperature of 25°C in a 14 light: 10 dark photoperiod [20].
RNA probes and whole mount in situ hybridization (ISH)
Antisense evx1 probe was generated from 1µg of a PCR-generated linear template containing a T3 RNA polymerase binding site (F-TAATACGACTCACTATAG;
R-T3- GGATCCATTAACCCTCACTAAAGGGAAGAGCTATGACGTCGCAT where the T3 binding site is underlined). Antisense digoxigenin-labeled shh, lef1, mmp9, and dlx5a probes were generated from lef1 cDNA [21], shh cDNA [21], mmp9 cDNA [22], and dlx5a cDNA [22]. The template for the col10a1bprobe was a generous gift from the lab of Dr. David Parichy (representing sequence with genbank ID # 68437010).
Whole-mount ISH was performed on 5 dpa regenerating fins as described [23]. Stained fins were examined on a Nikon Eclipse 80i microscope. Images were collected using a digital Nikon camera. At least 4 regenerating fins were assessed at a time, and all markers were examined in triplicate.
ISH on cryo-sections
Fin regenerates (5 dpa) were harvested and fixed overnight with 4% paraformaldehyde in PBS. Fins were dehydrated in 100% methanol at -20°C. Next, fins were rehydrated in a methanol-PBS series of washes before embedding in 1.5% agarose/5% sucrose and equilibrated overnight in 30% sucrose. Blocks were mounted in OCT and cryosectioned (15 µm sections) using a Reichert–Jung 2800 Frigocut cryostat. Sections were collected on Superfrost Plus slides (Fisher) and allowed air dry overnight at room temperature. Slides may be stored in a freezer box at -20°C for up to one year. A marking pen (ImmEdge™ Pen H-4000; PAP pen, Vector Laboratories) was used to circle the sections. Probe was pre-hybridized with a mixture of 1X salt solution (NaCl, Tris HCl, Tris Base, Na2HPO4.7H20, NaH2PO4, and 0.5 M EDTA) with 50% deionized formamide (Sigma), 10% dextran sulfate, 1mg/mL tRNA, and 1X Denhart’s (Fisher) at 70°C for 5 mins. Hybridization with digoxigenin-labeled antisense probes was completed overnight at 65°C. The next day, slides underwent series of washes in a solution that has 1X SSC, 50% formamide and 0.1% Tween-20 at 65°C. Slides were then transferred to room temperature for extensive washes in MABT (100 mM Maleic acid, 150 mM NaCl, and 0.1% Tween-20) before incubation in blocking solution (MABT, Goat serum and 10% milk) for at least 2 hours or overnight. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5000 overnight at 4°C. After incubation, slides were washed in MABT four times followed by two short washes in staining buffer (100mM Tris, 9.5, 50 mM MgCl2, 100mM NaCl, and 0.1% Tween20). Slides were next transferred to 10% polyvinyl alcohol (PVA; MW: 86,000) staining solution plus NBT/BCIP stock solution (Roche) and development proceeded overnight at 37°C. Once observing purple staining on the sections, the reaction was stopped by washing the slides with PBST for at least 3 hours. Sections were mounted in 100% glycerol and examined on a Nikon Eclipse 80i microscope. Images were collected using a digital Nikon camera. A minimum of 10 sections were evaluated for each of a minimum of two different fins.
Morpholino-mediated gene knockdown and electroporation
Injection and electroporation experiments were performed as described previously [7,16,23,24]. Targeting morpholinos were designed against the start codon and modified with fluorescein at the 3’ end (Gene Tools, LLC) to provide a charge and for detection. Control morpholinos were either custom mismatch morpholinos containing five mismatches to the targeted gene or were the Gene Tools ‘standard control’ morpholino, which does not recognize any zebrafish genes. Following injection and electroporation, fins were harvested at 1day post electroporation (dpe) to evaluate changes in gene expression. At least 4 regenerating fins were treated per morpholino (targeting or control), and all knockdown experiments were completed in triplicate.Morpholino sequences for cx43 were described previously [16]. Morpholinos used here include: evx1-MO, CTTTCCGTGCTTCGGCGAGCCCATT; evx1-MM, CTTTGCCTGGTTCGGCCACCCCATT; mmp9-MO, AAACGCCAGGACTCCAAGTCTCATC; dlx5a-MO, CGAATACTCCAGTCATAGTTTGGAT (also used in [25]); Standard control MO, CCTCTTACCTCAGTTACAATTTTATA.
Measurements
Measurements of the distal boundaries of ISH expression domains to the distal end of the fin were taken from the third fin ray (V+3) since that was established as a standard [19]. Student’s t-tests were performed to determine if data sets were statistically different (p < 0.05). At least 8 fin rays per marker (evx1, dlx5a, mmp9, col10a1b, shh, and lef1) were measured.
Results and Discussion
dlx5a and mmp9 are expressed downstream of evx1
Since our studies suggest that Cx43 influences joint formation, we were interested in identifying additional genes that function together to regulate this process. Unlike osteoblast genes which are expressed in broad domains throughout the lateral compartment in the regenerating fin [8], genes expressed during joint formation tend to be expressed in a discrete group of cells. The expression pattern typically appears as a band of cells following whole mount in situ hybridization (ISH), and these cells are located within the lateral population of skeletal precursor cells (i.e. see 1). Thus, we identified evx1, dlx5a, mmp9, and col10a1b as candidate joint genes based on their location of expression [1,22]. We first confirmed the location of gene expression of this set of genes using whole mount ISH on 5 days-post-amputation (dpa) regenerating fins and by ISH on cryo-sectioned tissue of 5 dpa caudal fins. As expected, we found evx1, dlx5a, mmp9 and col10a1b are strongly expressed in a discrete group of cells in the lateral compartment where the skeletal precursor cells reside (Figure 1).
Figure 1. Expression of joint genes in regenerating fins.

(Left) Whole mount ISH shows evx1, dlx5a, mmp9, and col10a1b are expressed in 5 dpa wild type fins. (Right) ISH on wild-type 5 dpa cryosections reveal expression of joint genes in the lateral skeletal precursor cells. Arrows point to gene expression in the skeletal precursor compartment. (e) epithelium; (m) mesenchyme.
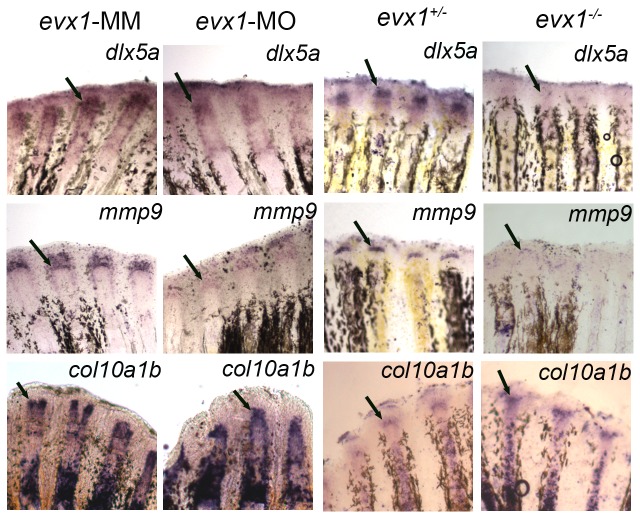
It has been proposed that evx1 is one of the earliest joint gene markers [1]. Indeed, evx1-/- mutants lack fin ray joints, demonstrating that evx1 is required for joint formation [15]. We have also found that morpholino-mediated knockdown of evx1 is sufficient to cause joint failure (data not shown). We next investigated if expression of dlx5a, mmp9, and col10a1b depend upon evx1 for their expression by taking advantage of both morpholino-mediated knockdown of evx1 and the evx1-/- mutant fins. We expected to find that expression of evx1-dependent genes is reduced in the knockdown fins and completely absent in the evx1-/- mutant fins. Indeed, we found that expression signals of dlx5a and mmp9 are reduced in evx1-knockdown fins, while col10a1b expression appeared unaffected (Figure 2). One possibility for failure to observe a knockdown effect on col10a1b expression is that the evx1-morpholino did not target the col10a1b-expressing cells located in the lateral mesenchyme. However, it was not possible to evaluate cells doubly-labeled for the fluorescein-tagged morpholino and for gene expression since the fluorescein signal is labile following in situ hybridization (Figure 3). Examination of the location of fluorescein signal prior to in situ hybridization reveals that the evx1-morpholino targets all compartments of the regenerating fin, including the lateral compartment of skeletal precursor cells (Figure 3). Together with the finding that the evx1-morpholino targets the dlx5a and mmp9-expressing cells located in the same compartment, these findings strongly suggest that morpholinos regularly target the lateral mesenchymal cells during gene-knockdown. We next evaluated gene expression in evx1-/- regenerating fins. Similar to our findings using the evx1-morpholino, we find that expression of dlx5a and mmp9 are more severely reduced in evx1 -/- regenerating fins, while col10a1b is also not affected in those fins (Figure 2). Interestingly, dlx5a and mmp9 are not completely abolished in the evx1 -/- mutants, suggesting that an alternate, non-evx1-dependent pathway may also contribute to expression of these genes. Taken together, these data suggest that dlx5a and mmp9 are expressed downstream of evx1, while col10a1b is not. Continued studies therefore focused on dlx5a and mmp9. Both evx1 and dlx5a encode for homeobox domain-containing transcription factors, although their direct targets are largely unknown. The mmp9 gene codes for a matrix metalloprotease enzyme, which is responsible for degradation of extracellular matrix proteins. During the process of joint morphogenesis, the previously uninterrupted bone matrix separates into two bony elements [7]. It is possible that Mmp9 activity contributes to this articulation event through digestion of the bone matrix.
Figure 2. dlx5a and mmp9 are genes downstream of evx1.
(A) Whole mount ISH shows levels of dlx5a expression and mmp9 expression are reduced in the evx1-morpholino (evx1-MO) injected side compared with the evx1-mismatch (evx1-MM) injected side, while the level of col10a1b expression is unchanged. (B) Whole mount ISH on evx1 -/- mutants displays similar results seen in the evx1-MO injected fins, except that a stronger reduction in dlx5a and mmp9 is observed. Arrows identify regions of the fin where staining is present and/or expected (i.e. in the cases where reduced evx1 influences expression levels).
Figure 3. Morpholinos target all cellular compartments of the regenerating fin.
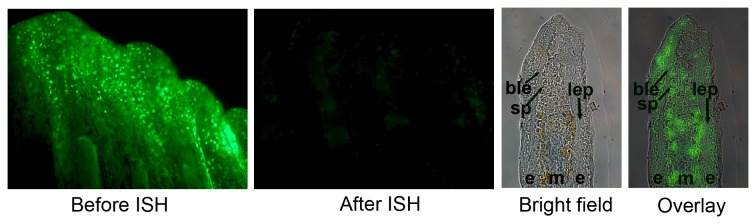
Fluorescein-positive cells successfully took up the morpholino (1 day post electroporation). The two left panels reveal loss of fluorescein signal following in situ hybridization. The two right panels demonstrate that fluorescein-positive cells are observed in all cellular compartments in freshly harvested and cryosectioned fins. The basal layer of the epidermis (ble) is identified as a row of cuboidal cells between the epithelium (e) and the mesenchyme (m). The skeletal precursor cells (sp) are located adjacent to the ble. Morpholino uptake is observed in the outer epithelial layers, in the skeletal precursors, and in the medial mesenchyme.
We next wished to distinguish between the dlx5a and mmp9 genes acting simply as joint markers or as providing a function during joint formation. Reduced function of genes required for joint formation is predicted to cause either complete joint failure or a delay of joint formation (i.e. longer segments). We find that morpholino-mediated knockdown of dlx5a and mmp9 both cause increased segment length (Figure 4). Importantly, the knockdown of these genes represents the first example of a manipulation causing longer segments in the fin. Indeed, the only other example of long fin ray segments is the alf dty86 mutant. We cannot rule out the possibility that dlx5a and mmp9 may also influence the rate of fin growth. However, changes in growth rate are not sufficient to influence segment length. Fish grown in crowded conditions grow slower than fish grown in sparse conditions, but segment length is not different between these groups [19]. Moreover, abrogation of either Fgfr1 or Shh, while influencing the rate of cell proliferation and fin length, do not influence segment length [26,27].Thus, our findings support a model where dlx5a and mmp9 contribute to correct joint placement, irrespective of any putative role in regulating fin growth.
Figure 4. dlx5a and mmp9 are necessary for correct joint placement.
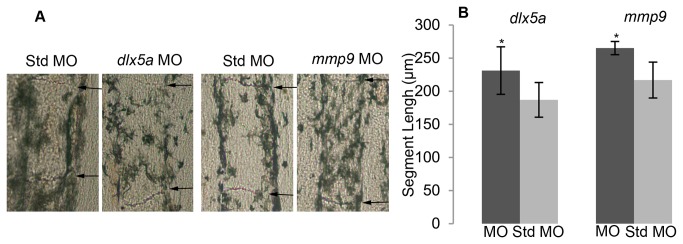
(A) Segment length is increased following targeted gene knockdown of dlx5a and mmp9 compared with standard control (std) morpholino knockdown (negative control). (B) Segment length is increased in dlx5a-knockdown and mmp9-knockdown fins compared with standard control morpholino. Statistically different data sets (*) were determined by the student’s t-test where p<0.05. The p-value for the comparison of segment length for the dlx5a-treated fins was p = 0.0047. The p-value for the comparison of segment length for the mmp9-treated fins was p = 0.0018. Error bars represent the standard deviation. MO, morpholino.
Placing the genes of the joint pathway in a linear order
Previously, our lab showed that the early genes required for osteoblast differentiation initiated in the more distal, less mature osteoblasts, while onset of expression of late osteoblast genes was observed in the more proximal, more mature osteoblasts [8]. Here we applied a similar approach for this set of joint-forming genes in an attempt to reveal a preliminary order of the evx1-dependent genes. We completed whole mount ISH at 5 dpa and measured the distance of expression domains of evx1, dlx5a, and mmp9 to the distal end of the fin. As anticipated, evx1 is expressed in the most distal domain of skeletal precursor cells, consistent with this gene acting the earliest (Figure 5). Since dlx5a and mmp9 appear downstream of evx1, we expected to find their gene expression more proximally. We find that dlx5a is expressed more proximally than evx1, while mmp9 is expressed more proximally than both evx1 and dlx5a (Figure 5). These findings support the hypothesis that dlx5a and mmp9 are expressed downstream of evx1 and further suggests the following linear pathway: evx1 followed by dlx5a followed by mmp9.
Figure 5. Expression domains of joint genes expressed during fin regeneration.
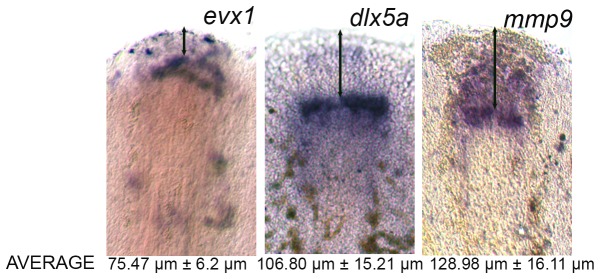
The double arrow identifies the measured distance between the expression domains and the distal tip of the fin. The measurement was taken from the third ray of each fin and the average and standard deviation (±) were calculated.
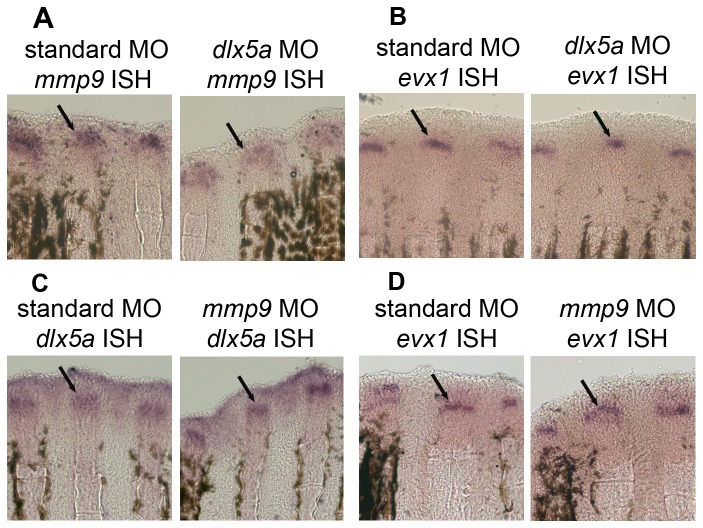
To confirm this predicted order of gene expression, we examined changes in gene expression following dlx5a -knockdown and mmp9 -knockdown by whole-mount ISH (Figure 6). We found that mmp9 expression is reduced in fins treated for dlx5a-knockdown, consistent with the hypothesis that mmp9 is expressed downstream of dlx5a. In contrast, dlx5a expression is not affected by mmp9 -knockdown. Similarly, evx1 expression is not affected by either dlx5a-knockdown or by mmp9 -knockdown. Together with our earlier findings that loss of evx1 causes reduced expression of dlx5a and mmp9 (i.e. Figure 2), these results confirm the relative order of gene expression predicted by the expression patterns along the proximal-distal axis and further suggests that dlx5a and mmp9 function does not feedback on expression of evx1.
Figure 6. Confirmation of the predicted evx1-dependent joint pathway.
Morpholino-mediated gene knockdown followed by whole mount ISH show that dlx5a-knockdown causes reduced mmp9 expression (A) but does not influence evx1 expression (B). mmp9-knockdown does not influence dlx5a expression (C) or evx1 expression (D). Arrows point to the in situ hybridization staining.
Cx43 regulates the evx1-dependent joint pathway
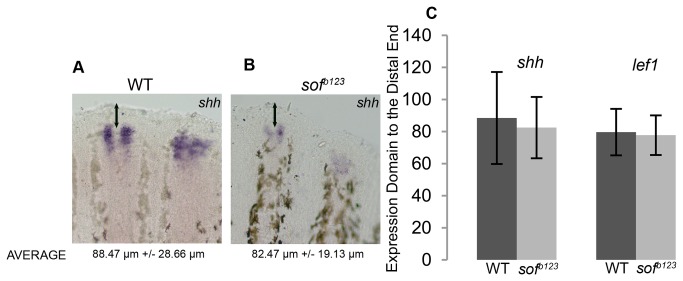
Based on our previous findings, we have suggested that Cx43 activity in the medial compartment, adjacent to the population of skeletal precursor cells, suppresses joint formation [7]. For example, the sof b123 mutant (reduced cx43) exhibits short segments/premature joints. Therefore, we predicted that the expression of the joint genes would initiate sooner, or more distally, in sof b123 fins than in wild type. Indeed, expression of evx1, dlx5a, and mmp9 genes are each initiated more distally in sof b123 regenerating fins compared with wild type (Table 1). These findings are consistent with the reduced level of Cx43 activity causing premature activation of the evx1-dependent joint pathway, and premature joints. It may be suggested that the reduced growth rate of sof b123 causes the shift of gene-expression domains to more distal locations. However, such a shift in patterning due to differential growth rates has not been observed. For example, fin amputations at more proximal locations regenerate more rapidly than fin amputations at more distal locations. However, when comparing these conditions for four different genes located in the basal layer of the epidermis (i.e. lef1, shh, wnt5b, pea3), the distance of expression of the gene domain to the distal end did not appear altered, although the strength of expression and/or size of the expression domain can change [21]. Therefore, to confirm that the reduced growth rate of sof b123 does not influence the patterning of gene expression in general, we compared the expression of both shh and lef1 between sof b123 and wild-type regenerating fins. Importantly, we found no significant changes in the distance of the distal expression domains for either gene to the distal end of the fin between wild type and sof b123 (although we do see that overall expression levels are slightly reduced) (Figure 7). Thus, the reduced growth rate of sof b123 is likely not the cause of the distal shift in joint gene expression. Rather, we suggest that reduced cx43 in sof b123 leads to premature expression of the joint genes.
Table 1. Expression domains of genes contributing to joint formation in the regenerating fin.
| WT | sof b123 | alf dty86 | |
|---|---|---|---|
| evx1 | (+) 75.47 μm ± 6.2 μm N=9 | (+) 64.1 μm ± 4.36 μm N=9 | (+/-) 109.33 μm ± 20.7 μm N=9 |
| dlx5a | (+) 106.80 μm ± 15.21 μm N=8 | (+) 79.16 μm ± 10.26 μm N=8 | (+) 110.52 μm ± 22.15 μm N=15 |
| mmp9 | (+) 128.98 μm ± 16.11 μm N=8 | (+) 86.34 μm ± 14.65 μm N=8 | (+) 110.73 μm ± 19.55 μm N=15 |
The distance is measured from the expression domain (determined by whole mount ISH) to the distal end of the fin, using the third fin ray as a standard, as previously established [19]. Irregular (+/-) expression of evx1 was observed in the alf dty86 fins, while dlx5a and mmp9 gene expression was present in all fin rays (+). The evx1 expression domain in the alf dty86 fins was measured from the subset of evx1-positive fin rays. N, number of fins.
Figure 7. Reduced growth rate of sof b123 mutants does not influence patterning of gene expression.
(A) Whole mount ISH of shh on WT fins (A) and on sof b123 fins (B). (C) The distance of expression of shh and lef1 to the distal end of the fin is not influenced by the reduced growth rate of sof b123 since the distances of the expression domains for shh and lef1 are not statistically different by the student’s t-test (p>0.05). The p-value for comparison of the shh domain was p = 0.19. The p-value for the comparison of the lef1 domain was p = 0.30. Arrows identify the region of the fin that was measured, from the distal expression domain to the distal end of the fin. Error bars represent the standard deviation.
We next examined joint gene expression in the alf dty86 mutants, which exhibit stochastic joint failure and overlong segments on average due to increased expression levels of cx43 [7,18]. Thus, in alf dty86 we expected to observe an irregular pattern (on/off) of joint gene expression and/or more proximal expression of the joint genes compared with wild type. Indeed, evx1 is expressed in a stochastic pattern and also initiates more proximally (Figure 8 and Table 1). Since evx1 is required for joint formation [15], these findings suggest that the stochastic nature of evx1 expression is the underlying cause of stochastic joint failure in alf dty86. Moreover, we suggest that the increased level of cx43 in alf dty86 is the underlying cause of stochastic evx1 expression (i.e. since cx43-knockdown rescues joint formation in alf dty86, [7]). Therefore, we next wished to determine if cx43-knockdown rescues evx1 expression. We tested this by injecting either a cx43-targeting morpholino or a cx43-mismatch morpholino across all fin rays in alf dty86 regenerating fins. Next, the percentage of evx1-positive fin rays was determined for each fin. We find that cx43-knockdown in alf dty86 regenerating fins significantly increases the percentage of evx1-positive fin rays compared with the cx43-mismatch morpholino and compared with uninjected alf dty86 regenerating fins (Figure 9). These findings reveal that cx43-knockdown relieves the suppression of evx1 expression, thereby permitting joint formation. Therefore, we suggest that cx43 suppresses joint formation by suppressing evx1 expression.
Figure 8. Expression of joint genes in alf dty86.
Whole mount ISH of evx1 shows that evx1 is expressed consistently in wild-type but irregularly in alf dty86 (i.e. “on” vs. “off”) Astericks were placed just proximal to the evx1-positive staining in alf dty86, which are present even though expression is weak. In contrast, dlx5a and mmp9 are expressed in all fin rays in both wild-type and alf dty86 fins.
Figure 9. Knockdown of cx43 rescues evx1 expression in alf dty86.
(A) Whole mount ISH of evx1 shows that evx1 is expressed in most fin rays in cx43-morpholino (MO) fins (on) but not in cx43-mismatch (MM) fins (off). Arrows identify evx1-positive signal. (B) evx1 expression is present in a much higher percentage of fin rays in alf dty86 following injection/electroporation of cx43-MO compared with either the cx43-MM control fins or the uninjected fin rays. Statistical significance (*) was determined by student’s t-test where p<0.05 show significant differences. The p-value for the comparison of cx43-MO and cx43-MM was p = 0.0015. The p-value for the comparison of cx43-MO and uninjected fin rays was p = 0.0036. The bars represent the standard deviation.
It was anticipated that the stochastic nature of evx1 expression would lead to stochastic expression of both dlx5a and mmp9. However, this was not observed (Figure 8). Instead, we find their expression is activated in all fin rays in alf dty86, consistent with the observation that dlx5a and mmp9 expression are not completely eliminated in the evx1 -/- mutant, and therefore appear to be activated even in the absence of evx1. Since joint failure occurs despite expression of dlx5a and mmp9, these data also suggest that dlx5a and mmp9 cannot mediate joint formation without the additional expression of evx1. Thus, since evx1 is required for joint formation but dlx5a and mmp9 are required but not sufficient for joint formation, evx1 must activate at least one other pathway to establish fin ray joints. Continued studies are required to identify this pathway.
Model of joint differentiation during fin regeneration
Our analyses of joint gene expression suggest a model for joint formation that requires evx1, which is expressed the earliest, followed by expression of dlx5a and mmp9 (Figure 10). Each of these genes is expressed in the population of skeletal precursor cells, adjacent to the Cx43-positive medial mesenchyme, and all three genes contribute to joint formation. We further suggest that Cx43 influences joint formation by influencing evx1 expression. When cx43 activity is reduced, as in sof b123, expression of all evx1-dependent joint genes is shifted distally, consistent with the observation that sof b123 produces premature joints. When cx43 activity is increased, as in alf dty86, expression of evx1 is irregular, consistent with the stochastic joint failure observed in alf dty86 regenerating fins. Indeed, cx43-knockdown rescues both evx1 expression and joint formation [7] in alf dty86. Interestingly, expression of dlx5a and mmp9 are not randomized, but instead are consistently expressed in all fin rays. Continued studies will be necessary to identify additional possible evx1-dependent pathways, and to understand how dlx5a and mmp9 expression is maintained in the absence of evx1.
Figure 10. Model of the identified joint pathway.
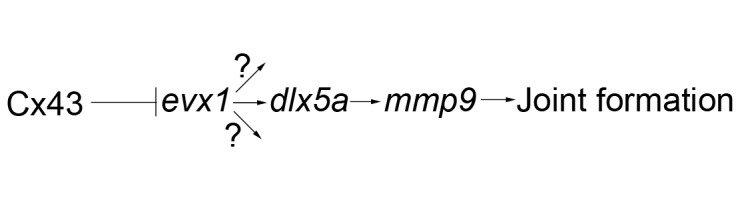
Both dlx5a and mmp9 appear to be regulated by both evx1-dependent and non-evx1-dependent manners. In addition, evx1 may activate additional genes required for joint formation. Cx43 appears to regulate joint differentiation by influencing evx1 expression.
Acknowledgments
The authors would like to thank Rebecca Jefferis for maintenance and care of the fish colony, and members of the Iovine lab for critical discussions regarding this manuscript. The authors also wish to thank Dr. Kate Lewis for sharing the evx1 -/- mutant line.
Funding Statement
This work was funded by the NSF (IOS1145582). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Borday V, Thaëron C, Avaron F, Brulfert A, Casane D et al. (2001) evx1 transcription in bony fin rays segment boundaries leads to a reiterated pattern during zebrafish fin development and regeneration. Dev Dyn 220: 91-98. doi: 10.1002/1097-0177(2000)9999:9999. PubMed: 11169842. [DOI] [PubMed] [Google Scholar]
- 2. Pacifici M, Koyama E, Shibukawa Y, Wu C, Tamamura Y et al. (2006) Cellular and molecular mechanisms of synovial joint and articular cartilage formation. Ann N Y Acad Sci 1068: 74-86. doi: 10.1196/annals.1346.010. PubMed: 16831907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Landis WJ, Géraudie J (1990) Organization and development of the mineral phase during early ontogenesis of the bony fin rays of the trout Oncorhynchus mykiss. Anat Rec 228: 383-391. doi: 10.1002/ar.1092280404. PubMed: 2285157. [DOI] [PubMed] [Google Scholar]
- 4. Mari-Beffa M, Carmona MC, Becerra J (1989) Elastoidin turn-over during tail fin regeneration in teleosts. A morphometric and radioautographic study. Anat Embryol (Berl) 180: 465-470. doi: 10.1007/BF00305121. [DOI] [PubMed] [Google Scholar]
- 5. Poss KD, Keating MT, Nechiporuk A (2003) Tales of regeneration in zebrafish. Dev Dyn 226: 202-210. doi: 10.1002/dvdy.10220. PubMed: 12557199. [DOI] [PubMed] [Google Scholar]
- 6. Santamaría JA, Marí-Beffa M, Becerra J (1992) Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation 49: 143-150. doi: 10.1111/j.1432-0436.1992.tb00662.x. PubMed: 1377652. [DOI] [PubMed] [Google Scholar]
- 7. Sims K Jr., Eble DM, Iovine MK (2009) Connexin43 regulates joint location in zebrafish fins. Dev Biol 327: 410-418. doi: 10.1016/j.ydbio.2008.12.027. PubMed: 19150347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown AM, Fisher S, Iovine MK (2009) Osteoblast maturation occurs in overlapping proximal-distal compartments during fin regeneration in zebrafish. Dev Dyn 238: 2922-2928. doi: 10.1002/dvdy.22114. PubMed: 19842180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akimenko MA, Marí-Beffa M, Becerra J, Géraudie J (2003) Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn 226: 190-201. doi: 10.1002/dvdy.10248. PubMed: 12557198. [DOI] [PubMed] [Google Scholar]
- 10. Knopf F, Hammond C, Chekuru A, Kurth T, Hans S et al. (2011) Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell 20: 713-724. doi: 10.1016/j.devcel.2011.04.014. PubMed: 21571227. [DOI] [PubMed] [Google Scholar]
- 11. Singh SP, Holdway JE, Poss KD (2012) Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell 22: 879-886. doi: 10.1016/j.devcel.2012.03.006. PubMed: 22516203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sousa S, Afonso N, Bensimon-Brito A, Fonseca M, Simões M et al. (2011) Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138: 3897-3905. doi: 10.1242/dev.064717. PubMed: 21862555. [DOI] [PubMed] [Google Scholar]
- 13. Tu S, Johnson SL (2011) Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 20: 725-732. doi: 10.1016/j.devcel.2011.04.013. PubMed: 21571228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faiella A, D'Esposito M, Rambaldi M, Acampora D, Balsofiore S et al. (1991) Isolation and mapping of EVX1, a human homeobox gene homologous to even-skipped, localized at the 5' end of HOX1 locus on chromosome 7. Nucleic Acids Res 19: 6541-6545. doi: 10.1093/nar/19.23.6541. PubMed: 1684419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schulte CJ, Allen C, England SJ, Juárez-Morales JL, Lewis KE (2011) Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev Dyn 240: 1240-1248. doi: 10.1002/dvdy.22534. PubMed: 21509898. [DOI] [PubMed] [Google Scholar]
- 16. Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR et al. (2008) Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol 317: 541-548. doi: 10.1016/j.ydbio.2008.02.051. PubMed: 18406403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL (2005) Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol 278: 208-219. doi: 10.1016/j.ydbio.2004.11.005. PubMed: 15649473. [DOI] [PubMed] [Google Scholar]
- 18. van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M et al. (1996) Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123: 255-262. PubMed: 9007245. [DOI] [PubMed] [Google Scholar]
- 19. Iovine MK, Johnson SL (2000) Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics 155: 1321-1329. PubMed: 10880491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Westerfield M (1993) The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio). Eugene, OR: University of Oregon Press. [Google Scholar]
- 21. Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA et al. (2009) Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol 331: 270-280. doi: 10.1016/j.ydbio.2009.05.545. PubMed: 19445916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoshinari N, Ishida T, Kudo A, Kawakami A (2009) Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev Biol 325: 71-81. doi: 10.1016/j.ydbio.2008.09.028. PubMed: 18950614. [DOI] [PubMed] [Google Scholar]
- 23. Ton QV, Iovine MK (2012) Semaphorin3d mediates Cx43-dependent phenotypes during fin regeneration. Dev Biol 366: 195-203. doi: 10.1016/j.ydbio.2012.03.020. PubMed: 22542598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thummel R, Bai S, Sarras MP Jr., Song P, McDermott J et al. (2006) Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn 235: 336-346. doi: 10.1002/dvdy.20630. PubMed: 16273523. [DOI] [PubMed] [Google Scholar]
- 25. Talbot JC, Johnson SL, Kimmel CB (2010) hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137: 2507-2517. doi: 10.1242/dev.049700. PubMed: 20573696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD (2005) Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132: 5173-5183. doi: 10.1242/dev.02101. PubMed: 16251209. [DOI] [PubMed] [Google Scholar]
- 27. Quint E, Smith A, Avaron F, Laforest L, Miles J et al. (2002) Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A 99: 8713-8718. doi: 10.1073/pnas.122571799. PubMed: 12060710. [DOI] [PMC free article] [PubMed] [Google Scholar]