Abstract
Native extracellular matrix (ECM) that is secreted and maintained by resident cells is of great interest for cell culture and cell delivery. We hypothesized that specialized bioengineered niches for stem cells can be established using ECM-derived scaffolding materials. Kidney was selected as a model system because of the high regional diversification of renal tissue matrix. By preparing the ECM from three specialized regions of the kidney (cortex, medulla, and papilla; whole kidney, heart, and bladder as controls) in three forms: (i) intact sheets of decellularized ECM, (ii) ECM hydrogels, and (iii) solubilized ECM, we investigated how the structure and composition of ECM affect the function of kidney stem cells (with mesenchymal stem cells, MSCs, as controls). All three forms of the ECM regulated KSC function, with differential structural and compositional effects. KSCs cultured on papilla ECM consistently displayed lower proliferation, higher metabolic activity, and differences in cell morphology, alignment, and structure formation as compared to KSCs on cortex and medulla ECM, effects not observed in corresponding MSC cultures. These data suggest that tissue- and region-specific ECM can provide an effective substrate for in vitro studies of therapeutic stem cells.
Keywords: extracellular matrix, biomaterials, decellularization, acellular matrix, hydrogels, kidney, papilla, stem cells, niche, regional specificity
INTRODUCTION
The extracellular matrix (ECM), the native scaffolding material secreted and maintained by residents cells, provides an ideal microenvironment for cells, with tissue-specific physical and molecular cues mediating cell proliferation, differentiation, gene expression, migration, orientation, and assembly [1]. Functional and structural components within the ECM contribute to the extracellular environment specific to each tissue and organ [2–4]. The complexity of the ECM has proven difficult to recapitulate in its entirety, with attempts often limited to mimicking only ECM structure using synthetic biomaterials [5] or mimicking composition by adding purified ECM components [6]. And although they may offer structural mimics, synthetic biomaterials can potentially generate cytotoxic degradation by-products at the site of implantation, leading to poor wound healing or an inflammatory environment [7].
An alternative to synthetic biomaterials is to directly isolate native ECM from the tissue of interest via the removal of cells and cellular remnants. ECM scaffolds have been derived from a variety of soft tissues such as the small intestine, bladder, heart, liver, and lung [8–10]. ECM-derived biomaterials can be processed into scaffolds with appropriate compositions and structures for cell cultivation and tissue engineering. Furthermore, ECM scaffolds gradually degrade while promoting tissue remodeling at the site of implantation. Due to their biocompatibility and their ability to modulate the host tissue response, ECM scaffolds have shown promise for tissue engineering and regenerative medicine applications [8–10].
ECM-based scaffolds can also be used to regulate the differentiation and maintenance of stem cells and their differentiated progeny. Stem cells normally reside within a unique and highly regulated ECM, which contributes to the stem cell niche [11–15]. Historically, complex tissues such as heart and lung have been decellularized to obtain native-ECM scaffolds without particular regard for any specific region of the organ or preservation of potential stem cell niches [16, 17]. Previous studies have shown that cells native to a particular region of the organ (e.g., vascular endothelium, liver sinusoidal cells) display ECM recognition and specificity [2–4]. If this site-specific recognition could be extended to stem cells, the choice of matrix would become an important consideration. However, there has been no study investigating differences in ECM derived from specific regions of an organ and how these regional differences could be preserved and harnessed for the in vitro culture of resident stem cell populations.
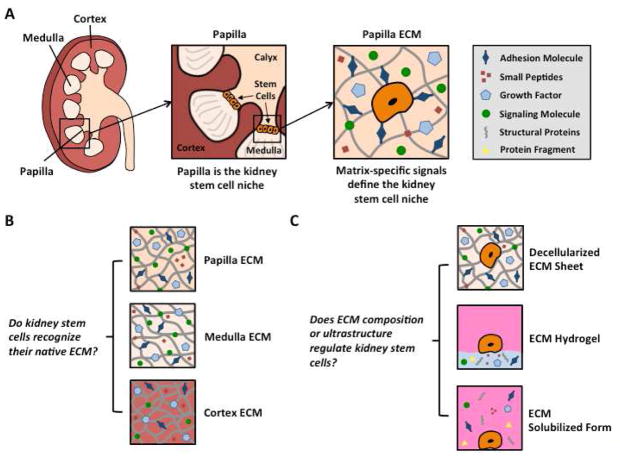
The kidney is a suitable organ for studying effects of regional ECM on the resident stem cell population. A cross-sectional view of the kidney reveals three distinct regions: cortex, medulla, and papilla (Figure 1A), with each region displaying its unique structure, function, and composition, and residing in environments with very different osmolalities and oxygen tensions.
Figure 1. Stem cell niche of the kidney.
A The renal papilla is the stem cell niche in the kidney, defined by region-specific cues in the extracellular matrix of the papilla. B The renal cortex, medulla, and papilla were dissected and decellularized separately to obtain kidney region-specific ECM. C ECM was prepared in sheets, hydrogels, and solubilized forms for cultivation of two types of stem cells (kidney stem cells and mesenchymal stem cells). To obtain kidney region-specific ECM sheets, kidney regions were dissected before decellularization or the regional matrix was punched from decellularized whole kidney sections. Alternatively, pre-sectioned regions were decellularized, snap-frozen in liquid nitrogen, ground into a fine powder, lyophilized, and pepsin-digested to yield kidney region-specific ECM digests that were neutralized to obtain solubilized ECM forms. By treating these digests with salt, base, and heat, we obtained kidney region-specific ECM hydrogels. Kidney stem cells (KSCs) that are native to the papilla were cultured on ECM sheets, hydrogels, or solubilized forms derived from the three kidney regions (cortex, medulla, papilla) and compared to mesenchymal cells (MSCs) cultured under the same conditions.
The cortex contains renal corpuscles, associated convoluted and straight tubules, collecting tubules and ducts, as well as an extensive vascular network. The medulla is arranged into pyramids, and characterized by straight tubules, collecting ducts, and the vasa recta, a specialized capillary system involved in the concentration of urine. At the apex of each medullary pyramid, where the collecting ducts converge and empty into the renal calyx, is the papilla. Recent studies have shown that the renal papillae contain a putative population of adult stem cells that remains quiescent after development is complete and that is mobilized after injury [11, 13, 18]. This stem cell population has been isolated in mice and expanded in culture, making the kidney an excellent model to study interactions between a native stem cell population and the matrix derived from distinct regions within the organ.
The present study describes a method to derive region-specific ECM biomaterials (ECM sheets, ECM hydrogels, and solubilized ECM) for stem cell culture from the three regions of the kidney (cortex, medulla, papilla). Our objective was to determine if there were region-specific effects of kidney ECM on the growth and metabolism of kidney stem cells, how these effects depend on the preservation of ECM structure versus composition alone, and if these effects extend to exogenous (non-kidney) stem cells, such as mesenchymal stem cells (MSCs).
MATERIALS AND METHODS
Decellularization
Porcine bladders, hearts, and kidneys were procured from Yorkshire pigs (65–70kg) immediately following euthanasia, excess tissue was trimmed, and the blood and debris removed with water. The organs were stored at −80°C for at least 24 hrs, thawed and then sliced into <2mm thin cross-sections. Cross-sections from the middle third of the kidney were separated into cortical, medullary, and papillary regions. Whole kidneys and kidney regions were decellularized using a modification of a previously established method [17]. Briefly, the slices were washed with 2X phosphate-buffered saline (PBS) for 15 min, followed by 2 hrs of 0.02% trypsin, 2 hrs of 3% Tween 20, and 2 hrs of 4% sodium deoxycholate treatment. After each step, kidney sections were washed with 2X PBS for 15 min. Kidney ECM slices were treated for 1 hr with 0.1% peracetic acid and subjected to alternating sterile 1X PBS and dH2O washes.
Histological analysis
Native and decellularized tissue samples were fixed in formalin, embedded in paraffin, sectioned at 5μm thickness, stained with hematoxylin and eosin, Trichrome, or Alcian Blue, and imaged using an Olympus IX81 microscope at 10X.
DNA quantification
DNA content of decellularized tissue was quantified using Quanti-iT PicoGreen dsDNA Assay kit (Invitrogen) according to the manufacturer’s instructions. Briefly, tissue samples were weighed, digested overnight with Proteinase K in TEX buffer at 56°C, and mixed with PicoGreen reagent. Fluorescence emission was measured at 520nm with excitation at 480nm, and DNA was quantified using a standard curve.
ECM characterization
Collagen content of kidney region sheets/digests was determined using the Sircol™ collagen assay kit (Biocolor). Samples were digested in 0.1mg pepsin/mL overnight at room temperature (25°C), and the Sircol™ assay was performed according to the manufacturer’s instructions. Sulfated glycosaminoglycan (sGAG) content of kidney region sheets/digests was determined using the 1, 9-dimethylene blue (DMB) dye binding assay. Samples were digested in 125μg papain/mL (Sigma) overnight at 60°C. sGAG content was quantified by mixing ECM digest samples with DMB dye in a 1:5 ratio and reading spectrophotometric absorbance at 595nm and 540nm. The difference in absorbance at these wavelengths was used with a chondroitin-6-sulphate standard curve to quantify sGAG content. Pepsin digests of the kidney region ECM and Collagen I (BD, Biosciences) were electrophoresed on 7.5% polyacrylamide gels (BioRad) under reducing conditions (5% 2-mercaptoethanol). The proteins were visualized with Coomassie Brilliant Blue (BioRad) and imaged by scanning the polyacrylamide gel.
Scanning electron microscopy (SEM)
Native and decellularized sections of regional kidney ECM were fixed in formalin, rinsed in 70% EtOH, frozen, lyophilized, and gold-coated (5nm thickness). Sections were imaged on a Hitachi S-4700 FE-SEM with accelerating voltage 2.5kV.
Immunohistochemical staining
Sections of native and decellularized kidney ECM were fixed in formalin for 30min, embedded in paraffin, cut to 5μm, and mounted on slides. Sections were deparaffinized and subjected to boiling citrate buffer (pH = 6.0) for 16 minutes for antigen retrieval, and blocked with 10% Normal Goat Serum in PBS for 2hrs at room temperature. Primary antibody staining was performed for 2hrs at 4°C using the following primary antibodies and dilutions: Collagen IV (Rb pAb to Coll IV, ab6586) diluted 1:200 and Fibronectin (Rb pAb to Fibronectin, ab23750) diluted 1:200. For all stains, the secondary antibody (Goat pAb to Rb IgG, ab98464) was diluted 1:200 and incubated for 1hr at room temperature. Sections were mounted in Vectashield Mounting Medium with DAPI, cover slipped, and imaged with an Olympus IX81 microscope at 10X.
Mouse kidney stem cells
Mouse kidney stem cells (KSCs) were obtained from mouse kidneys as previously described [18], cultured in Dulbecco’s Modified Eagle Medium (DMEM) with high glucose supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin under standard culture conditions (37°C and 5% CO2).
Mouse mesenchymal stem cells
Mouse mesenchymal stem cells (MSCs) were obtained from Texas A&M Health Science Center College of Medicine Institute for Regenerative Medicine and cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS, 10% horse serum, and 1% penicillin/streptomycin under standard culture conditions (37°C and 5% CO2).
Preparation of ECM sheets, ECM hydrogels, and solubilized ECM
Decellularized whole organ and kidney region slices were either perforated with a 7 mm biopsy punch into sheets or snap-frozen in liquid nitrogen. Sheets were stored in 1X PBS at 4°C until use while frozen pieces were pulverized into a fine powder using a mortar and pestle, and lyophilized for 24hrs. Lyophilized ECM powder was digested as previously described [19]. Briefly, 1g of lyophilized ECM powder was mixed with 0.1g pepsin (Sigma, ~2500U/mg) in 0.01M HCl. The solution was allowed to digest for 48 hrs at room temperature (25°C) under constant stirring. Final digests were aliquotted and stored at −80°C until use. The soluble ECM was obtained by neutralizing ECM stock digests and added to cell culture media directly (Typically 1 mg dry ECM/mL media). Hydrogels were prepared as previously described by mixing ECM stock digests with 1X PBS, 10X PBS, and 0.1M NaOH to yield a hydrogel with a final concentration of 6mg/mL at 4°C [19].
Solubilized mitogenicity assay
KSCs and MSCs were seeded on tissue culture (TC) plastic at 2.5×104 cells/mL, cultured for 24 hrs in media supplemented with 10% FBS, and starved for 24hrs in media containing 0.5% FBS. Next, ECM digests were added to the media (0.1 mg/mL) with 0.5% FBS for 48hrs. On the fourth day, culture media was replaced with media containing 10% Alamar Blue® (Invitrogen) and the cells were incubated for 14 hrs. Culture media were transferred into new 96-well plates and absorbance was measured at 570 nm and normalized to 600 nm.
DNA quantification
DNA content of seeded ECM sheets and hydrogels was quantified using Quanti-iT PicoGreen dsDNA Assay kit (Invitrogen) according to the manufacturer’s instructions. After 48hrs of culture, samples were digested in 125μg papain/mL overnight at 60°C and mixed with PicoGreen reagent. Fluorescence emission was measured at 520nm with excitation at 480nm, and DNA was quantified using a standard curve.
Metabolic activity
ECM sheets were glued to the bottom of 96-well plates using 2% fibrin. ECM and Collagen I hydrogels were prepared in 96-well plates by adding 50μL of hydrogel (neutralized and brought to the concentration of 6 mg/ml) at 4°C. The plates containing the hydrogels were incubated for 40 minutes at 37°C until gelation was observed. KSCs and MSCs were grown under standard culture conditions, trypsinized, seeded into the ECM sheets or hydrogels at 2.5×104 cells/mL, and cultured for 48 hrs or 7 days. After a 48-hr incubation, the culture media was replaced with media containing 10% Alamar Blue® (Invitrogen). After 14-hr incubation, media was transferred into new 96-well plates and absorbance was measured at 570nm and normalized to 600nm.
Confocal imaging
KSCs and MSCs grown under standard culture conditions were seeded into ECM sheets or hydrogels at 2.5×104 cells/mL and cultured for 48hrs or 7 days, at which times they were stained with Live/Dead Viability Kit (Invitrogen) or fixed with 3.7% formaldehyde and stained with rhodamine-phalloidin (Invitrogen) and DAPI according to the manufacturer’s instructions. Confocal imaging was performed using an Olympus Fluoview FV1000 Confocal Microscope.
Gelation kinetics
Regional kidney ECM hydrogels and Collagen I hydrogels were prepared as previously described [19]. Gelation kinetics were determined spectrophotometrically as previously described [19]. Briefly, gel solutions at 4°C were transferred to a cold 96-well plate by adding 100μL/well in triplicate. The SpectraMax spectrophotometer was preheated to 37°C, plate was loaded, and turbidity measured at 405nm every 2min for 1.5hrs. Absorbance values were recorded for each well and averaged. Three separate tests were performed on two separate batches of kidney region ECM hydrogels.
Chemotaxis (transwell) assay
KSCs were cultured for 24 hrs in 0.5% FBS, trypsinized, and seeded onto transwells with 8μm pores. Solubilized kidney region ECM was added to the media at a concentration of 0.1mg/mL. After 6 hrs, transwells were collected, attached cells removed from the top of the membrane using a Q-tip, and membranes were detached. DNA from cells attached to the bottoms of the detached membranes was quantified with CyQuant® Direct Cell Proliferation Assay Kit according to the manufacturer’s instructions. Fluorescence emission was measured at 535 nm with excitation at 480 nm, and DNA was quantified using a standard curve.
Statistical analysis
One-way ANOVA test with Tukey’s multiple comparison post hoc test and two-way ANOVA test with Bonferroni post hoc test were performed using Prism v6 (GraphPad, La Jolla CA). p < 0.05 was considered statistically significant.
RESULTS
Composition and Gelation Properties of Decellularized Kidney Region ECM
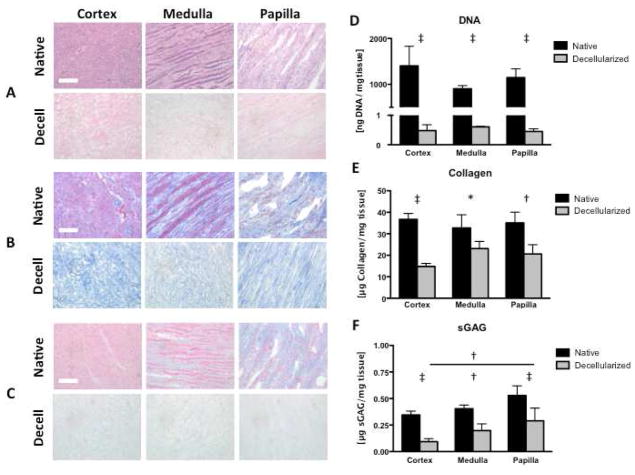
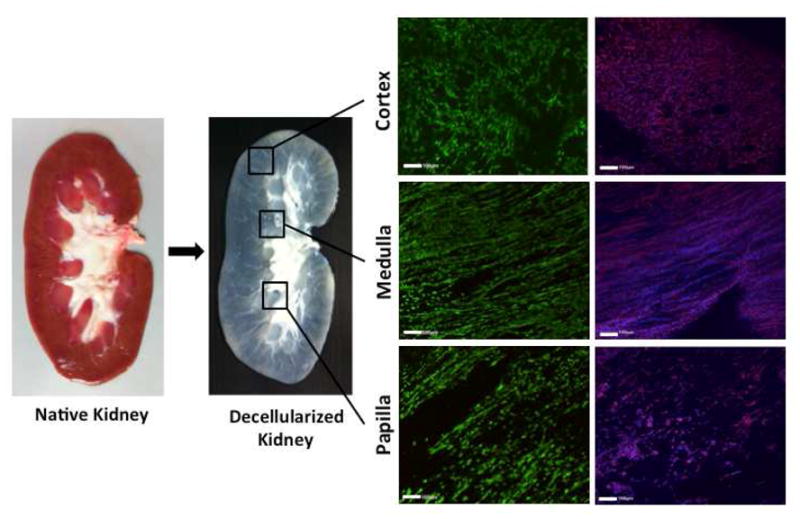
Decellularization of kidney regions (cortex, medulla, papilla) by a four-step method (trypsin, Tween 20, sodium deoxycholate, peracetic acid) resulted in the removal of >99% nuclear material as shown by H&E staining and DNA quantification (Figure 2A, 2D). Collagen content of decellularized kidney regions was reduced in all three regions, and most significantly in the cortex (Figure 2B, 2E). A similar trend was observed with sulfated glycosaminoglycan (sGAG) content (Figure 2C, 2F). Histological sections of kidney regions stained with H&E (Figure 2A), Trichrome (Figure 2B), and Alcian Blue (Figure 2C) show complete removal of cellular nuclei with some preservation of ECM structure and distribution of remaining collagen (blue) (Figure 2B) and glycosaminoglycans (light blue) (Figure 2C).
Figure 2. Removal of cellular material and preservation of ECM in decellularized kidney regions.
Histology confirms decellularization with preservation of matrix proteins in kidney regions. A H&E stain shows the absence of cell nuclei. B Trichrome stain shows the preservation of collagen (blue), and C Alcian Blue stain shows loss of proteoglycans (light blue). D DNA quantification indicates >99% removal of nuclear material after decellularization. E Collagen quantification shows comparable retention of collagen among decellularized kidney regions. F Sulfated glycosaminoglycan (sGAG) quantification indicates papilla retains significantly more sGAG than cortex. Scale bars: 100μm. * p < 0.05, † p < 0.01, ‡ p < 0.001.
Electrophoresed kidney region ECM digests and purified Collagen I showed major bands at similar locations, indicating that Collagen I is a large component of the kidney region ECM digests, with other bands distinct from pure Collagen I (Figure S1A). In digested (solubilized) kidney region ECM, the amounts of collagen per unit ECM protein were comparable among the three regions, while the amount of sGAG per unit ECM protein was lower for the cortex region (Figure S1B). The measurements of gelation kinetics showed sigmoidal curves for ECM derived from all kidney regions and the Collagen I hydrogel, with kidney hydrogels having delayed kinetics relative to Collagen I (Figure S1C). Among the kidney hydrogels, the time for gelation increased from papilla to medulla and cortex. Polymerized kidney region hydrogels had similar macroscopic appearance (Figure S1D).
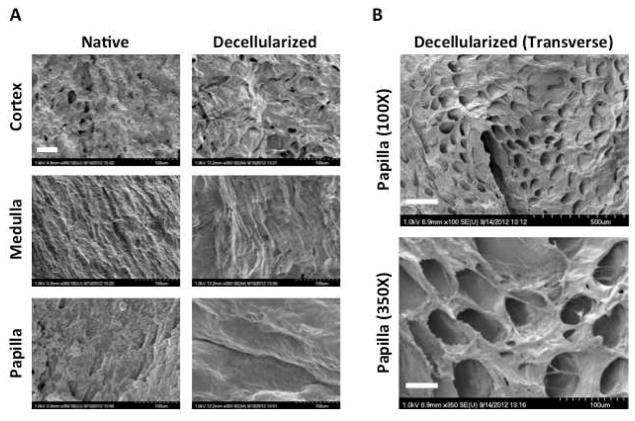
Ultrastructure of Native and Decellularized Kidney Region ECM
Native and decellularized kidney regions were imaged via SEM to investigate preservation of the ultrastructure after decellularization (Figure 3). Increased magnification at 350X reveals large differences in the native structures of the ECM in various regions of the kidney as well as distinct topographical differences retained in decellularized kidney regions (Figure 3A). A transverse section of decellularized papilla reveals preservation of tubular ultrastucture in the KSC niche (Figure 3B).
Figure 3. Ultrastructure of native and decellularized kidney regions.
A Scanning electron microscopy at 350X reveals differences in ECM topography between kidney regions before and after decellularization. Scale bar: 50μm. B Transverse sections of decellularized papilla at 100X and 350X indicate that tubular ultrastructure is preserved in the KSC niche after decellularization. Scale bars: 250μm (top) and 50μm (bottom).
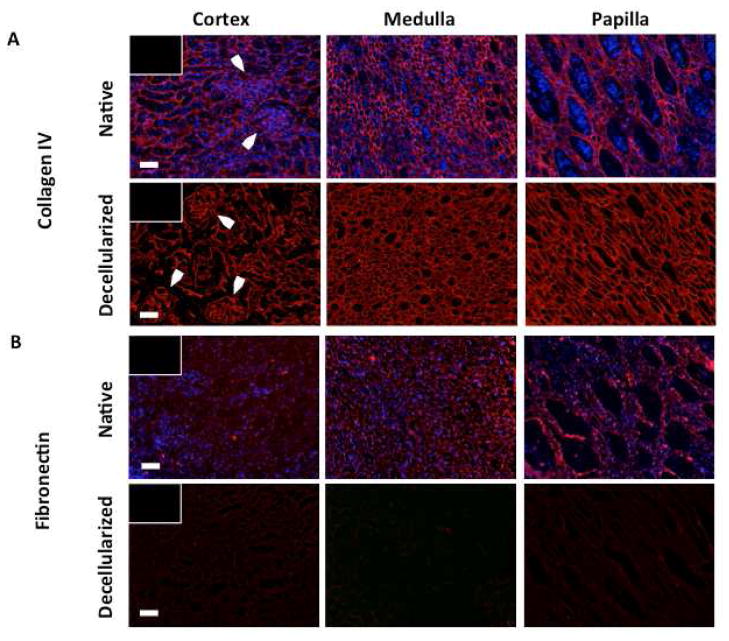
Collagen IV and Fibronectin in Native and Decellularized Kidney Region ECM
Native and decellularized kidney regions were immunostained to reveal the amounts and distributions of Collagen IV and fibronectin in kidney regions before and after decellularization (Figure 4). Immunostaining for Collagen IV indicates a significant amount of Collagen IV is retained after decellularization as well as the retention of renal corpuscular structures in the cortex (Figure 4A). Immunostaining for fibronectin indicates a significant loss of fibronectin after decellularization (Figure 4B).
Figure 4. Collagen IV and fibronectin immunostaining of native and decellularized kidney regions.
Immunostaining reveals A significant retention of Collagen IV in the basement membrane of decellularized kidney regions, including preservation of glomerular structures in decellularized cortex (arrows), and B depletion of fibronectin in kidney regions after decellularization. Scale bars: 100μm.
DNA and Metabolic Activity of KSCs in Solubilized Kidney Region ECM
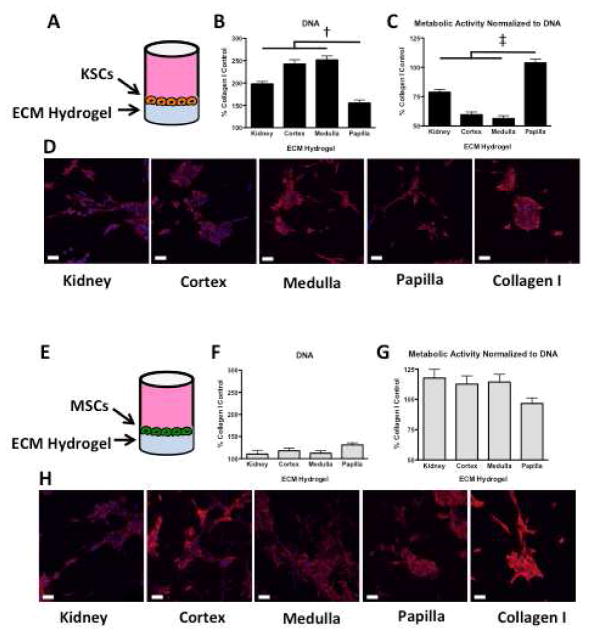
KSCs and MSCs were cultured for 48hrs on tissue culture plastic in media supplemented with solubilized ECM derived from the three kidney regions or the whole kidney. DNA and metabolic activity were measured and expressed relative to the corresponding values measured for cells grown in media supplemented with purified solubilized Collagen I (Figure 5A, 5D).
Figure 5. DNA content and metabolic activity of KSCs and MSCs in the presence of kidney region-specific solubilized ECM.
A KSCs cultured on tissue culture plastic in the presence of solubilized whole organ or kidney region-specific ECM for 48hrs. B DNA quantification of KSCs revealed less DNA in the presence of solubilized papilla ECM than in the presence of solubilized ECM from cortex, medulla, or whole kidney. C KSCs cultured in the presence of solubilized papilla ECM were significantly more metabolically active than KSCs cultured in the presence of solubilized ECM from cortex, medulla, or whole kidney. D MSCs cultured on tissue culture plastic in the presence of solubilized whole kidney and kidney region ECM for 48hrs. E DNA quantification revealed no significant differences between MSCs cultured in the presence of whole organ or kidney region solubilized ECM. F Metabolic activity of MSCs showed no differences between MSCs cultured in the presence of whole organ or kidney region solubilized ECM. * p < 0.05.
The number of KSCs in cultures with solubilized papilla ECM was significantly lower than in the solubilized ECM from any other region or the whole kidney (Figure 5B), suggesting that papillary ECM suppresses cell cycling. Interestingly, the whole kidney ECM showed an intermediate value between the ECMs of the three regions. Metabolic activity per unit DNA indicates that KSCs grown in solubilized papilla, although fewer in number, were significantly more metabolically active than KSCs grown in solubilized whole kidney, cortex, or medulla ECM.
No significant differences in metabolism were observed between KSCs in solubilized cortex and medulla ECM (Figure 5C). MSCs in solubilized kidney region ECM showed no significant differences in DNA or metabolic activity (Figure 5E, 5F).
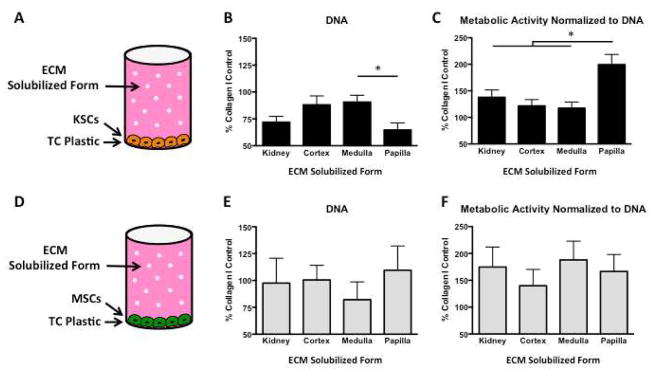
DNA, Metabolic Activity, and Phenotype of KSCs on Kidney Region ECM Hydrogels
KSCs and MSCs were seeded at equal densities on decellularized kidney region ECM hydrogels (Figure 6A, 6E), cultured for 48hrs, assayed for DNA and metabolic activity, and data were normalized to those measured for Collagen I hydrogel. DNA quantification of KSCs revealed significant differences between whole kidney, cortex, medulla, and papilla regions, with papilla hydrogel again yielding significantly fewer KSCs (Figure 6B). The whole kidney hydrogel showed an intermediate value approximating an average for the three regions. Equal initial seeding densities resulted in significantly more KSCs on kidney region ECM hydrogels than MSCs after 48 hrs (Figure 6B, 6F).
Figure 6. DNA content, metabolic activity, and rhodamine-phalloidin/DAPI staining of KSCs and MSCs on kidney region-specific ECM hydrogels.
A KSCs seeded onto ECM hydrogels and cultured for 48hrs. B DNA quantification of KSCs shows that papilla hydrogel contained significantly fewer cells than ECM derived from other kidney regions or the whole kidney. C Metabolic activity normalized to DNA reveals that KSCs are significantly more metabolically active on papilla ECM hydrogel than on ECM hydrogels from other kidney regions or the whole kidney. D Confocal imaging of KSCs on kidney region-specific ECM hydrogels and Collagen I hydrogel shows longitudinal cell alignment in kidney ECM hydrogels but not in Collagen I. E MSCs seeded onto ECM hydrogels and cultured for 48hrs. F DNA quantification of MSCs also shows no significant differences between kidney region hydrogels. G Metabolic activity per unit DNA for MSCs was comparable for all ECM hydrogels, derived regionally or from the whole kidney. H Confocal imaging of MSCs on kidney region hydrogels and Collagen I shows similar cell morphology in all kidney ECM hydrogels and Collagen I hydrogel. Scale bars: 50μm. † p < 0.01, ‡ p < 0.001.
Metabolic activity per unit DNA again indicated that KSCs on papilla ECM hydrogel were significantly more metabolically active than were KSCs on whole kidney, cortex or medulla ECM hydrogel. No significant differences were observed in metabolism between KSCs on cortex and medulla hydrogels (Figure 6C). Morphology of KSCs on whole kidney and regional kidney hydrogels appeared consistent (Figure 6D). No significant differences in DNA, metabolic activity, or morphology were observed for MSCs on kidney region ECM hydrogels (Figure 6F, 6G, 6H).
DNA, Metabolic Activity, and Phenotype of KSCs on Kidney Region ECM Sheets
KSCs and MSCs were seeded at equal densities on decellularized kidney ECM sheets (Figure 7A, 7E), cultured for 48hrs, assayed for DNA and metabolic activity, and data were normalized to those measured for cells grown on tissue culture plastic.
Figure 7. DNA content, metabolic activity, and rhodamine-phalloidin/DAPI staining of KSCs and MSCs on kidney region-specific decellularized ECM sheets.
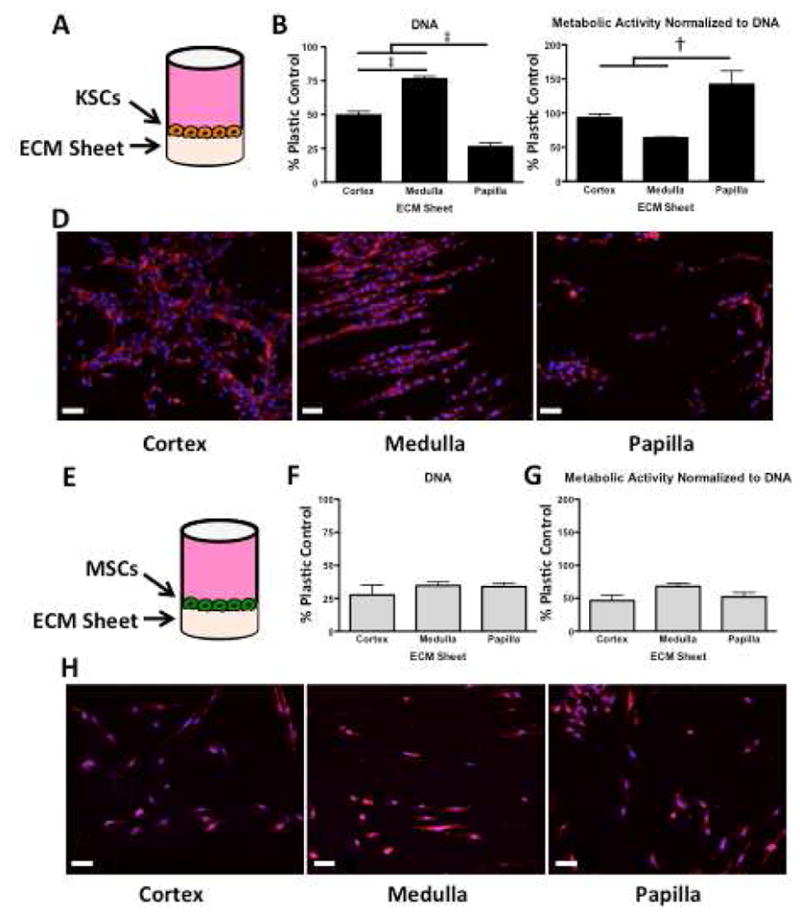
A KSCs seeded onto ECM sheets and cultured for 48hrs. B DNA quantification reveals that papilla ECM contained significantly fewer cells, and that medulla contained significantly more cells than either cortex or papilla. C Metabolic activity per unit DNA indicates that the fewer KSCs on papilla are significantly more metabolically active than KSCs on cortex or medulla ECM. D Rhodamine-phalloidin/DAPI staining shows clear differences in morphology, orientation, and structure formation between KSCs on cortex, medulla, and papilla ECM sheets. KSCs on cortex show star-like morphology with random orientation, whereas KSCs on medulla exhibit elongated morphology with significant aligning and the formation of tubular structures. KSCs on papilla show clusters with periodic rounded morphology. E MSCs seeded onto ECM sheets and cultured for 48hrs. F DNA quantification shows no significant differences in MSC cultures on ECM from different kidney regions. G Metabolic activity per unit of DNA reveals no differences in metabolic activity of MSCs. H Rhodamine-phalloidin/DAPI staining shows consistency in MSC number and phenotype in ECM from all kidney regions. Scale bars: 50μm. † p < 0.01, ‡ p < 0.001.
DNA quantification of KSCs cultured on decellualrized ECM sheets revealed significant differences between cortex, medulla, and papilla regions, with papilla ECM again yielding the fewest KSCs (Figure 7B). Metabolic activity per unit DNA confirms that KSCs on papilla were significantly more metabolically active per cell than were KSCs on either cortex or medulla (Figure 7C). No significant differences in metabolism between KSCs on cortex and medulla ECM sheets were observed.
In addition to differences in cell number, distinct morphologies and orientation of KSCs were observed in cortex, medulla, and papilla ECM (Figure 7D). KSCs on cortex showed aggregate, star-like morphology with random orientations, whereas KSCs on medulla exhibited elongated morphology with significant aligning and the formation of tubular structures. KSCs on papilla showed clusters with periodic rounded morphology as well as some end-to-end alignment in the upper papilla. No significant differences in DNA, metabolic activity, or morphology were observed for MSCs on kidney region ECM sheets (Figure 7F, 7G, 7H). Significantly more KSCs were observed on decellularized kidney region ECM when compared to MSCs after 48 hrs (Figure 7B, 7F).
Structure Formation by KSCs on Kidney Region ECM Sheets
KSCs were seeded at equal densities onto ECM sheets derived from decellularized kidney regions (cortex, medulla, and papilla), cultured for 48hrs or 7days, and imaged. KSCs showed clear differences when cultured on ECM sheets from different kidney regions in cell morphology, orientation, and structure formation already by 48 hrs of cultivation (Figure 8). On decellularized cortex sheets, KSCs consistently showed distinct morphology and regional aggregations (Figure 8, top). On decellularized medulla, KSCs displayed elongated morphology, end-to-end alignment, and tubular formations distinctly not seen in decellularized cortex at 48hrs (Figure 8, middle). On decellularized papilla, KSCs appeared morphologically different from KSCs on cortex, with some alignment in the upper papilla similar to KSCs on medulla (Figure 8, bottom). Additionally, KSCs with a rounded morphology were periodically observed in decellularized papilla sheets, only rarely in medulla, and not in cortex. KSCs on cortex displayed structures resembling renal corpuscles similar to those seen in native cortex H&E histological sections, while KSCs on medulla displayed many straight tubular bundles similar to the medullary rays seen in medulla H&E histological sections (Figure 2A).
Figure 8. Live/Dead and rhodamine-phalloidin staining of KSCs and MSCs on kidney region-specific decellularized ECM sheets.
KSCs cultured on decellularized kidney ECM from different regions display significantly different cell number, morphology, orientation, and structure formation at 48 hours (left). KSCs on cortex sheets show star-like morphology in random aggregations, whereas KSCs on medulla exhibit elongated morphology with significant aligning and formation of tubular structures. KSCs on papilla show some aligning but also periodic rounded morphology not seen in cortex or medulla. Rhodamine-phalloidin staining (right) highlights significant differences in KSC morphology and orientation between kidney regions after 7 days. The corresponding cultures of MSCs show no differences cell number, morphology, or alignment (not shown). Scale bars: 100μm.
Metabolic Activity of KSCs on Whole Organ ECM
Kidney stem cells (KSCs) were seeded onto tissue culture plastic and cultured for 48hrs in three different forms of ECM (decellularized sheets, hydrogels, and solubilized forms) obtained from porcine hearts, bladders, and kidneys. KSCs grown on decellularized whole kidney sheets and whole kidney hydrogel showed significantly higher metabolic activity at 48hrs when compared to KSCs grown on bladder and heart ECM sheets and hydrogels (Figure S2A, S2B). Furthermore, KSCs cultured on tissue culture plastic in the presence of solubilized whole kidney ECM were more metabolically active than KSCs cultured in the presence of solubilized bladder and heart ECM, with a significant difference between cells cultured in the presence of kidney and bladder ECM (Figure S2C). These results indicate that KSCs cultured on or in the presence of whole kidney ECM in various forms – decellularized sheets, hydrogels, and solubilized forms – are significantly more metabolically active than KSCs cultured in various forms of ECM from other organs, suggesting a degree of recognition or specificity by endogenous kidney stem cells to the ECM of their native organ.
Chemotaxis (Transwell) Assay
KSCs seeded onto transwells with 8μm pores were cultured in the presence of solubilized kidney region ECM (Figure S3A). KSCs cultured in the presence of solubilized papilla ECM demonstrated the least chemotaxis across the membrane, while KSCs cultured in solubilized cortex ECM demonstrated the most chemotaxis (Figure S3B).
DISCUSSION
We investigated whether ECM biomaterials derived from specific regions of the kidney, i.e., cortex, medulla, and papilla, modulate KSCs in a region-specific manner. Data show that there is a significant degree of recognition and specificity between adult kidney stem cells and their extracellular environment. KSCs showed significantly higher proliferation and higher metabolic activity in kidney ECM when compared to KSCs in ECM from other organs (Figure S2). In addition, KSCs showed lower proliferation and higher metabolic activity when cultured in papilla ECM (kidney stem cell niche) compared to medulla and cortex ECM. The decrease in cell proliferation by ECM is of great interest since in vivo the stem cells of the renal papilla (KSCs) show little cycling activity [18]. These effects were not observed with bone marrow-derived MSCs cultured under the same conditions, and the observed differences were independent of the form of ECM, i.e., sheet, hydrogel, and solubilized forms (Figure 5, 6, 7).
Kidney stem cells cultured on whole kidney ECM were compared to KSCs cultured on ECM derived from the urinary bladder and heart to determine if there was recognition between KSCs and the ECM at the organ level. ECM from whole bladder, heart, and kidney was prepared in three different forms: decellularized sheets, hydrogels, and solubilized forms. KSCs were significantly more proliferative and metabolically active in all three forms of kidney ECM when compared to respective forms of bladder or heart ECM (Figure S2).
The organ specificity of KSCs found in the present study remains consistent with previous studies that demonstrated specificity of liver sinusoidal endothelial cells to liver ECM [3] and suggests that decellularized kidney ECM sheets contain organ-specific cues. Interestingly, higher KSC metabolism was observed in whole kidney ECM hydrogel and solubilized ECM, where ECM ultra-structure is absent and only a homogenous mix of digested ECM proteins (cross-linked in the hydrogel or dissolved in the solubilized form) comprises the extracellular environment (Figure S1A, S1B). Taken together, these data suggest that the interactions responsible for cell-matrix recognition might not be limited to structural cues from decellularized matrix but may also rely on signaling from small molecules or protein fragments.
With a degree of kidney stem cell-matrix specificity having been shown at the organ level, the next step was to develop methods to isolate and prepare ECM biomaterials from three distinct regions of the kidney – the cortex, medulla, and papilla – to investigate cell-matrix interactions at the regional level. Each region of the kidney harbors a variety of cell types and structures, including extensive networks of tubules, collecting ducts, and capillaries, necessary for filtering blood or concentrating urine. In the adult mouse kidney, label-retaining cells (KSCs) have been shown to remain quiescent in the renal papilla (stem cell niche) and to migrate to the site of injury following renal ischemia [11, 13, 18]. Consequently, this adult kidney stem cell population was chosen to investigate region-specific effects of kidney ECM on the proliferation and metabolism of KSCs.
Characterization of the ECM in native cortex, medulla, and papilla revealed significant differences in structure and composition, many of which are retained after decellularization and further processing. Following the removal of >99% nuclear material (Figure 2A, 2D), some ECM proteins - such as Collagens I and IV, were preserved similarly across regions of the kidney (Figure 2B, 2E, 4A), while the amount of sGAG (Figure 2C, 2F) as well as overall ECM ultrastructure (Figure 3) differed significantly across regions. In all regions, the cell adhesion molecule fibronectin was significantly depleted after decellularization (Figure 4B), consistent with previous studies [20].
Scanning electron micrographs of kidney region ECM showed comparable topographies between native and decellularized sections, indicating that many ultra-structural features of the ECM are retained after cells are removed. Additionally, large tubular collecting ducts approximately 50μm in diameter were seen in decellularized papilla sections (Figure 3B) and demonstrate distinguishing ultra-structural features of the KSC niche not found in medulla or cortex. Given the sensitivity of stem cells to their environment, such differences in matrix architecture between kidney regions may account for differences observed in KSC activity and morphology.
While structural cues may account for some organ- or even region-specific signaling to kidney stem cells, compositional cues from the ECM may also play a key role in informing KSCs about their extracellular environment. Previous studies have shown the ability of decellularized whole mouse kidney ECM to direct the differentiation of embryonic stem cells into specialized cells types as well as to encourage proliferation along the basement membrane, suggesting that the basement membrane or one or more of its components may promote signaling for proliferation [21]. Differences in the composition and distribution of the basement membrane in different regions of the kidney may thus account for some of the region-specific differences observed in KSC proliferation and metabolism. Future studies will determine if kidney region ECM biomaterials can be used to selectively differentiate KSCs into region-specific cell types.
As shown in Figure 7D, KSCs showed significant differences in cell number, morphology, and arrangement as a function of the region from which the ECM was derived. Such differences were not observed with MSCs (Figure 7H), suggesting that structural and/or compositional differences in the kidney ECM are recognized by KSCs. In Figure 8, KSCs seeded on the cortex show arrangement into distinct circular shapes similar to the renal corpuscular structures seen in the immunostaining of native cortex sections (Figure 4A). Together these data suggest that the KSCs were able to recognize the type of matrix and adopt morphology similar to that of the native tissue. Further studies will determine whether or not KSCs are differentiating into one or multiple cells types in situ and the mechanism behind the matrix recognition.
Across all kidney region ECMs investigated, KSCs cultured in papilla ECM consistently showed significantly lower cell number (DNA content) when compared to KSCs in cortex and medulla ECM (Figure 5B, 6B, 7B). Similar trends in mitochondrial activity and cell number were observed at two different time points (48 hours and 7 days) when cultured in three different forms of kidney ECM: sheets, hydrogels, and solubilized forms (Figure 5C, 6C, 7C). This trend suggests that the effect of the ECM on KSCs’ growth and metabolic activity does not depend on the structural form of the ECM but rather on the composition. This is further supported by the trend in metabolic activity when cultured in hydrated or solubilized forms of the ECM where high metabolic activity was found in the papilla ECM and low metabolic activity in the cortex and medulla ECM.
When cultured in ECM obtained from entire kidney sections (containing cortex, medulla, and papilla), the metabolic activity was found to be within the values obtained for the individual regions, suggesting a dose effect. In addition, factors may be more readily available in the solubilized form but may still be locked into place or obscured by other proteins in an intact decellularized sheet. What specific components are responsible for this ECM regional specificity remains to be investigated in future studies.
One aspect of this work is the development of tissue-specific biomaterials and the potential for kidney tissue regeneration using region-specific ECM biomaterials to direct the differentiation of reparative stem cells to address renal pathologies such as diabetes or kidney failure. This approach may also translate into other regionalized organs as well. A recent study showed that cultivation of epithelial and endothelial cells on fully decellularized rat kidney scaffolds in a whole-organ perfused bioreactor resulted in a bioengineered kidney that produced rudimentary urine in vitro (in the bioreactor) and in vivo (following orthotopic implantation into rat) [22]. At this point, we can envision a number of cell and tissue engineering applications that may be developed in conjunction with region-specific ECMs. For example, since the renal papilla is the KSC niche, it may be used to maintain the cells in a stem-like state in vitro, while cortex and medulla ECM may be used to differentiate KSCs into other renal cell types.
Previous studies have shown that ischemic conditions in the cortex encourage mobilization, migration, and differentiation of quiescent KSCs in the papilla [18]. The data in Figure S3B support these findings. Solubilized factors from damaged matrix could provide chemotactic cues for KSCs, signaling them to migrate to the site of injury and differentiate into cell types for repair and regeneration. In addition, ECM hydrogels allow for production in large quantities when compared to ECM sheets (by pooling ECM from a large number of kidneys) as well as the use of ECM derived from regions that are small in size or volume, such as the renal papilla. Further studies are needed to determine the mechanism of KSC-matrix interactions and subsequent changes in gene expression after longer culture times.
CONCLUSIONS
ECM biomaterials derived from the kidney affect the growth and metabolism of KSCs with regional specificity. Region-specific ECM may thus provide an effective substrate for the in vitro cultivation or the delivery of therapeutic kidney stem cells and their derivatives. The significance of this work lies in the potential future development of biomaterials or applications in kidney repair and regeneration using region-specific ECM biomaterials to deliver and direct the differentiation of reparative stem cells to address renal pathologies such as diabetes or kidney failure.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the NIH support of this work (grant EB002520 to GVN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 2.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sellaro TL, Ravindra AK, Stolz DB, Badylak SF. Maintenance of hepatic sinusoidal endothelial cell phenotype in vitro using organ-specific extracellular matrix scaffolds. Tissue Eng. 2007;13:2301–10. doi: 10.1089/ten.2006.0437. [DOI] [PubMed] [Google Scholar]
- 4.Tu Q, Yang Z, Zhu Y, Xiong K, Maitz MF, Wang J, et al. Effect of tissue specificity on the performance of extracellular matrix in improving endothelialization of cardiovascular implants. Tissue Eng Part A. 2013;19:91–102. doi: 10.1089/ten.tea.2011.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han D, Gouma PI. Electrospun bioscaffolds that mimic the topology of extracellular matrix. Nanomedicine. 2006;2:37–41. doi: 10.1016/j.nano.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Geckil H, Xu F, Zhang X, Moon S, Demirci U. Engineering hydrogels as extracellular matrix mimics. Nanomedicine (Lond) 2010;5:469–84. doi: 10.2217/nnm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikos AG, McIntire LV, Anderson JM, Babensee JE. Host response to tissue engineered devices. Adv Drug Deliv Rev. 1998;33:111–39. doi: 10.1016/s0169-409x(98)00023-4. [DOI] [PubMed] [Google Scholar]
- 8.Badylak SF. Regenerative medicine and developmental biology: the role of the extracellular matrix. Anat Rec B New Anat. 2005;287:36–41. doi: 10.1002/ar.b.20081. [DOI] [PubMed] [Google Scholar]
- 9.Badylak SF. The extracellular matrix as a biologic scaffold material. Biomaterials. 2007;28:3587–93. doi: 10.1016/j.biomaterials.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 10.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Al-Awqati Q, Oliver JA. The kidney papilla is a stem cells niche. Stem Cell Rev. 2006;2:181–4. doi: 10.1007/s12015-006-0046-3. [DOI] [PubMed] [Google Scholar]
- 12.Michelini M, Franceschini V, Sihui Chen S, Papini S, Rosellini A, Ciani F, et al. Primate embryonic stem cells create their own niche while differentiating in three-dimensional culture systems. Cell Prolif. 2006:217–29. doi: 10.1111/j.1365-2184.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver JA, Maarouf O, Cheema FH, Martens TP, Al-Awqati Q. The renal papilla is a niche for adult kidney stem cells. J Clin Invest. 2004;114:795–804. doi: 10.1172/JCI20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26:4744–55. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006:1075–9. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 16.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 17.Wainwright JM, Czajka CA, Patel UB, Freytes DO, Tobita K, Gilbert TW, et al. Preparation of cardiac extracellular matrix from an intact porcine heart. Tissue Eng Part C Methods. 2010;16:525–32. doi: 10.1089/ten.tec.2009.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver JA, Klinakis A, Cheema FH, Friedlander J, Sampogna RV, Martens TP, et al. Proliferation and migration of label-retaining cells of the kidney papilla. J Am Soc Nephrol : JASN. 2009;20:2315–27. doi: 10.1681/ASN.2008111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freytes DO, Martin J, Velankar SS, Lee AS, Badylak SF. Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix. Biomaterials. 2008;29:1630–7. doi: 10.1016/j.biomaterials.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Godier-Furnemont AF, Martens TP, Koeckert MS, Wan L, Parks J, Arai K, et al. Composite scaffold provides a cell delivery platform for cardiovascular repair. Proc Natl Acad Sci U S A. 2011;108:7974–9. doi: 10.1073/pnas.1104619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross EA, Williams MJ, Hamazaki T, Terada N, Clapp WL, Adin C, et al. Embryonic stem cells proliferate and differentiate when seeded into kidney scaffolds. J Am Soc Nephrol. 2009;20:2338–47. doi: 10.1681/ASN.2008111196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19(5):646–51. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.