Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules, which participate in diverse biological processes and may regulate tumor suppressor genes or oncogenes. Single nucleotide polymorphisms (SNPs) in miRNA may contribute to diverse functional consequences, including cancer development, by altering miRNA expression. Numerous studies have shown the association between miRNA SNPs and cancer risk; however, the results are generally debatable and inconclusive, mainly due to limited statistical power. To assess the relationship between the five most common SNPs (miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, miR-149 rs2292832, and miR-27a rs895919) and the risk cancer development, we performed a meta-analysis of 66 published case-control studies. Crude odds ratios at 95% confidence intervals were used to investigate the strength of the association. No association was observed between rs2910164 and cancer risk in the overall group. However, in stratified analysis, we found that either the rs2910164 C allele or the CC genotype was protective against bladder cancer, prostate cancer, cervical cancer, and colorectal cancer, whereas it was a risk factor for papillary thyroid carcinoma and squamous cell carcinoma of the head and neck (SCCHN). Further, rs11614913 was found to be significantly associated with decreased cancer risk, in particular, for bladder cancer, gastric cancer, and SCCHN. For miR-499, a significant association was found between the rs3746444 polymorphism and cancer risk in pooled analysis. In subgroup analysis, similar results were mainly observed for breast cancer. Finally, no association was found between rs2292832 and rs895919 polymorphisms and cancer risk in the overall group and in stratified analysis. In summary, miR-196a2 rs11614913, miR-146a rs2910164, and miR-499 rs3746444 are risk factors for cancer development, whereas mir-149 rs2292832 and miR-27a rs895919 are not associated with cancer risk.
Introduction
Cancer is an outcome of unregulated expression of genes involved in development, cell growth, and differentiation. Many studies have shown that cancer is not only related to environmental factors, but also to individuals’ genetic susceptibility (predisposition). Recently, a new mechanism of microRNA (miRNA)-mediated transcriptional regulation was elucidated [1]. MiRNAs are a class of single-stranded short (21∼25 nt) RNAs, which are evolutionarily well conserved but are non–protein-coding. These RNAs regulate a broad range of biologic and pathologic process, including apoptosis, proliferation, differentiation, angiogenesis, and immune response, which are known to play critical roles in carcinogenesis [1]–[3]. MiRNAs bind to the 3′-untranslated region of the target mRNAs, leading to their degradation or translational suppression, thereby regulating the expression of target genes at the post-transcriptional level [2]. Estimates suggest that a single miRNA can target hundreds of mRNAs, and approximately 50% miRNA genes are located in cancer-related chromosomal regions [4]–[7]. Studies have shown that mature miRNAs regulate the expression of roughly 10–30% of all human genes [8]. Moreover, recent studies have suggested that miRNAs may participate in the carcinogenesis, progression (proliferation, migration, and invasion), and prognosis of multiple human malignancies by regulating the expression of tumor suppressor genes or proto-oncogenes [9]–[12].
Single nucleotide polymorphisms (SNPs) are the most common type of variation in the human genome, affecting sequence coding and splicing, which can influence the population diversity, disease susceptibility, and individual response to medicine [13]. SNPs can alter miRNA expression and/or maturation to affect function in three ways: through the transcription of the primary transcript, through pri-miRNA and pre-miRNA processing, and by affecting miRNA–mRNA interactions [14].
Many epidemiological studies have demonstrated the association of SNPs in miRNAs with the development and progression of cancer [14], [15]. MiR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, miR-149 rs2292832, and miR-27a rs895919 are well-established miRNA polymorphisms [16]–[28] that have been reported to be associated with cancer risk [14]. However, conclusions of these studies remain inconsistent due to heterogeneity of the cancer subtype, limited sample size, and differences in the ethnicity of patients. To better assess the association of miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, miR-149 rs2292832, and miR-27a rs895919 in the miRNA genes with cancer risk, we conducted a meta-analysis of all eligible published case-control studies and evaluated the effect of the five SNPs on overall cancer risk. The effects of tumor type, ethnicity, source of controls, and sample size were also evaluated.
Materials and Methods
Publication Search
To identify all potentially eligible studies on miRNA polymorphisms and cancer risk, we carried out a systematic search on PubMed, Web of Science, Science Direct, and Embase, covering all papers published up to June 30, 2013, by using the search terms: “microRNA 146a/196a2/499/149/27a”, “mir-146a/196a2/499/27a”, “polymorphism”, and “cancer”. References of the retrieved articles and review articles were also screened. Eligible studies had to meet all of the following criteria: (a) full-text study, (b) evaluation of the association between miRNA polymorphisms and cancer risk, (c) unrelated case-control design, and (d) sufficient data for estimating the odds ratio (OR) with 95% confidence interval (CI) and a P-value. Studies containing two or more case-control groups were considered as two or more independent studies.
Data Extraction
Two investigators independently reviewed and extracted information from all publications that met the inclusion criteria. In the case of a conflict, an agreement was reached by discussion between the two reviewers. The following information was sought from each publication: first author’s surname, year of publication, country of origin, ethnicity, cancer type, genotyping method, source of control groups, numbers of cases and controls for each genotype.
Statistical Analysis
We first assessed the departure of frequencies of miRNA polymorphisms from expectation under Hardy-Weinberg equilibrium (HWE) for each study by using the goodness-of-fit test (chi-square or Fisher exact test) in controls. Crude OR corresponding to 95% CI was used to assess the strength of the association between miRNA polymorphisms and cancer risk according to the methods published by Woolf et al [29]. The statistical significance of the pooled OR was determined by the Z-test, and a P-value of <0.05 was considered statistically significant. For miR-146a G/C, we investigated the association between genetic variants and cancer risk in allelic contrast (C vs. G), homozygote comparisons (CC vs. GG), heterozygote comparisons (GC vs. GG), dominant model (CC+GC vs. GG) and recessive models (CC vs. GC+GG), respectively. The same method was applied to analyze other polymorphisms. Subgroup analyses were also conducted by ethnicity (Caucasian and Asian), cancer types (if one cancer type contained only one individual study, it was combined into other cancer subgroups), source of control (population-based and hospital-based), and sample size (small sample: the total number of controls and cases less than 1000; large sample: the total number of controls and cases not less than 1000).
Statistical heterogeneity between studies was checked by Cocharan’s chi-square based Q-test [30]. However, as the Q test was insensitive in cases where studies were small or few, I2 values were also calculated, which represent the percentage of total variation across studies and provide a result of heterogeneity rather than chance. If the P-value for heterogeneity was <0.05, or if I2 was ≥50%, indicating substantial heterogeneity among studies, then a random-effect model using the DerSimonian and Laird method [31], which yielded wider CIs, was chosen to calculate the pooled OR; otherwise, a fixed-effect model using the Mantel-Haenszel method [32] was used. One-way sensitivity analyses were performed to assess the stability of the meta-analysis results [33]. Potential publication bias was estimated using Egger’s linear regression test by visual inspection of the Funnel plot. A P value <0.05 was used as an indication of potential publication bias [34]. All statistical analyses were carried out with the STATA software package version 10.0 (Stata Corporation, College Station, TX).
Results
Study Identification
In total, 66 published articles [15]–[20], [22]–[28], [35]–[87] (Table 1), with 127 comparisons, were identified through literature search with different combinations of key terms and were selected based on the inclusion criteria (Figure 1). During data extraction, 85 out of 151 articles were excluded, including 34 articles on meta-analysis, 35 articles that were not about cancer, 12 articles that were concerned with cancer prognosis, 1 article that provided incomplete polymorphism distribution data, and 3 articles that lacked full text. Two articles [41], [80] that did not provide the distribution of all three genotypes in detail, but presented genotypes as CC+GC and GG were still kept in our analysis. In two studies [70], [85], genotype frequencies were presented separately according to the country of origin of the study subjects, and thus each of these studies was treated as a separate study. In addition, Zhang et al. [46] investigated two types of cancers in one study. Each type of cancer in this article was considered separately for meta-analysis.
Table 1. Main characteristics of studies included in the meta-analysis.
| Author | Ref | Year | Country | Ethnicity | Cancer type | Design | Genotyping methods | Number of cases/controls | Genotypes distribution of cases/controls | HWE (P) | |||
| miR-146a rs2910164 | |||||||||||||
| GG | GC | CC | |||||||||||
| 1 | Horikawa | [22] | 2008 | USA | Caucasian | Renal Cell Cancer | PB | SNPlex assay | 261/235 | 144/126 | 103/94 | 14/15 | 0.65 |
| 2 | Jazdzewski | [85] | 2008 | Finland | Caucasian | PTC | PB | SNuPE Assay | 206/274 | 99/150 | 104/105 | 3/19 | 0.91 |
| 3 | Jazdzewski | [85] | 2008 | Poland | Caucasian | PTC | PB | SNuPE Assay | 201/475 | 115/286 | 82/163 | 4/26 | 0.66 |
| 4 | Jazdzewski | [85] | 2008 | USA | Caucasian | PTC | PB | SNuPE Assay | 201/152 | 91/90 | 101/52 | 9/10 | 0.51 |
| 5 | Xu | [84] | 2008 | China | Asian | Liver Cancer | HB | PCR-RFLP | 479/504 | 80/58 | 241/249 | 158/197 | 0.12 |
| 6 | Yang | [86] | 2008 | USA | Caucasian | Bladder Cancer | PB | SNPlex assay | 691/674 | 414/385 | 242/258 | 35/31 | 0.14 |
| 7 | Hoffman | [83] | 2009 | USA | Caucasian | Breast Cancer | PB/HB | massARRAY | 439/478 | 234/273 | 176/178 | 29/27 | 0.77 |
| 8 | Hu | [81] | 2009 | China | Asian | Breast Cancer | PB | PCR-RFLP | 1009/1093 | 165/180 | 515/551 | 329/362 | 0.22 |
| 9 | Tian | [82] | 2009 | China | Asian | Lung Cancer | PB | PCR-RFLP | 1058/1035 | 360/364 | 510/502 | 188/169 | 0.85 |
| 10 | Catucci | [70] | 2010 | Italy | Caucasian | Breast Cancer | PB | Sequencing | 754/1243 | 409/650 | 286/520 | 59/73 | 0.02 |
| 11 | Catucci | [70] | 2010 | Germany | Caucasian | Breast Cancer | PB | Sequencing | 805/904 | 451/536 | 304/318 | 50/50 | 0.75 |
| 12 | Guo | [66] | 2010 | China | Asian | ESCC | PB | SNaPshot | 444/468 | 234/206 | 190/220 | 20/42 | 0.12 |
| 13 | Liu | [71] | 2010 | USA | Caucasian | SCCHN | HB | PCR-RFLP | 1109/1130 | 630/655 | 411/405 | 68/70 | 0.49 |
| 14 | Okubo | [20] | 2010 | Japan | Asian | Gastric Cancer | HB | PCR-RFLP | 552/697 | 73/121 | 243/322 | 236/254 | 0.28 |
| 15 | Pastrello | [68] | 2010 | Italy | Caucasian | Mix(breast and ovarian cancer) | PB | Sequencing | 101/155 | 60/90 | 36/59 | 5/6 | 0.33 |
| 16 | Srivastava | [79] | 2010 | India | Asian | Gallbladder Cancer | PB | PCR-RFLP | 230/224 | 129/138 | 90/81 | 11/5 | 0.08 |
| 17 | Xu | [65] | 2010 | China | Asian | Prostate Cancer | HB | PCR-RFLP | 251/280 | 68/54 | 135/150 | 48/76 | 0.19 |
| 18 | Zeng | [69] | 2010 | China | Asian | Gastric Cancer | HB | PCR-RFLP | 304/304 | 62/53 | 153/132 | 89/119 | 0.12 |
| 19 | Akkiz | [59] | 2011 | Turkey | Caucasian | Liver Cancer | HB | PCR-RFLP | 222/222 | 137/144 | 75/67 | 10/11 | 0.38 |
| 20 | Garcia | [56] | 2011 | French | Caucasian | Breast Cancer | PB | TaqMan | 1130/596 | 676/352 | 388/220 | 66/24 | 0.15 |
| 21 | George | [62] | 2011 | India | Asian | Prostate Cancer | PB | PCR-RFLP | 159/230 | 4/7 | 79/107 | 76/116 | 0.00 |
| 22 | Hishida | [60] | 2011 | Japan | Asian | Gastric Cancer | HB | PCR-CTPP | 583/1637 | 82/229 | 271/775 | 230/633 | 0.74 |
| 23 | Mittal | [15] | 2011 | India | Asian | Bladder Cancer | PB | PCR-RFLP | 212/250 | 127/135 | 79/108 | 6/7 | 0.01 |
| 24 | Permuth-Wey | [55] | 2011 | USA | Caucasian | Glioma | PB | GoldenGate | 593/614 | 345/375 | 198/214 | 50/25 | 0.42 |
| 25 | Vinci | [61] | 2011 | Italy | Caucasian | NSCLC | NR | HRMA | 101/129 | 44/73 | 48/45 | 9/11 | 0.29 |
| 26 | Yue | [18] | 2011 | China | Asian | Cervical Cancer | HB | PCR-RFLP | 447/443 | 118/87 | 224/206 | 105/150 | 0.29 |
| 27 | Zhang | [58] | 2011 | China | Asian | Liver Cancer | HB | PIRA–PCR | 925/1593 | 156/291 | 450/725 | 319/577 | 0.02 |
| 28 | Zhou | [19] | 2011 | China | Asian | CSCC | HB | PCR-RFLP | 226/309 | 43/34 | 113/159 | 70/116 | 0.06 |
| 29 | Alshatwi | [40] | 2012 | Saudi | Asian | Breast Cancer | PB | TaqMan | 100/100 | 2/3 | 50/46 | 48/51 | 0.05 |
| 30 | Chu | [42] | 2012 | China | Asian | Oral Cancer | HB | PCR-RFLP | 470/425 | 54/54 | 242/196 | 174/175 | 0.94 |
| 31 | Hezova | [51] | 2012 | Czech | Caucasian | Colorectal Cancer | HB | TaqMan | 197/212 | 115/124 | 70/79 | 12//9 | 0.41 |
| 32 | Kim | [46] | 2012 | Korea | Asian | Liver Cancer | PB | PCR-RFLP | 286/201 | 27/24 | 159/103 | 100/74 | 0.19 |
| 33 | Lung | [54] | 2012 | China | Asian | Nasopharyngeal Carcinoma | PB | Tm-shift | 229/3631 | 24/497 | 88/1721 | 117/1413 | 0.46 |
| 34 | Mihalache | [47] | 2012 | Italy and Germany | Caucasian | Cholangiocarcinoma | HB | TaqMan | 182/350 | 118/211 | 53/122 | 11/17 | 0.91 |
| 35 | Min | [39] | 2012 | Korea | Asian | Colorectal Cancer | HB | PCR-RFLP | 446/502 | 62/69 | 233/245 | 151/188 | 0.44 |
| 36 | Wang | [41] | 2012 | China | Asian | Bladder Cancer | HB | TaqMan | 1017/1179 | 369/340 | 456/571 | 192/268 | 0.34 |
| 37 | Xiang | [38] | 2012 | China | Asian | Liver Cancer | HB | PCR-RFLP | 100/200 | 27/45 | 45/100 | 28/55 | 0.97 |
| 38 | Zhou | [37] | 2012 | China | Asian | Liver Cancer | PB | PCR-RFLP | 186/483 | 33/71 | 86/254 | 67/158 | 0.06 |
| 39 | Zhou | [36] | 2012 | China | Asian | Gastric Cancer | HB | TaqMan | 1686/1895 | 578/551 | 822/951 | 286/393 | 0.64 |
| 40 | Ma | [32] | 2013 | China | Asian | TNBC | HB | massARRAY | 192/191 | 35/34 | 94/93 | 63/64 | 0.98 |
| 41 | Ma | [34] | 2013 | China | Asian | Colorectal Cancer | HB | TaqMan | 1147/1203 | 444/397 | 534/614 | 169/192 | 0.08 |
| 42 | ORSÓS | [29] | 2013 | Hungary | Caucasian | SCCHN | PB | PCR-RFLP | 468/468 | 284/323 | 168/136 | 16/9 | 0.22 |
| 43 | Song | [35] | 2013 | USA | Caucasian | OSCC | HB | PCR-RFLP | 325/335 | 184/203 | – | – | – |
| 44 | Vinci | [33] | 2013 | Italy | Caucasian | Colorectal Cancer | NR | HRMA | 160/178 | 86/100 | 57/65 | 17/13 | 0.59 |
| 45 | Wei | [31] | 2013 | China | Asian | PTC | PB | massARRAY | 753/760 | 136/138 | 323/345 | 294/277 | 0.09 |
| 46 | Wei | [87] | 2013 | China | Asian | ESCC | HB | massARRAY | 368/370 | 67/67 | 184/181 | 117/122 | 0.99 |
| 47 | Yamashita | [30] | 2013 | Japan | Asian | Malignant melanoma | NR | PCR-RFLP | 50/107 | 0/3 | 35/53 | 15/51 | 0.01 |
| miR-196a2 rs11614913 | |||||||||||||
| CC | CT | TT | |||||||||||
| 1 | Horikawa | [22] | 2008 | USA | Caucasian | Renal Cell cancer | PB | SNPlex assay | 276/277 | 105/101 | 126/117 | 45/59 | 0.02 |
| 2 | Yang | [86] | 2008 | USA | Caucasian | Bladder Cancer | PB | SNPlex assay | 736/731 | 255/257 | 348/342 | 133/132 | 0.32 |
| 3 | Hoffman | [83] | 2009 | USA | Caucasian | Breast Cancer | PB/HB | massARRAY | 426/466 | 181/166 | 209/229 | 36/71 | 0.58 |
| 4 | Hu | [81] | 2009 | China | Asian | Breast Cancer | PB | PCR-RFLP | 1009/1093 | 239/218 | 483/517 | 287/358 | 0.21 |
| 5 | Tian | [82] | 2009 | China | Asian | Lung Cancer | PB | PCR-RFLP | 1058/1035 | 253/209 | 512/519 | 293/307 | 0.70 |
| 6 | Catucci | [70] | 2010 | Italy | Caucasian | Breast Cancer | PB | TaqMan | 751/1243 | 334/532 | 330/550 | 87/161 | 0.32 |
| 7 | Catucci | [70] | 2010 | Germany | Caucasian | Breast Cancer | PB | TaqMan | 1101/1496 | 432/584 | 512/696 | 157/216 | 0.71 |
| 8 | Christensen | [80] | 2010 | USA | Caucasian | SCCHN | PB | Taqman | 484/555 | 182/188 | – | – | – |
| 9 | Dou | [77] | 2010 | China | Asian | Glioma | HB | PCR-LDR | 643/656 | 111/143 | 343/305 | 189/208 | 0.12 |
| 10 | Kim | [75] | 2010 | Korea | Asian | Lung Cancer | HB | PCR-FRET | 654/640 | 187/155 | 305/300 | 162/185 | 0.13 |
| 11 | Li | [67] | 2010 | China | Asian | Liver Cancer | HB | PCR-RFLP | 310/222 | 78/42 | 150/102 | 82/78 | 0.40 |
| 12 | Liu | [71] | 2010 | USA | Caucasian | SCCHN | HB | PCR-RFLP | 1109/1130 | 350/383 | 565/545 | 194/202 | 0.74 |
| 13 | Okubo | [20] | 2010 | Japan | Asian | Gastric Cancer | HB | PCR-RFLP | 552/697 | 105/124 | 281/350 | 166/223 | 0.51 |
| 14 | Peng | [78] | 2010 | China | Asian | Gastric Cancer | HB | PCR-RFLP | 213/213 | 76/161 | 94/107 | 43/50 | 0.94 |
| 15 | Qi | [72] | 2010 | China | Asian | Liver Cancer | HB | PCR-LDR | 361/590 | 82/125 | 179/304 | 100/161 | 0.40 |
| 16 | Srivastava | [79] | 2010 | India | Asian | Gallbladder Cancer | PB | PCR-RFLP | 230/230 | 119/136 | 95/75 | 16/19 | 0.07 |
| 17 | Wang | [76] | 2010 | China | Asian | ESCC | HB | SNaPshot | 458/489 | 148/128 | 262/250 | 48/111 | 0.60 |
| 18 | Akkiz | [59] | 2011 | Turkey | Caucasian | Liver Cancer | HB | PCR-RFLP | 185/185 | 77/58 | 86/87 | 22/40 | 0.49 |
| 19 | George | [62] | 2011 | India | Asian | Prostate Cancer | PB | PCR-RFLP | 159/230 | 55/106 | 101/114 | 3/10 | 0.00 |
| 20 | Hong | [64] | 2011 | Korea | Asian | Lung Cancer | HB | Taqman | 406/428 | 86/96 | 224/198 | 96/134 | 0.16 |
| 21 | Jedlinski | [63] | 2011 | Australia | Caucasian | Breast Cancer | PB | PCR-RFLP | 187/171 | 68/58 | 86/82 | 33/31 | 0.83 |
| 22 | Mittal | [15] | 2011 | India | Asian | Bladder Cancer | PB | PCR-RFLP | 212/250 | 76/109 | 131/127 | 5/14 | 0.00 |
| 23 | Vinci | [61] | 2011 | Italy | Caucasian | NSCLC | NR | HRMA | 101/129 | 35/58 | 54/61 | 12/10 | 0.27 |
| 24 | Zhan | [57] | 2011 | China | Asian | Colorectal Cancer | HB | PCR-RFLP | 252/543 | 68/113 | 128/267 | 56/163 | 0.85 |
| 25 | Zhang | [58] | 2011 | China | Asian | Liver Cancer | HB | PIRA–PCR | 934/1622 | 208/328 | 449/817 | 277/477 | 0.52 |
| 26 | Zhou | [19] | 2011 | China | Asian | CSCC | HB | PCR-RFLP | 226/309 | 46/58 | 123/169 | 57/82 | 0.08 |
| 27 | Alshatwi | [40] | 2012 | Saudi | Asian | Breast Cancer | PB | TaqMan | 100/100 | 35/46 | 63/50 | 2/4 | 0.03 |
| 28 | Chen | [49] | 2012 | China | Asian | CRC | HB | PCR–LDR | 126/407 | 27/94 | 64/206 | 35/107 | 0.79 |
| 29 | Chu | [42] | 2012 | China | Asian | Oral Cancer | HB | PCR-PFLP | 470/425 | 57/87 | 277/206 | 136/132 | 0.69 |
| 30 | Hezova | [51] | 2012 | Czech | Caucasian | Colorectal Cancer | HB | TaqMan | 197/212 | 82/87 | 89/103 | 26/22 | 0.29 |
| 31 | Kim | [46] | 2012 | Korea | Asian | Liver Cancer | PB | PCR-RFLP | 286/201 | 58/45 | 154/107 | 74/49 | 0.36 |
| 32 | Linhares | [52] | 2012 | Brazil | Caucasian | Breast Cancer | HB | TaqMan | 325/274 | 83/94 | 148/114 | 94/66 | 0.00 |
| 33 | Min | [39] | 2012 | Korea | Asian | Colorectal Cancer | HB | PCR-RFLP | 446/502 | 120/100 | 201/254 | 125/148 | 0.63 |
| 34 | Zhang | [44] | 2012 | China | Asian | Breast Cancer | PB | PCR-RFLP | 248/243 | 1/17 | 89/93 | 148/133 | 0.89 |
| 35 | Zhu | [48] | 2012 | China | Asian | Colorectal Cancer | HB | TaqMan | 573/588 | 140/121 | 303/295 | 130/172 | 0.79 |
| 36 | Song | [35] | 2013 | USA | Caucasian | OSCC | HB | PCR-RFLP | 325/335 | 95/96 | – | – | – |
| 37 | Vinci | [33] | 2013 | Italy | Caucasian | CRC | NR | HRMA | 160/178 | 62/83 | 86/84 | 12/11 | 0.09 |
| 38 | Wei | [87] | 2013 | China | Asian | ESCC | HB | massARRAY | 367/370 | 65/87 | 196/170 | 106/113 | 0.14 |
| miR-499 rs3746444 | |||||||||||||
| TT | TC | CC | |||||||||||
| 1 | Hu | [81] | 2009 | China | Asian | Breast Cancer | PB | PCR-RFLP | 1093/1009 | 707/816 | 258/248 | 44/29 | 0.06 |
| 2 | Tian | [82] | 2009 | China | Asian | Lung Cancer | PB | PCR-RFLP | 1035/1058 | 781/755 | 253/254 | 24/26 | 0.40 |
| 3 | Catucci | [70] | 2010 | Italy | Caucasian | Breast Cancer | PB | Sequencing | 1242/756 | 414/704 | 295/452 | 47/86 | 0.25 |
| 4 | Catucci | [70] | 2010 | Germany | Caucasian | Breast Cancer | PB | Sequencing | 925/823 | 536/601 | 250/290 | 37/34 | 0.89 |
| 5 | Liu | [71] | 2010 | USA | Caucasian | SCCHN | HB | PCR-RFLP | 1130/1109 | 745/710 | 309/366 | 55/54 | 0.44 |
| 6 | Okubo | [20] | 2010 | Japan | Asian | Gastric Cancer | HB | PCR-RFLP | 697/552 | 364/466 | 151/198 | 37/33 | 0.05 |
| 7 | Srivastava | [79] | 2010 | India | Asian | Gallbladder Cancer | PB | PCR-RFLP | 230/230 | 112/121 | 97/94 | 21/15 | 0.57 |
| 8 | Akkiz | [59] | 2011 | Turkey | Caucasian | Liver Cancer | HB | PCR-RFLP | 222/222 | 45/47 | 87/93 | 90/82 | 0.04 |
| 9 | George | [62] | 2011 | India | Asian | Prostate Cancer | PB | PCR-RFLP | 230/159 | 48/104 | 98/92 | 13/34 | 0.07 |
| 10 | Mittal | [15] | 2011 | India | Asian | Bladder Cancer | PB | PCR-RFLP | 250/212 | 95/121 | 92/94 | 25/35 | 0.02 |
| 11 | Vinci | [61] | 2011 | Italy | Caucasian | Lung Cancer | NR | HRMA | 129/101 | 53/70 | 41/48 | 7/11 | 0.50 |
| 12 | Zhou | [19] | 2011 | China | Asian | CSCC | HB | PCR-RFLP | 309/226 | 134/223 | 84/71 | 8/15 | 0.00 |
| 13 | Alshatwi | [40] | 2012 | Saudi | Asian | Breast Cancer | PB | TaqMan | 100/100 | 30/45 | 62/40 | 8/15 | 0.23 |
| 14 | Chu | [42] | 2012 | China | Asian | Oral Cancer | HB | PCR-PFLP | 425/270 | 339/356 | 119/66 | 12/3 | 0.98 |
| 15 | Kim | [46] | 2012 | Korea | Asian | Liver Cancer | PB | PCR-RFLP | 201/286 | 200/120 | 81/74 | 5/7 | 0.28 |
| 16 | Min | [39] | 2012 | Korea | Asian | Colorectal Cancer | HB | PCR-RFLP | 502/446 | 292/334 | 142/154 | 12/14 | 0.45 |
| 17 | Xiang | [38] | 2012 | China | Asian | Liver Cancer | HB | PCR-RFLP | 200/100 | 36/106 | 40/71 | 24/23 | 0.04 |
| 18 | Zhou | [37] | 2012 | China | Asian | Liver Cancer | PB | PCR-RFLP | 483/186 | 141/371 | 41/100 | 4/12 | 0.10 |
| 19 | Song | [35] | 2013 | USA | Caucasian | OSCC | HB | PCR-RFLP | 325/335 | 184/214 | – | – | – |
| 20 | Vinci | [33] | 2013 | Italy | Caucasian | CRC | NR | HRMA | 178/160 | 93/105 | 32/56 | 35/17 | 0.03 |
| 21 | Wei | [87] | 2013 | China | Asian | ESCC | HB | massARRAY | 358/376 | 291/289 | 60/76 | 7/11 | 0.14 |
| miR-149 rs2292832 | |||||||||||||
| CC | CT | TT | |||||||||||
| 1 | Hu | [81] | 2009 | China | Asian | Breast Cancer | PB | PCR-RFLP | 1009/1093 | 450/482 | 460/503 | 99/108 | 0.16 |
| 2 | Tian | [82] | 2009 | China | Asian | Lung Cancer | PB | PCR-RFLP | 1058/1035 | 123/112 | 472/453 | 463/470 | 0.86 |
| 3 | Liu | [71] | 2010 | USA | Caucasian | SCCHN | HB | PCR-RFLP | 1109/1130 | 580/586 | 441/445 | 88/99 | 0.27 |
| 4 | Vinci | [61] | 2011 | Italy | Caucasian | NSCLC | NR | HRMA | 101/129 | 44/65 | 41/53 | 16/11 | 0.97 |
| 5 | Chu | [42] | 2012 | China | Asian | Oral Cancer | HB | PCR-PFLP | 470/425 | 37/26 | 88/84 | 345/315 | 0.00 |
| 6 | Kim | [46] | 2012 | Korea | Asian | Liver Cancer | PB | PCR-RFLP | 286/201 | 24/21 | 113/97 | 149/83 | 0.34 |
| 7 | Min | [39] | 2012 | Korea | Asian | Colorectal Cancer | HB | PCR-RFLP | 446/502 | 48/51 | 177/219 | 221/232 | 0.95 |
| 8 | Zhang | [43] | 2012 | China | Asian | Colorectal Cancer | PB | PCR-RFLP | 443/435 | 50/46 | 190/202 | 203/187 | 0.43 |
| 9 | Zhang | [43] | 2012 | China | Asian | Gastric Cancer | PB | PCR-RFLP | 274/269 | 41/35 | 101/120 | 132/114 | 0.70 |
| 10 | Zhang | [44] | 2012 | China | Asian | Breast Cancer | PB | PCR-RFLP | 245/229 | 23/24 | 102/113 | 120/92 | 0.21 |
| 11 | Song | [35] | 2013 | USA | Caucasian | OSCC | HB | PCR-RFLP | 325/335 | 158/162 | – | – | – |
| 12 | Vinci | [33] | 2013 | Italy | Caucasian | CRC | NR | HRMA | 160/178 | 79/86 | 58/75 | 23/17 | 0.91 |
| miR-27a rs895919 | |||||||||||||
| AA | AG | GG | |||||||||||
| 1 | Hoffman | [83] | 2009 | USA | Mixed | Breast Cancer | PB/HB | massARRAY | 434/477 | 184/220 | 200/211 | 50/46 | 0.65 |
| 2 | Sun | [73] | 2010 | China | Asian | Gastric Cancer | HB | PCR-RFLP | 304/304 | 115/145 | 135/119 | 54/40 | 0.05 |
| 3 | Yang | [74] | 2010 | Germany | Caucasian | Breast Cancer | PB | Sequencing | 1189/1416 | 576/605 | 486/660 | 127/151 | 0.14 |
| 4 | Catucci | [53] | 2012 | Italy | Caucasian | Breast Cancer | PB | TaqMan | 1025/1593 | 547/803 | 388/633 | 90/157 | 0.05 |
| 5 | Hezova | [51] | 2012 | Czech | Caucasian | Colorectal Cancer | HB | TaqMan | 197/212 | 88/93 | 86/94 | 23/25 | 0.29 |
| 6 | Shi | [45] | 2012 | China | Asian | Renal Cell Cancer | HB | TaqMan | 594/600 | 334/288 | 213/262 | 47/50 | 0.37 |
| 7 | Zhang | [44] | 2012 | China | Asian | Breast Cancer | PB | PCR-RFLP | 245/243 | 60/75 | 144/109 | 41/59 | 0.12 |
| 8 | Zhou | [50] | 2012 | China | Asian | Gastric Cancer | HB | massARRAY | 295/413 | 166/214 | 122/167 | 7/32 | 0.94 |
| 9 | Wei | [87] | 2013 | China | Asian | ESCC | HB | massARRAY | 379/377 | 216/208 | 143/139 | 20/30 | 0.14 |
HB: hospital based; PB: population based; Mixed: hospital and population based; NR: not reported; PTC: papillary thyroid carcinoma; ESCC: esophageal squamous cell carcinoma; SCCHN: squamous cell carcinoma of the head and neck; NSCLC: non-small cell lung cancer; CSCC: cervical cancer; TNBC: triple negative breast cancer; OSCC: oral squamous cell carcinoma; CRC: colorectal cancer; PCR-RFLP: polymerase chain reaction–restriction fragment length polymorphism; HRMA: high-resolution melting analysis; PIRA–PCR: primer-introduced restriction analysis-polymerase chain reaction; PCR-LDR: polymerase chain reaction-ligation detection reaction; PCR-FRET: polymerase chain reaction-fluorescence resonance energy transfer; Tm-shift: melting-temperature –shift allele-specific genotyping; HWE: Hardy-Weinberg equilibrium; P: p value.
Figure 1. Flow chart of the study selection process.
Overall, 47, 38, 21, 12, and 9 studies were pooled for meta-analysis of the rs2910164, rs11614913, rs3746444, rs2292832, and rs895919, respectively. Among all the included articles, there were 11 articles on liver cancer and breast cancer each, 8 studies on gastric cancer and colorectal cancer each, 5 studies on squamous cell carcinoma of the head and neck (SCCHN), 4 studies on lung cancer, 3 studies on bladder cancer and esophageal squamous cell carcinoma (ESCC) each, 2 studies on prostate cancer, glioma cancer, renal cell cancer, papillary thyroid carcinoma (PTC) and cervical cancer each, and 1 study each on gallbladder cancer, malignant melanoma and breast/ovarian cancer. The ethnicity of subjects in 42 studies and 24 studies were Asian and Caucasian, respectively. The controls from 37 studies came from a hospital-based population, whereas 25 studies had population-based controls. One study included both population-based and hospital-based controls [83], while three studies lacked the information of control source [36], [39], [61]. To determine the SNPs, multiple genotyping methods were employed including polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), TaqMan assay, SNPlex, SNuPE Assay, high-resolution melting analysis (HRMA), polymerase chain reaction-ligation detection reaction (PCR-LDR), direct sequencing, SNaPshot, Sequenom’s MassARRAY, fluorescence labeled hybridization (PCR-FRET), polymerase chain reaction with confronting two-pair primers (PCR-CTTP), Illumina’s GoldenGate, primer introduced restriction analysis- polymerase chain reaction (PIRA-PCR) and Tm-shift allele-specific genotyping. Genotypic distribution of most of the studied SNPs was in agreement with HWE (P>0.05) in controls.
Quantitative Synthesis
miR-146a rs2910164
For miR-146a rs2910164 polymorphism, our study contained 47 comparisons with 22,055 cases and 29,138 controls. The frequency of the rs2910164 C allele had a significantly higher representation in the Asian population compared to the Caucasian population (Asian: 54.3%, 95% CI = 49.1–59.4%; Caucasian: 24.2%, 95% CI = 22.9–25.4%; P<0.001).
The results of the meta-analysis on rs2910164 and cancer risk are shown in Table 2. Overall, no significant association was found between rs2910164 and cancer risk under any genetic model when all the eligible studies were pooled into the meta-analysis. After exclusion of four studies [15], [36], [58], [70], whose genotypic distributions in controls were not in agreement with HWE, the results did not significantly change.
Table 2. Meta-analysis of miR-146a rs2910164 polymorphism with cancer risk.
| Variables | na | C vs. G | CC vs. GG | GC vs. GG | CC+GC vs. GG | CC vs. GC+GG | |||||||||||||||
| OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | ||
| Total | 47 | 0.978(0.931–1.027) | 0.375 | <0.001 | 63.6 | 0.952(0.851–1.065) | 0.393 | <0.001 | 60.5 | 0.982(0.921–1.048) | 0.588 | <0.001 | 45.9 | 0.983(0.919–1.051) | 0.614 | <0.001 | 55.4 | 0.959(0.880–1.045) | 0.339 | <0.001 | 58.9 |
| Cancer type | |||||||||||||||||||||
| Bladder Cancer | 3 | 0.838(0.762–0.921) | 0.001 | 0.324 | 11.2 | 0.724(0.587–0.893) | 0.003 | 0.241 | 29.7 | 0.789(0.689–0.904) | 0.001 | 0.526 | 0.0 | 0.781(0.687–0.889) | <0.001 | 0.290 | 19.3 | 0.836(0.693–1.010) | 0.063 | 0.446 | 0.0 |
| Breast Cancer | 7 | 1.032(0.966–1.102) | 0.353 | 0.864 | 0.0 | 1.138(0.970–1.335) | 0.112 | 0.818 | 0.0 | 0.999(0.907–1.099) | 0.976 | 0.485 | 0.0 | 1.025(0.935–1.123) | 0.601 | 0.682 | 0.0 | 1.073(0.944–1.219) | 0.282 | 0.504 | 0.0 |
| Cervical Cancer | 2 | 0.719(0.620–0.835) | <0.001 | 0.796 | 0.0 | 0.503(0.370–0.684) | <0.001 | 0.814 | 0.0 | 0.721(0.545–0.953) | 0.022 | 0.254 | 23.1 | 0.632(0.485–0.823) | 0.001 | 0.382 | 0.0 | 0.654(0.520–0.822) | <0.001 | 0.359 | 0.0 |
| Colorectal Cancer | 4 | 0.912(0.833–0.999) | 0.047 | 0.324 | 13.6 | 0.873(0.716–1.064) | 0.179 | 0.281 | 21.5 | 0.854(0.740–0.985) | 0.030 | 0.376 | 3.4 | 0.859(0.750–0.984) | 0.028 | 0.294 | 19.2 | 0.926(0.785–1.091) | 0.357 | 0.393 | 0.0 |
| ESCC | 2 | 0.841(0.631–1.121) | 0.237 | 0.047 | 74.6 | 0.648(0.288–1.457) | 0.294 | 0.021 | 81.1 | 0.834(0.667–1.042) | 0.109 | 0.235 | 29.2 | 0.815(0.585–1.134) | 0.224 | 0.142 | 53.7 | 0.700(0.360–1.362) | 0.294 | 0.033 | 77.9 |
| Gastric Cancer | 4 | 0.953(0.782–1.162) | 0.633 | <0.001 | 86.4 | 0.915(0.625–1.339) | 0.648 | <0.001 | 84.1 | 0.907(0.806–1.020) | 0.104 | 0.136 | 45.8 | 0.960(0.742–1.240) | 0.753 | 0.011 | 73.1 | 0.919(0.700–1.206) | 0.543 | <0.001 | 83.5 |
| Lung Cancer | 2 | 1.079(0.959–1.214) | 0.205 | 0.209 | 36.7 | 1.139(0.891–1.455) | 0.300 | 0.710 | 0.0 | 1.264(0.753–2.122) | 0.375 | 0.068 | 69.9 | 1.246(0.799–1.945) | 0.332 | 0.095 | 64.2 | 1.104(0.885–1.377) | 0.381 | 0.912 | 0.0 |
| Primary Liver Cancer | 7 | 0.950(0.879–1.027) | 0.199 | 0.444 | 0.0 | 0.919(0.778–1.086) | 0.320 | 0.313 | 15.3 | 0.969(0.840–1.118) | 0.666 | 0.103 | 43.2 | 0.951(0.831–1.088) | 0.463 | 0.139 | 38.0 | 0.924(0.820–1.040) | 0.191 | 0.641 | 0.0 |
| Prostate Cancer | 2 | 0.801(0.660–0.971) | 0.024 | 0.200 | 39.1 | 0.565(0.354–0.900) | 0.016 | 0.234 | 29.5 | 0.761(0.509–1.137) | 0.182 | 0.384 | 0.0 | 0.685(0.466–1.007) | 0.054 | 0.340 | 0.0 | 0.757(0.568–1.008) | 0.057 | 0.235 | 29.1 |
| PTC | 4 | 1.070(0.958–1.196) | 0.230 | 0.520 | 0.0 | 0.639(0.321–1.272) | 0.202 | 0.040 | 63.9 | 1.319(0.985–1.768) | 0.063 | 0.042 | 63.4 | 1.189(1.009–1.402) | 0.039 | 0.164 | 41.2 | 0.547(0.244–1.227) | 0.143 | 0.006 | 75.6 |
| SCCHN | 5 | 1.160(0.956–1.407) | 0.133 | 0.005 | 76.3 | 1.223(0.981–1.526) | 0.074 | 0.134 | 46.3 | 1.147(1.003–1.311) | 0.045 | 0.366 | 5.3 | 1.165(1.035–1.310) | 0.011 | 0.349 | 10.0 | 1.187(0.807–1.744) | 0.384 | 0.003 | 78.4 |
| other | 5 | 1.103(0.969–1.255) | 0.136 | 0.173 | 37.3 | 1.673(1.163–2.408) | 0.006 | 0.285 | 20.4 | 1.026(0.867–1.215) | 0.763 | 0.758 | 0.0 | 1.093(0.930–1.285) | 0.281 | 0.705 | 0.0 | 1.174(0.603–2.285) | 0.638 | 0.007 | 71.8 |
| Ethnicity | |||||||||||||||||||||
| Caucasian | 19 | 1.069(1.015–1.126) | 0.011 | 0.476 | 0.0 | 1.183(1.030–1.359) | 0.017 | 0.193 | 22.0 | 1.077(0.978–1.185) | 0.131 | 0.017 | 46.2 | 1.074(1.009–1.142) | 0.024 | 0.135 | 27.0 | 1.162(1.014–1.331) | 0.030 | 0.076 | 34.4 |
| Asian | 28 | 0.926(0.870–0.986) | 0.017 | <0.001 | 68.5 | 0.868(0.762–0.989) | 0.033 | <0.001 | 64.2 | 0.899(0.848–0.953) | <0.001 | 0.06 | 31.3 | 0.899(0.822–0.983) | 0.020 | 0.001 | 52.5 | 0.907(0.827–0.995) | 0.039 | 0.001 | 62.9 |
| Design | |||||||||||||||||||||
| HB | 21 | 0.903(0.845–0.964) | 0.002 | <0.001 | 62.8 | 0.821(0.716–0.942) | 0.005 | <0.001 | 60.3 | 0.908(0.829–0.995) | 0.038 | 0.026 | 41.8 | 0.893(0.809–0.986) | 0.025 | 0.001 | 57.4 | 0.869(0.795–0.951) | 0.002 | 0.015 | 45.4 |
| PB | 22 | 1.046(0.983–1.113) | 0.159 | 0.010 | 46.0 | 1.108(0.940–1.306) | 0.223 | 0.008 | 47.1 | 1.027(0.941–1.122) | 0.548 | 0.023 | 41.4 | 1.044(0.961–1.134) | 0.307 | 0.033 | 39.0 | 1.087(0.940–1.256) | 0.261 | 0.001 | 56.4 |
| Sample size | |||||||||||||||||||||
| ≥1000 | 16 | 1.015(0.947–1.088) | 0.676 | <0.001 | 75.0 | 1.074(0.924–1.247) | 0.352 | <0.001 | 73.1 | 0.949(0.880–1.022) | 0.167 | 0.024 | 45.6 | 0.976(0.895–1.064) | 0.583 | <0.001 | 63.6 | 1.085(0.967–1.218) | 0.164 | 0.001 | 69.8 |
| <1000 | 31 | 0.947(0.882–1.016) | 0.128 | 0.001 | 51.6 | 0.836(0.712–0.982) | 0.029 | 0.010 | 41.3 | 1.015(0.913–1.129) | 0.783 | 0.005 | 44.9 | 0.993(0.897–1.100) | 0.898 | 0.001 | 50.4 | 0.841(0.752–0.940) | 0.002 | 0.042 | 33.1 |
| HWE | |||||||||||||||||||||
| Yes | 42 | 0.982(0.931–1.037) | 0.517 | <0.001 | 66.1 | 0.940(0.833–1.060) | 0.315 | <0.001 | 62.4 | 0.986(0.920–1.056) | 0.683 | 0.001 | 47.0 | 0.981(0.912–1.057) | 0.619 | <0.001 | 58.2 | 0.959(0.875–1.051) | 0.368 | <0.001 | 59.7 |
ESCC: esophageal squamous cell carcinoma; PTC: papillary thyroid carcinoma; SCCHN: squamous cell carcinoma of the head and neck; HB: hospital based; PB: population based; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; P: p value; P-H:P value of Q for heterogeneity test; I2: 0–25%, no heterogeneity; 25–50%, modest heterogeneity; 50%, high heterogeneity;
Number of studies involved Random effects model was used when P value of Q for heterogeneity test (P-H)<0.05 or I2>50%; otherwise, fixed effect model was used.
However, in the stratified analysis by cancer type, the C allele and CC genotype of rs2910164 were found to be associated with an inverse risk of bladder cancer under all genetic models, except for the recessive model (C vs. G: OR = 0.838, 95% CI = 0.762–0.921, P H = 0.324; CC vs. GG: OR = 0.724, 95% CI = 0.587–0.893, P H = 0.241; GC vs. GG: OR = 0.789, 95% CI = 0.689–0.904, P H = 0.526; CC+GC vs. GG: OR = 0.781, 95% CI = 0.687–0.889, P H <0.290), cervical cancer under all genetic models (C vs. G: OR = 0.719, 95% CI = 0.620–0.839, P H = 0.796; CC vs. GG: OR = 0.503, 95% CI = 0.370–0.684, P H = 0.814; GC vs. GG: OR = 0.721, 95% CI = 0.545–0.953, P H = 0.254; CC+GC vs. GG: OR = 0.632, 95% CI = 0.485–0.823, P H = 0.382; CC vs. GC+GG: OR = 0.654, 95% CI = 0.520–0.822, P H = 0.359), colorectal cancer under allelic contrast, heterozygote comparison and the dominant model (C vs. G: OR = 0.912, 95% CI = 0.833–0.999, P H = 0.324; GC vs. GG: OR = 0.854, 95% CI = 0.740–0.985, P H = 0.376; CC+GC vs. GG: OR = 0.859, 95% CI = 0.750–0.984, P H = 0.294) and prostate cancer under allelic contrast and homozygote comparison (C vs. G: OR = 0.801, 95% CI = 0.660–0.971, P H = 0.200; CC vs. GG: OR = 0.565, 95% CI = 0.354–0.900, P H = 0.234). In addition, rs2910164 was found to be associated with risks of PTC and SCCHN in the heterozygote comparison (CC+GC vs. GG: OR = 1.189, 95% CI = 1.009–1.402, P H = 0.164) and the dominant model (GC vs. GG: OR = 1.147, 95% CI = 1.003–1.311, P H = 0.366). Nevertheless, the direction of ORs in the two cancers was opposite to that of the former four cancers.
When stratified analysis was performed by ethnicity of study population, rs2910164 C allele and CC genotype were shown to be associated with substantial decrease in cancer risk in Asian populations under all genetic models. On the contrary, Caucasian C or CC carriers were more susceptible to cancers under all genetic models, except for heterozygote comparison. Further subgroup analysis revealed the C allele or CC genotype to be associated with decreased cancer risk in studies of hospital-based study design for all genetic models, but not in studies of population based study design. When stratified on the basis of sample size, the CC genotype had an effect of decreased cancer risk among small size subgroups compared with GG genotype or G allele carriers.
miR-196a2 rs11614913
The miR-196a2 rs11614913 polymorphism was analyzed in 38 comparisons with 16,414 cases and 19,465 controls. We also observed a wide variation of the T allele frequency across different ethnicities (Asian: 49.8%, 95% CI = 45.3%–54.3%; Caucasian: 38.8%, 95% CI = 35.9%–41.7%; P = 0.002).
Table 3 summarizes the results from the meta-analysis of miR-196a2 rs11614913 and cancer risk. In the overall analysis, we found a significant association between rs11614913 and reduced cancer risk in the allelic contrast (OR = 0.949, 95% CI = 0.902–0.998, P H <0.001), homozygote comparison (OR = 0.861, 95% CI = 0.772–0.959 P H<0.001) and recessive model (OR = 0.865, 95% CI = 0.802–0.934, P H = 0.002). Removing four studies with genotype frequencies in controls that deviated from HWE did not alter the pooled results [15], [43], [53], [62].
Table 3. Meta-analysis of miR-196a2 rs11614913 polymorphism with cancer risk.
| Variables | na | T vs. C | TT vs. CC | CT vs. CC | TT+CT vs. CC | TT vs. CT+CC | |||||||||||||||
| OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | ||
| Total | 38 | 0.949(0.902–0.998) | 0.044 | <0.001 | 58.2 | 0.861(0.772–0.959) | 0.007 | <0.001 | 58.7 | 1.033(0.951–1.123) | 0.441 | <0.001 | 56.6 | 0.984(0.909–1.065) | 0.685 | <0.001 | 60.0 | 0.865(0.802–0.934) | <0.001 | 0.002 | 45.2 |
| Cancer type | |||||||||||||||||||||
| Bladder Cancer | 2 | 1.032(0.906–1.174) | 0.639 | 0.562 | 0.0 | 0.961(0.724–1.277) | 0.786 | 0.224 | 32.4 | 1.192(0.837–1.696) | 0.331 | 0.106 | 61.7 | 1.102(0.915–1.327) | 0.307 | 0.171 | 46.5 | 0.738(0.320–1.701) | 0.476 | 0.100 | 63.1 |
| Breast Cancer | 8 | 0.978(0.868–1.102) | 0.716 | 0.002 | 68.3 | 0.903(0.699–1.167) | 0.436 | 0.004 | 66.9 | 0.976(0.888–1.074) | 0.623 | 0.12 | 38.9 | 0.989(0.841–1.164) | 0.898 | 0.014 | 60.3 | 0.915(0.765–1.095) | 0.334 | 0.031 | 54.5 |
| Colorectal Cancer | 6 | 0.910(0.794–1.043) | 0.177 | 0.061 | 52.6 | 0.754(0.627–0.907) | 0.003 | 0.108 | 44.6 | 0.878(0.755–1.021) | 0.091 | 0.168 | 35.9 | 0.848(0.735–0.979) | 0.025 | 0.082 | 48.9 | 0.838(0.721–0.974) | 0.021 | 0.165 | 36.3 |
| ESCC | 2 | 0.863(0.551–1.351) | 0.518 | 0.001 | 90.6 | 0.685(0.209–2.245) | 0.532 | <0.001 | 93.9 | 1.166(0.692–1.962) | 0.564 | 0.030 | 78.7 | 1.020(0.537–1.935) | 0.953 | 0.005 | 87.3 | 0.610(0.268–1.390) | 0.240 | 0.001 | 91.4 |
| Gastric Cancer | 2 | 0.893(0.778–1.024) | 0.104 | 0.230 | 30.5 | 0.803(0.608–1.062) | 0.125 | 0.306 | 4.5 | 0.839(0.653–1.077) | 0.167 | 0.163 | 48.5 | 0.819(0.647–1.037) | 0.097 | 0.162 | 48.8 | 0.894(0.722–1.107) | 0.305 | 0.698 | 0.0 |
| Lung Cancer | 4 | 0.893(0.821–0.971) | 0.008 | 0.149 | 43.8 | 0.794(0.672–0.938) | 0.007 | 0.259 | 25.5 | 0.991(0.771–1.274) | 0.945 | 0.059 | 59.7 | 0.935(0.745–1.175) | 0.565 | 0.075 | 56.6 | 0.842(0.737–0.962) | 0.011 | 0.201 | 0.201 |
| Primary Liver Cancer | 5 | 0.890(0.767–1.032) | 0.123 | 0.034 | 61.7 | 0.790(0.589–1.061) | 0.117 | 0.041 | 59.8 | 0.873(0.754–1.010) | 0.068 | 0.776 | 0.0 | 0.859(0.748–0.986) | 0.030 | 0.334 | 12.5 | 0.871(0.690–1.100) | 0.248 | 0.043 | 59.4 |
| SCCHN | 4 | 1.067(0.965–1.179) | 0.205 | 0.442 | 0.0 | 1.241(0.841–1.831) | 0.276 | 0.099 | 63.3 | 1.490(0.835–2.658) | 0.177 | 0.006 | 86.7 | 1.123(0.851–1.481) | 0.413 | 0.006 | 76.1 | 0.948(0.797–1.127) | 0.544 | 0.683 | 0.0 |
| other | 5 | 1.026(0.928–1.135) | 0.613 | 0.352 | 9.5 | 0.966(0.776–1.201) | 0.754 | 0.491 | 0.0 | 1.306(1.106–1.542) | 0.002 | 0.188 | 34.9 | 1.212(1.035–1.419) | 0.017 | 0.159 | 39.3 | 0.853(0.716–1.017) | 0.076 | 0.720 | 0.0 |
| Ethnicity | |||||||||||||||||||||
| Caucasian | 14 | 0.981(0.894–1.076) | 0.683 | 0.002 | 61.8 | 0.934(0.766–1.138) | 0.496 | 0.003 | 61.7 | 1.023(0.946–1.108) | 0.565 | 0.3 | 14.7 | 0.989(0.895–1.092) | 0.825 | 0.048 | 42.3 | 0.918(0.788–1.070) | 0.276 | 0.035 | 47.1 |
| Asian | 24 | 0.934(0.879–0.991) | 0.025 | 0.001 | 55.0 | 0.827(0.727–0.940) | 0.004 | 0.001 | 55.5 | 1.043(0.924–1.177) | 0.500 | <0.001 | 65.9 | 0.986(0.878–1.107) | 0.808 | <0.001 | 66.6 | 0.845(0.773–0.923) | <0.001 | 0.011 | 44.1 |
| Design | |||||||||||||||||||||
| HB | 21 | 0.918(0.855–0.986) | 0.019 | <0.001 | 64.5 | 0.849(0.726–0.993) | 0.040 | <0.001 | 69.5 | 0.997(0.881–1.127) | 0.956 | <0.001 | 63.4 | 0.946(0.842–1.064) | 0.355 | <0.001 | 65.6 | 0.848(0.763–0.942) | 0.002 | 0.001 | 56.4 |
| PB | 14 | 0.963(0.916–1.012) | 0.132 | 0.152 | 29.2 | 0.869(0.783–0.966) | 0.009 | 0.553 | 0.0 | 1.064(0.943–1.201) | 0.314 | 0.024 | 48.9 | 1.011(0.907–1.127) | 0.843 | 0.022 | 48.5 | 0.908(0.832–0.991) | 0.031 | 0.525 | 0.0 |
| Sample size | |||||||||||||||||||||
| ≥1000 | 11 | 0.941(0.904–0.979) | 0.003 | 0.131 | 33.5 | 0.880(0.811–0.955) | 0.002 | 0.143 | 32.0 | 0.964(0.902–1.031) | 0.287 | 0.085 | 39.6 | 0.936(0.857–1.021) | 0.137 | 0.048 | 45.8 | 0.904(0.845–0.966) | 0.003 | 0.593 | 0.0 |
| <1000 | 27 | 0.966(0.889–1.050) | 0.416 | <0.001 | 65.0 | 0.854(0.709–1.029) | 0.098 | <0.001 | 65.4 | 1.092(0.959–1.243) | 0.183 | <0.001 | 60.3 | 0.835(0.769–0.907) | <0.001 | <0.001 | 64.4 | 0.833(0.729–0.952) | 0.007 | <0.001 | 55.3 |
| HWE | |||||||||||||||||||||
| Yes | 32 | 0.929(0.884–0.977) | 0.004 | <0.001 | 55.1 | 0.851(0.763–0.948) | 0.003 | <0.001 | 58.7 | 0.990(0.914–1.073) | 0.815 | 0.001 | 50.9 | 0.948(0.874–1.028) | 0.196 | <0.001 | 56.8 | 0.863(0.800–0.931) | <0.001 | 0.004 | 44.7 |
ESCC: esophageal squamous cell carcinoma; SCCHN: squamous cell carcinoma of the head and neck; HB: hospital based; PB: population based; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; P: p value; P-H: P value of Q for heterogeneity test; I2: 0–25%, no heterogeneity; 25–50%, modest heterogeneity; 50%, high heterogeneity;
Number of studies involved.
Random effects model was used when P value of Q for heterogeneity test (P-H) <0.05 or I2>50%; otherwise, fixed effect model was used.
In subgroup analysis by cancer type, significant association between rs11614913 and decreased cancer risk was found for lung cancer (T vs. C: OR = 0.893, 95% CI = 0.821–0.971, P H = 0.149; TT vs. CC: OR = 0.794, 95% CI = 0.627–0.938, P H = 0.259; TT vs. CT+CC: OR = 0.842, 95% CI = 0.737–0.962, P H = 0.201) and colorectal cancer (TT vs. CC: OR = 0.754, 95% CI = 0.627–0.907, P H = 0.108; TT+CT vs. CC: OR = 0.848, 95% CI = 0.735–0.979, P H = 0.082; TT vs. CT+CC: OR = 0.838, 95% CI = 0.721–0.974, P H = 0.165). For liver cancer, T allele carriers showed decreased cancer susceptibility compared with homozygote CC (OR = 0.859, 95% CI = 0.748–0.986, P H = 0.334). However, no association was found between rs11614913 and bladder cancer, breast cancer, ESCC, gastric cancer, or SCCHN.
In ethnic subgroup analysis, a strong association was found between rs11614913 and cancer risk in the allelic contrast, the homozygote comparison, and the recessive model among Asians, whereas negative results were obtained for Caucasians in all genetic models. With respect to the control source, decreased risk was observed in both the hospital- and population-based controls for the homozygote comparison and the recessive model. We also found a reduced risk for allelic contrast in hospital-based studies. In stratified analysis by sample size, significant association of decreased cancer risk was found in both of the subgroups.
miR-499 rs3746444
For miR-499 rs3746444, 21 comparisons with 8,888 cases and 10,292 controls were included. No significant difference in C allele frequency between Asians and Caucasians was observed (Asian: 22.2%, 95% CI = 16.7%–27.7%; Caucasian: 29.9%, 95% CI = 14.4%–45.4%; P = 0.178).
The results of the meta-analysis for miR-499 rs3746444 and the risk of cancer are presented in Table 4. Overall, we observed that rs3746444 could decrease the cancer risk in the allelic contrast (OR = 1.106, 95% CI = 1.005–1.218, P H <0.001) and the dominant model (OR = 1.148, 95% CI = 1.020–1.292, P H <0.001). However, this association disappeared after the exclusion of six studies [15], [35], [40], [52], [62], [80], whose genotypic distribution in controls was derived from HWE.
Table 4. Meta-analysis of miR-499 rs3746444 polymorphism with cancer risk.
| Variables | na | C vs. T | CC vs. TT | TC vs. TT | CC+TC vs. TT | CC vs. TC+TT | |||||||||||||||
| OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | ||
| Total | 21 | 1.106(1.005–1.218) | 0.040 | <0.001 | 67.2 | 1.167(0.969–1.405) | 0.103 | 0.042 | 38.3 | 1.126(0.985–1.288) | 0.081 | <0.001 | 71.9 | 1.148(1.020–1.292) | 0.022 | <0.001 | 69.0 | 1.100(0.903–1.339) | 0.344 | 0.007 | 49.4 |
| Cancer type | |||||||||||||||||||||
| Breast Cancer | 4 | 1.101(1.006–1.204) | 0.036 | 0.214 | 33.0 | 1.165(0.915–1.482) | 0.215 | 0.189 | 37.1 | 1.163(0.952–1.420) | 0.140 | 0.047 | 62.3 | 1.150(0.973–1.359) | 0.102 | 0.102 | 51.7 | 1.065(0.712–1.595) | 0.758 | 0.059 | 59.8 |
| Colorectal Cancer | 2 | 1.136(0.938–1.375) | 0.192 | 0.161 | 49.1 | 1.557(0.670–3.621) | 0.304 | 0.096 | 63.9 | 0.867(0.541–1.390) | 0.554 | 0.100 | 63.0 | 1.045(0.831–1.314) | 0.705 | 0.964 | 0.0 | 1.645(0.611–4.428) | 0.325 | 0.047 | 74.6 |
| Lung Cancer | 2 | 0.963(0.822–1.129) | 0.643 | 0.828 | 0.0 | 0.880(0.538–1.439) | 0.610 | 0.919 | 0.0 | 0.981(0.812–1.185) | 0.843 | 0.595 | 0.0 | 0.970(0.809–1.163) | 0.742 | 0.682 | 0.0 | 0.874(0.537–1.424) | 0.589 | 0.836 | 0.0 |
| Primary Liver Cancer | 4 | 1.094(0.737–1.623) | 0.656 | <0.001 | 83.3 | 1.187(0.560–2.516) | 0.655 | 0.017 | 70.5 | 1.007(0.696–1.458) | 0.970 | 0.048 | 62.1 | 1.074(0.685–1.683) | 0.757 | 0.004 | 77.2 | 1.201(0.675–2.136) | 0.533 | 0.065 | 58.6 |
| SCCHN | 3 | 1.290(0.593–2.804) | 0.521 | <0.001 | 95.4 | 1.774(0.429–7.328) | 0.429 | 0.030 | 78.6 | 1.220(0.527–2.821) | 0.643 | <0.001 | 94.8 | 1.289(0.751–2.215) | 0.357 | <0.001 | 91.9 | 1.685(0.503–5.643) | 0.398 | 0.061 | 71.5 |
| other | 6 | 1.103(0.988–1.231) | 0.081 | 0.124 | 42.2 | 1.078(0.823–1.413) | 0.585 | 0.494 | 0.0 | 1.280(0.935–1.753) | 0.123 | <0.001 | 77.7 | 1.223(0.947–1.579) | 0.124 | 0.006 | 69.7 | 0.946(0.730–1.226) | 0.675 | 0.121 | 42.6 |
| Ethnicity | |||||||||||||||||||||
| Caucasian | 7 | 1.003(0.926–1.086) | 0.951 | 0.197 | 31.8 | 1.110(0.912–1.352) | 0.299 | 0.233 | 26.9 | 0.939(0.846–1.042) | 0.236 | 0.143 | 39.4 | 0.997(0.908–1.095) | 0.952 | 0.166 | 34.3 | 1.139(0.948–1.368) | 0.166 | 0.088 | 47.8 |
| Asian | 14 | 1.142(0.999–1.305) | 0.052 | <0.001 | 71.5 | 1.169(0.894–1.529) | 0.253 | 0.036 | 44.7 | 1.234(1.035–1.471) | 0.019 | <0.001 | 73.9 | 1.220(1.032–1.442) | 0.020 | <0.001 | 73.7 | 1.039(0.781–1.381) | 0.794 | 0.009 | 53.4 |
| Design | |||||||||||||||||||||
| HB | 9 | 1.188(0.962–1.465) | 0.109 | <0.001 | 82.6 | 1.283(0.910–1.809) | 0.156 | 0.036 | 53.3 | 1.163(0.899–1.504) | 0.250 | <0.001 | 80.5 | 1.216(0.965–1.533) | 0.097 | <0.001 | 80.7 | 1.224(1.004–1.491) | 0.045 | 0.101 | 39.2 |
| PB | 10 | 1.055(0.985–1.130) | 0.127 | 0.088 | 40.4 | 1.061(0.881–1.278) | 0.533 | 0.369 | 7.9 | 1.142(0.969–1.346) | 0.114 | 0.001 | 67.4 | 1.110(0.961–1.282) | 0.156 | 0.006 | 60.8 | 0.968(0.808–1.160) | 0.726 | 0.097 | 41.5 |
| Sample size | |||||||||||||||||||||
| ≥1000 | 6 | 1.031(0.930–1.143) | 0.560 | 0.039 | 57.4 | 1.135(0.946–1.362) | 0.173 | 0.259 | 23.3 | 0.992(0.913–1.077) | 0.840 | 0.076 | 49.9 | 1.015(0.901–1.142) | 0.809 | 0.049 | 55.1 | 1.135(0.948–1.359) | 0.167 | 0.293 | 18.5 |
| <1000 | 15 | 1.157(0.996–1.343) | 0.056 | <0.001 | 68.7 | 1.165(0.869–1.562) | 0.307 | 0.030 | 46.1 | 1.225(0.985–1.523) | 0.068 | <0.001 | 73.7 | 1.241(1.038–1.485) | 0.018 | <0.001 | 68.5 | 1.043(0.762–1.429) | 0.791 | 0.003 | 58.5 |
| HWE | |||||||||||||||||||||
| Yes | 14 | 1.049(0.939–1.172) | 0.397 | <0.001 | 67.8 | 1.063(0.902–1.252) | 0.465 | 0.275 | 16.4 | 1.109(0.952–1.293) | 0.184 | <0.001 | 74.0 | 1.094(0.947–1.265) | 0.222 | <0.001 | 73.2 | 1.006(0.857–1.181) | 0.938 | 0.111 | 33.0 |
SCCHN: squamous cell carcinoma of the head and neck; HB: hospital based; PB: population based; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; P: p value; P-H: P value of Q for heterogeneity test; I2: 0–25%, no heterogeneity; 25–50%, modest heterogeneity; 50%, high heterogeneity;
Number of studies involved.
Random effects model was used when P value of Q for heterogeneity test (P-H)<0.05 or I2>50%; otherwise, fixed effect model was used.
In stratified analysis by cancer type, significant associations were only maintained in breast cancer under allelic contrast (OR = 1.101, 95% CI = 1.006–1.204, P H = 0.214), but no significant association was observed with colorectal cancer, lung cancer, liver cancer, SCCHN, and other cancers under any genetic model. Subgroup analysis by ethnicity showed a decreased cancer risk in the Asian population (TC vs. TT: OR = 1.234, 95% CI = 1.035–1.471, P H <0.001; TC+CC vs. TT: OR = 1.220, 95% CI = 1.032–1.442, P H <0.001), but not in the Caucasian population. Based on study design, studies with hospital-based controls showed elevated risk (CC vs. TC+TT: OR = 1.224, 95% CI = 1.004–1.491, P H = 0.045). However, studies with population-based controls showed no significant association. Further subgroup analysis by sample size revealed increased cancer risks only in a small sample group using the dominant model (TC+CC vs. TT: OR = 1.241, 95% CI = 1.038–1.485, P H <0.001).
miR-149 rs2292832
Twelve comparisons with 5926 cases and 5961 controls assessed for the association between miR-149 rs2292832 polymorphism and cancer risk. The frequency of T allele was significant higher in Asian population compared to that in Caucasian population (Asian: 65.1%, 95% CI = 53.2%–77.0%; Caucasian: 30.6%, 95% CI = 25.2%–36.0%; P = 0.003).
Overall, none of the genetic models produced significant association between rs2292832 and cancer risk. Similarly, no positive result was found in most of the subgroups, except that homozygote TT had an effect of increasing risk of other cancers compared with C allele carriers (OR = 1.388, 95% CI = 1.083–1.778, P H = 0.427) and significant association with increased cancer risk was also found in small sample group for allelic contrast (OR = 1.106, 95% CI = 1.012–1.209, P H = 0.461) and recessive model (OR = 1.217, 95% CI = 1.078–1.373, P H = 0.380). These results are summarized in Table 5.
Table 5. Meta-analysis of miR-149 rs2292832 polymorphism with cancer risk.
| Variables | na | T vs. C | TT vs. CC | CT vs. CC | TT+CT vs. CC | TT vs. CT+CC | |||||||||||||||
| OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | ||
| Total | 12 | 1.022(0.966–1.082) | 0.449 | 0.283 | 16.8 | 1.002(0.880–1.140) | 0.980 | 0.529 | 0.0 | 0.950(0.862–1.048) | 0.306 | 0.971 | 0.0 | 0.975(0.892–1.065) | 0.571 | 0.979 | 0.0 | 1.082(0.990–1.183) | 0.083 | 0.109 | 36.2 |
| Cancer type | |||||||||||||||||||||
| Breast Cancer | 2 | 1.078(0.860–1.350) | 0.515 | 0.121 | 58.4 | 1.043(0.794–1.369) | 0.763 | 0.361 | 0.0 | 0.977(0.821–1.162) | 0.789 | 0.907 | 0.0 | 0.991(0.839–1.169) | 0.911 | 0.656 | 0.0 | 1.170(0.819–1.670) | 0.388 | 0.123 | 58.0 |
| Colorectal Cancer | 3 | 1.063(0.935–1.207) | 0.352 | 0.979 | 0.0 | 1.072(0.807–1.424) | 0.631 | 0.619 | 0.0 | 0.856(0.660–1.109) | 0.238 | 0.996 | 0.0 | 0.942(0.738–1.202) | 0.629 | 0.995 | 0.0 | 1.161(0.972–1.386) | 0.100 | 0.628 | 0.0 |
| Lung Cancer | 2 | 1.090(0.762–1.560) | 0.638 | 0.073 | 68.8 | 1.259(0.547–2.902) | 0.588 | 0.058 | 72.1 | 0.986(0.764–1.273) | 0.915 | 0.562 | 0.0 | 0.995(0.782–1.266) | 0.966 | 0.238 | 28.2 | 1.234(0.598–2.545) | 0.569 | 0.071 | 69.4 |
| SCCHN | 3 | 0.957(0.853–1.074) | 0.458 | 0.688 | 0.0 | 0.863(0.661–1.126) | 0.277 | 0.619 | 0.0 | 0.976(0.826–1.154) | 0.776 | 0.323 | 0.0 | 0.966(0.839–1.111) | 0.626 | 0.651 | 0.0 | 0.930(0.753–1.149) | 0.503 | 0.742 | 0.0 |
| other | 2 | 1.200(0.997–1.444) | 0.054 | 0.259 | 21.6 | 1.181(0.790–1.767) | 0.417 | 0.271 | 17.3 | 0.825(0.550–1.239) | 0.354 | 0.409 | 0.0 | 0.992(0.677–1.452) | 0.965 | 0.312 | 2.0 | 1.388(1.083–1.778) | 0.010 | 0.427 | 0.0 |
| Ethnicity | |||||||||||||||||||||
| Caucasian | 4 | 1.014(0.904–1.139) | 0.810 | 0.220 | 33.9 | 1.261(0.749–2.123) | 0.383 | 0.102 | 56.2 | 0.991(0.848–1.160) | 0.914 | 0.688 | 0.0 | 1.000(0.876–1.143) | 0.996 | 0.767 | 0.0 | 1.286(0.761–2.171) | 0.348 | 0.082 | 59.9 |
| Asian | 8 | 1.025(0.960–1.095) | 0.463 | 0.255 | 22.0 | 0.986(0.851–1.143) | 0.852 | 0.745 | 0.0 | 0.925(0.817–1.048) | 0.222 | 0.949 | 0.0 | 0.955(0.849–1.075) | 0.447 | 0.945 | 0.0 | 1.085(0.986–1.193) | 0.094 | 0.154 | 34.3 |
| Design | |||||||||||||||||||||
| HB | 4 | 0.984(0.891–1.087) | 0.756 | 0.599 | 0.0 | 0.901(0.718–1.130) | 0.368 | 0.733 | 0.0 | 0.960(0.821–1.123) | 0.613 | 0.532 | 0.0 | 0.963(0.843–1.100) | 0.577 | 0.831 | 0.0 | 1.012(0.860–1.190) | 0.890 | 0.451 | 0.0 |
| PB | 6 | 1.029(0.957–1.107) | 0.437 | 0.156 | 37.6 | 1.007(0.854–1.187) | 0.934 | 0.645 | 0.0 | 0.943(0.826–1.077) | 0.387 | 0.923 | 0.0 | 0.970(0.855–1.101) | 0.636 | 0.917 | 0.0 | 1.092(0.979–1.217) | 0.116 | 0.079 | 49.4 |
| Sample size | |||||||||||||||||||||
| ≥1000 | 3 | 0.967(0.898–1.042) | 0.383 | 0.901 | 0.0 | 0.924(0.778–1.098) | 0.370 | 0.892 | 0.0 | 0.984(0.877–1.104) | 0.781 | 0.950 | 0.0 | 0.971(0.871–1.084) | 0.604 | 0.920 | 0.0 | 0.940(0.823–1.073) | 0.357 | 0.891 | 0.0 |
| <1000 | 9 | 1.106(1.012–1.209) | 0.027 | 0.461 | 0.0 | 1.111(0.914–1.350) | 0.292 | 0.442 | 0.0 | 0.870(0.724–1.046) | 0.138 | 0.957 | 0.0 | 0.981(0.845–1.139) | 0.802 | 0.902 | 0.0 | 1.217(1.078–1.373) | 0.001 | 0.380 | 0.0 |
| HWE | |||||||||||||||||||||
| Yes | 10 | 1.029(0.970–1.091) | 0.346 | 0.261 | 19.8 | 1.019(0.892–1.164) | 0.781 | 0.534 | 0.0 | 0.957(0.867–1.057) | 0.389 | 0.977 | 0.0 | 0.981(0.893–1.078) | 0.693 | 0.973 | 0.0 | 1.095(0.997–1.202) | 0.058 | 0.090 | 40.2 |
SCCHN: squamous cell carcinoma of the head and neck; HB: hospital based; PB: population based; HWE: Hardy-Weinberg equilibrium; OR: odds ratio; CI: confidence interval; P: p value; P-H: P value of Q for heterogeneity test; I2: 0–25%, no heterogeneity; 25–50%, modest heterogeneity; 50%, high heterogeneity;
Number of studies involved.
Random effects model was used when P value of Q for heterogeneity test (P-H)<0.05 or I2>50%; otherwise, fixed effect model was used.
miR-27a rs895919
For miR-27a rs895919, we collected nine comparisons with 4662 cases and 5625 controls. No significant difference in G allele frequency between Asians and Caucasians was observed (Asian: 32,4%, 95% CI = 21.2%–43.6%; Caucasian: 32.1%, 95% CI = 28.7%–35.6%; P = 0.949).
Overall, there was no significant association observed in all comparisons. However, in subgroup analysis, a decreased risk was found in other cancers (AG vs. AA: OR = 0.828, 95% CI = 0.698–0.982, P H = 0.030; GG+AG vs. AA: OR = 0.821, 95% CI = 0.698–0.966, P H = 0.017), large sample groups (G vs. A: OR = 0.875, 95% CI = 0.811–0.945, P H = 0.001; AG vs. AA: OR = 0.806, 95% CI = 0.726–0.895, P H <0.001; GG+AG vs. AA: OR = 0.815, 95% CI = 0.738–0.900, P H <0.001), the Caucasian population (AG vs. AA: OR = 0.879, 95% CI = 0.792–0.975, P H = 0.015) and population-based studies (G vs. A: OR = 0.900, 95% CI = 0.830–0.975, P H = 0.010) (Table 6).
Table 6. Meta-analysis of miR-27a rs895919 polymorphism with cancer risk.
| Variables | na | G vs. A | GG vs. AA | AG vs. AA | GG+AG vs. AA | GG vs. AG+AA | |||||||||||||||
| OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | OR(95% CI) | P | P-H | I2 | ||
| Total | 9 | 0.945(0.853–1.048) | 0.284 | 0.008 | 61.1 | 0.897(0.714–1.127) | 0.352 | 0.017 | 56.9 | 0.980(0.836–1.149) | 0.805 | 0.001 | 68.6 | 0.959(0.828–1.112) | 0.581 | 0.002 | 67.1 | 0.891(0.722–1.101) | 0.286 | 0.023 | 55.0 |
| Cancer type | |||||||||||||||||||||
| Breast Cancer | 4 | 0.930(0.864–1.002) | 0.056 | 0.155 | 42.7 | 0.915(0.775–1.081) | 0.295 | 0.419 | 0.0 | 1.009(0.788–1.292) | 0.944 | 0.003 | 78.5 | 0.977(0.797–1.199) | 0.825 | 0.014 | 71.6 | 0.929(0.793–1.087) | 0.357 | 0.165 | 41.0 |
| Gastric Cancer | 2 | 1.029(0.587–1.803) | 0.922 | 0.001 | 90.9 | 0.717(0.122–4.227) | 0.713 | <0.001 | 92.6 | 1.152(0.765–1.734) | 0.499 | 0.078 | 67.8 | 1.116(0.629–1.977) | 0.708 | 0.009 | 85.2 | 0.668(0.139–3.220) | 0.616 | 0.001 | 91.0 |
| other | 3 | 0.862(0.759–0.979) | 0.022 | 0.513 | 0.0 | 0.794(0.586–1.077) | 0.139 | 0.643 | 0.0 | 0.828(0.698–0.982) | 0.030 | 0.153 | 46.7 | 0.821(0.698–0.966) | 0.017 | 0.259 | 26.1 | 0.865(0.645–1.161) | 0.335 | 0.512 | 0.0 |
| Ethnicity | |||||||||||||||||||||
| Caucasian | 4 | 0.929(0.862–1.002) | 0.055 | 0.157 | 42.4 | 0.924(0.780–1.094) | 0.359 | 0.421 | 0.0 | 0.879(0.792–0.975) | 0.015 | 0.116 | 49.2 | 0.914(0.782–1.068) | 0.258 | 0.099 | 52.1 | 0.983(0.836–1.156) | 0.84 | 0.647 | 0.0 |
| Asian | 5 | 0.938(0.769–1.144) | 0.526 | 0.004 | 74.0 | 0.788(0.485–1.280) | 0.336 | 0.004 | 74.4 | 1.064(0.789–1.435) | 0.685 | 0.001 | 77.8 | 1.008(0.762–1.334) | 0.954 | 0.002 | 77.1 | 0.744(0.480–1.154) | 0.187 | 0.006 | 72.4 |
| Design | |||||||||||||||||||||
| HB | 5 | 0.938(0.765–1.149) | 0.536 | 0.004 | 73.9 | 0.801(0.483–1.329) | 0.390 | 0.003 | 0.003 | 0.964(0.760–1.223) | 0.762 | 0.022 | 65.0 | 0.946(0.739–1.212) | 0.662 | 0.008 | 70.8 | 0.819(0.529–1.270) | 0.373 | 0.013 | 68.5 |
| PB | 3 | 0.900(0.830–0.975) | 0.010 | 0.745 | 0.0 | 0.864(0.722–1.034) | 0.112 | 0.970 | 0.0 | 0.978(0.726–1.316) | 0.881 | 0.004 | 81.9 | 0.921(0.742–1.143) | 0.455 | 0.039 | 69.2 | 0.889(0.750–1.053) | 0.174 | 0.197 | 38.4 |
| Sample size | |||||||||||||||||||||
| ≥1000 | 3 | 0.875(0.811–0.945) | 0.001 | 0.575 | 0.0 | 0.855(0.717–1.018) | 0.078 | 0.936 | 0.0 | 0.806(0.726–0.895) | <0.001 | 0.200 | 37.9 | 0.815(0.738–0.900) | <0.001 | 0.296 | 17.8 | 0.944(0.798–1.117) | 0.503 | 0.790 | 0.0 |
| <1000 | 6 | 1.008(0.857–1.184) | 0.927 | 0.017 | 63.6 | 0.895(0.583–1.374) | 0.611 | 0.004 | 71.0 | 1.127(0.985–1.291) | 0.082 | 0.186 | 33.4 | 1.087(0.956–1.236) | 0.205 | 0.080 | 49.2 | 0.817(0.547–1.220) | 0.323 | 0.004 | 70.9 |
HB: hospital based; PB: population based; OR: odds ratio; CI: confidence interval; P: p value; P-H: P value of Q for heterogeneity test; I2:0–25%, no heterogeneity; 25–50%, modest heterogeneity; 50%, high heterogeneity;
Number of studies involved. Random effects model was used when P value of Q for heterogeneity test (P-H) <0.05 or I2>50%; otherwise, fixed effect model was used.
Test of Heterogeneity
Heterogeneity between studies was observed in overall comparisons and subgroup analyses across the studies of rs2910164, rs11614913, rs3746444, and rs895919. Then we evaluated the source of heterogeneity for allelic contrast by cancer type, ethnicity, source of controls and sample size. For rs2910164, cancer type (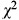 = 51.58, df = 11, P<0.001), ethnicity (
= 51.58, df = 11, P<0.001), ethnicity (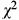 = 24.43, df = 1, P<0.001) and control type (
= 24.43, df = 1, P<0.001) and control type ( = 29.55, df = 3, P<0.001) provided potential sources of between-study heterogeneity. For rs11614913, cancer type (
= 29.55, df = 3, P<0.001) provided potential sources of between-study heterogeneity. For rs11614913, cancer type (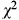 = 17.84, df = 8, P = 0.002) and control type (
= 17.84, df = 8, P = 0.002) and control type (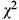 = 13.08, df = 3, P = 0.004) was found to contribute to substantial heterogeneity. For rs3746444, ethnicity (
= 13.08, df = 3, P = 0.004) was found to contribute to substantial heterogeneity. For rs3746444, ethnicity (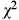 = 4.92, df = 1, P = 0.027) and sample size (
= 4.92, df = 1, P = 0.027) and sample size ( = 4.6, df = 1, P = 0.032) contributed substantially to heterogeneity. For miR-27a rs895919, sample size (
= 4.6, df = 1, P = 0.032) contributed substantially to heterogeneity. For miR-27a rs895919, sample size (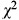 = 5.74, df = 1, P = 0.017) was the main source of between-study heterogeneity.
= 5.74, df = 1, P = 0.017) was the main source of between-study heterogeneity.
Sensitivity Analysis
Influence of each study involved in the meta-analysis on the pooled ORs for each of the studied SNPs was examined by repeating the meta-analysis and omitting each study one at a time. The corresponding pooled ORs were not materially altered.
Publication Bias
We conducted Begg’s funnel plot and Egger’s test to assess the publication bias of included studies for all the SNPs. For miR-146a rs2910164 (Figure S1), miR-196a2 rs11614913 (Figure S2) and miR-499 rs3746444 (Figure S3), no evidence of publication bias was suggested in the results from the Begg’s funnel plot and Egger’s test for allelic contrast. Similar results were observed in other models (data not shown). However, for miR-149 rs2292832 (Figure S4), significant publication bias was found in allelic contrast (P = 0.006), homozygote comparison (P = 0.005) and the recessive model (P = 0.007). For miR-27a rs895919 (Figure S5), no evidence of publication bias was detected for allelic contrast, but publication bias was found in the heterozygote comparison (P = 0.039), probably due to the small number of included studies.
Discussion
In the present study, we performed five independent meta-analyses to investigate the association between cancer risk and polymorphisms in miRNA (miR-146a rs2910164, miR-196a2 rs11614913, miR-499 rs3746444, miR-149 rs2292832, and miR-27a rs895919). The results demonstrated that the rs2910164 C allele or CC genotype was a protective factor for bladder cancer, prostate cancer, cervical cancer and colorectal cancer, but a risk factor for PTC and SCCHN. The significant association between rs2910164 and cancer risk was observed in both Asians and Caucasians, although in opposite directions. The miR-196a2 rs11614913 T allele was observed to be significantly associated with reduced cancer risk, especially for lung cancer and colorectal cancer, particularly in the Asian population. The miR-499 rs3746444 C allele increased cancer risk in the allelic contrast model and in the dominant model, especially in breast cancer. Nevertheless, this association was only observed in Asians, not in Caucasians. On the other hand, mir-149 rs2292832 and miR-27a rs895919 were not significantly related to cancer susceptibility.
Several meta-analyses have been conducted on a single miRNA SNP [88]–[110] or several miRNA SNPs [21], [111]–[120] associated with the risk of cancer(s). However, none of the meta-analyses have comprehensively covered all the studies on a particular miRNA SNP or all the commonly studied miRNA SNPs. In this study, we included all the papers published to date on the five commonly studied miRNA SNPs associated with cancer susceptibility, and in so doing incorporated more studies and cancer types than the previously published meta-analyses. For example, compared to the recently published meta-analysis by He et al. [112], our paper included several new studies for each of the miRNA SNPs. For mir-146a rs2910164, 19 new studies were added; for mir-196a rs11614913, 11 new studies were added; for mir-499 rs3746444, 6 new studies were added; and for mir-149 rs2292832, 5 new studies were added. In addition, we analyzed 9 case-control studies on miR-27a rs895919, which were not included in the meta-analysis by He et al. [112]. Thus, to the best of our knowledge, the present study is the most comprehensive and robust meta-analysis when compared to previously published meta-analyses in this field [21], [88]–[120].
The rs2910164 (miR-146a) locus resides at position +60 relative to the first nucleotide of the pre-miR-146a gene. This polymorphism presents as a change from G to C in the passenger strand, resulting in a change from the G:U pair to the C:U mismatch in the stem structure of the miR-146a precursor [17]. The C-allelic miR-146a precursor has lower transcriptional activity than the G-allele due to decreased nuclear primiR-146a processing efficiency; this leads to low levels of mature miR-146a and affects target mRNA binding [84], [85]. The decreased amount of miR-146a reduces the inhibition of target genes involved in the Toll-like receptor and cytokine signaling pathway (TRAF6, IRAK1) and impaired nuclear factor (NF)-κB activity [85], [121]. Studies have shown that miR-146a plays an important role in cell proliferation and metastatic ability in some cancers and that its deregulation is possibly involved in carcinogenesis [84], [85], [121]–[123]. However, the meta-analysis results suggested no significant association between this polymorphism and cancer susceptibility in the overall pooled result. In the case of subgroup analysis divided by cancer type, the rs2910164 C allele was associated with a decreased risk of bladder cancer, cervical cancer, colorectal cancer and prostate cancer, but an increased risk of PTC and SCCHN. In contrast to the previously published results by He et al. [112], no significant association was found between rs2910164 and HCC or ESCC. These results suggest that the association between the miR-146a rs2910164 polymorphism and cancer susceptibility was cancer-type dependent. The potential explanation for this phenomenon may be that different cancers have differing pathogenesis. In addition, we found that the association between the rs2910164 polymorphism and cancer risk was ethnicity dependent, as supported by Wang et al. [88]. This may be due to the difference in genetic backgrounds among races due to allele frequency or various carcinogenic mechanisms at tumor sites; another possibility may be that the polymorphism may be in linkage disequilibrium with the causal variant [124]. In contrast to our results, He et al. [112] found no association between this polymorphism and cancer risk among Caucasians.
MiR-196a2 is composed of two different mature miRNAs (miR-196a-5P and miR-196a-3P), which are processed from the same stem-loop [125]. rs11614913, located in the mature sequence of miR-196a-3P, could influence the production levels of mature miR-196a and could have an impact on the expression of its target gene. Therefore, the altered expression patterns of miR-196a could influence its potential targets, which may play a role in regulating carcinogenesis. Previous meta-analysis studies have suggested an association between rs11614913 and the risk of cancers [21], [91]–[92], [112]–[114]. The present meta-analysis also provides evidence that the miR-196a2 rs11614913 T allele is significantly associated with reduced cancer risk in the allelic contrast, the homozygote comparison, and the recessive models, similar to the findings of previous studies [112]–[114]. In the subgroup analysis that was divided by cancer type, homozygote TT had the effect of decreasing the risk of lung cancer and colorectal cancer compared with that for CC homozygote or C allele carriers. T allele carriers also showed decreased cancer susceptibility compared with homozygote CC carriers in liver cancer, whereas Wang et al. [111] and He et al. [112] reported that this polymorphism has no association with the risk of HCC. Moreover, no association was found between miR-196a2 rs11614913 and bladder cancer, breast cancer, gastric cancer, ESCC, or SCCHN. Guo et al. [89] and Wang et al. [93] found that the C allele could increase cancer risk in gastric cancer. In ethnic subgroup analysis, a strong association was found between rs11614913 and cancer risk among Asians but not among Caucasians, which was similar to the findings of previous studies [112]–[113]. In addition, biochemical studies on rs11614913 confirmed the results of our meta-analysis. It has been well established that Hox gene expression is deregulated in lung and prostate cancers [126]–[127], and members of the Hox family have been found to be significantly downregulated in cells treated with pre-miR-196a-C [127]. Two tumor suppressors (GADD45G and INHBB) were reported to be downregulated and several oncogenes (TP63 and genes encoding two calcium-binding proteins) were found to be upregulated in breast cancer cells after pre-miR-196a-C introduction, suggestive of the oncogenic activity of pre-miR-196a-C and protective role of pre-miR-196a-T [83], [128]. Our results provide compelling evidence that the miR-196a2 rs11614913 polymorphism plays a crucial role in the development of cancer. Screening patients harboring the miR-196a2 rs11614913 polymorphism may prove clinically useful for the prediction and prevention of cancer.
The miR-499T>C (rs3746444) polymorphism has been identified within the stem region of the mir-499 gene and results in an A:U to G:U mismatch in the stem structure of the miR-499 precursor. The presence of this mismatch would affect Sox6 and Rod1 genes, which are important for the etiology of cancers [72], [129]. Several studies have identified miR-499 rs3746444 as a possible biomarker for multiple cancers [20], [70], [71], [79], [82]; however, the mechanism by which this occurs remains unknown. Our results showed that the rs3746444 C allele could increase cancer risk in the allelic contrast model and in the dominant model, which was consistent with the results of Srivastava et al. [114]. In analysis stratified by cancer type, significant associations between the rs3746444 polymorphism and cancer risk were observed for breast cancer, which is in contrast to the results reported by Srivastava et al. [114] and He et al. [112]. However, no significant result was observed for other cancers under any genetic model. Subgroup analysis by ethnicity showed that the C allele was associated with increased cancer risk in the Asian population, but not in the Caucasian population.
For mir-149 rs2292832, a significant association was found only in some of the subgroup analyses but not in the pooled results. rs2292932 in miR-149 has been tested for several cancers but was not found to be associated with cancer risk [61], [71], [81], [82], [90], [112], [114]. This suggests that the molecular mechanisms underlying the genetic associations of miRNA-SNPs with cancer risk may be complex and variable. Our results should be interpreted with caution, considering that the influence of the T allele in miR-149 might be masked by the presence of other unidentified causal genes involved in cancer development [90] and the limited number (12) of studies on this polymorphism. More studies will need to be analyzed to confirm the results.
MiR-27a rs895919 is located in the terminal loop of pre-miRNA-27a (an intergenic region of chromosome 19), which is upregulated in many tumors [130] and has been considered to be an oncomir [131]–[133]. To date, several epidemiologic studies have been conducted to investigate the association between the rs895919 polymorphism and cancer risk [26]–[28], [73], [74]; however, the results remain inconsistent and inconclusive. The results of two previous meta-analyses have indicated that the G allele in miR-27a rs895819 may be associated with decreased risk for some cancers, as well as with reduced cancer risk in Caucasians to some extent [94], [95]. Based on our study, no association was observed between this polymorphism and cancer risk when all the data were pooled in the meta-analysis. Our results also showed that the rs895819 G allele was associated with decreased cancer risk in a Caucasian population, but was inconsistent with the abovementioned two articles on cancer type. Because of the limited number (9) of studies on this polymorphism, the results should be interpreted with caution.
Nevertheless, our study still has some limitations. First, relatively large heterogeneity was observed across some studies, which could be due to the difference in cancer types, the geographic areas (environmental factors), and genetic backgrounds of the samples. Second, the relatively small sample size of studies for some SNPs may lead to low statistical power, especially in stratified analysis. Third, lack of original data from the reviewed studies restricted further evaluation of potential interactions; this is of particular importance because gene–gene and gene–environment interactions may modulate various disease risks. Fourth, our analysis was limited to Asian and Caucasian ethnicities; therefore, it is uncertain whether these results can be generalized to other populations. Fifth, restriction to studies published in English or Chinese might confer potential language bias; moreover, publication bias might also exist because only published studies were included in this meta-analysis, and studies with no statistically significant results often have less chance for publication.
In conclusion, our results suggest that the miR-146a rs2910164 C allele is a protective factor for bladder cancer, prostate cancer, cervical cancer, and colorectal cancer in Asians, whereas it is a risk factor for PTC and SCCHN in Caucasians. mir-196a2 rs11614913 has significant association with overall cancer risk, especially for lung cancer, colorectal cancer, and other cancers in the Asian population. We also found that the mir-499 rs3746444 polymorphism could increase cancer risk in the Asian population. However, no significant association was observed between mir-149 rs2292832 and miR-27a rs895919 and overall cancer risk. Further studies with a larger sample size will be needed to clarify the possible roles of these polymorphisms in different kinds of cancers.
Supporting Information
Begg’s funnel plot of publication bias for miR-146a rs2910164 G>C: C vs. G. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-196a2 rs11614913 C>T: T vs. C. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-499 rs3746444 T>C: C vs. T. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-149 rs2292832 C>T: T vs. C. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-27a rs895919A>G: G vs. A. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Acknowledgments
We are grateful to Prof. Jiaxue-Wu and Xianmei-Yang, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai, China, for their critical review and scientific editing of the manuscript and constructive comments. We thank Dr. Yang Yang for her critical reading of this manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31100895 and 31071193), the National Natural Science Foundation of China for Creative Research Groups (30024001), http://www.nsfc.gov.cn/; the National Key Sci-Tech Special Project of China (2008ZX10002-020), http://www.nmp.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 2. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 3. Ruan K, Fang X, Ouyang G (2009) MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 285: 116–126. [DOI] [PubMed] [Google Scholar]
- 4. Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103: 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269. [DOI] [PubMed] [Google Scholar]
- 7. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838. [DOI] [PubMed] [Google Scholar]
- 8. Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, et al. (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120: 21–24. [DOI] [PubMed] [Google Scholar]
- 9. Cho WC (2010) MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 42: 1273–1281. [DOI] [PubMed] [Google Scholar]
- 10. Cho WC (2010) Recent progress in genetic variants associated with cancer and their implications in diagnostics development. Expert Rev Mol Diagn 10: 699–703. [DOI] [PubMed] [Google Scholar]
- 11. Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866. [DOI] [PubMed] [Google Scholar]
- 12. Garzon R, Croce CM (2011) MicroRNAs and cancer: introduction. Semin Oncol 38: 721–723. [DOI] [PubMed] [Google Scholar]
- 13. Shastry BS (2009) SNPs: impact on gene function and phenotype. Methods Mol Biol 578: 3–22. [DOI] [PubMed] [Google Scholar]
- 14. Ryan BM, Robles AI, Harris CC (2010) Genetic variation in microRNA networks: the implications for cancer research. NATURE REVIEWS CANCER 10: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mittal RD, Gangwar R, George GP, Mittal T, Kapoor R (2011) Investigative Role of Pre-MicroRNAs in Bladder Cancer Patients: A Case-Control Study in North India. DNA AND CELL BIOLOGY 30: 401–406. [DOI] [PubMed] [Google Scholar]
- 16. Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O (2011) Genetic Variation in the MicroRNA-499 Gene and Hepatocellular Carcinoma Risk in a Turkish Population: Lack of Any Association in a Case-Control Study. ASIAN PACIFIC JOURNAL OF CANCER PREVENTION 12: 3107–3112. [PubMed] [Google Scholar]
- 17. Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O, et al. (2011) No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene 486: 104–109. [DOI] [PubMed] [Google Scholar]
- 18. Yue C, Wang ML, Ding B, Wang W, Fu SL, et al. (2011) Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. GYNECOLOGIC ONCOLOGY 122: 33–37. [DOI] [PubMed] [Google Scholar]
- 19. Zhou B, Wang K, Wang Y, Xi M, Zhang Z, et al. (2011) Common genetic polymorphisms in pre-microRNAs and risk of cervical squamous cell carcinoma. Mol Carcinog 50: 499–505. [DOI] [PubMed] [Google Scholar]
- 20. Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, et al. (2010) Association Between Common Genetic Variants in Pre-microRNAs and Gastric Cancer Risk in Japanese Population. HELICOBACTER 15: 524–531. [DOI] [PubMed] [Google Scholar]
- 21. Xu W, Xu J, Liu S, Chen B, Wang X, et al. (2011) Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis. PLOS One 6: e20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horikawa Y, Wood CG, Yang H, Zhao H, Ye Y, et al. (2008) Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 14: 7956–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou J, Lv RX, Song XB, Li DD, Hu X, et al. (2012) Association Between Two Genetic Variants in miRNA and Primary Liver Cancer Risk in the Chinese Population. DNA AND CELL BIOLOGY 31: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiang Y, Fan S, Cao J, Huang SF, Zhang LP (2012) Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. MOLECULAR BIOLOGY REPORTS 39: 7019–7023. [DOI] [PubMed] [Google Scholar]
- 25. Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, et al. (2012) Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. MOLECULAR CARCINOGENESIS 511: E65–E73. [DOI] [PubMed] [Google Scholar]
- 26. Shi DN, Li P, Ma L, Zhong DY, Chu HY, et al. (2012) A Genetic Variant in pre-miR-27a Is Associated with a Reduced Renal Cell Cancer Risk in a Chinese Population. PLOS ONE 7: e46566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hezova R, Kovarikova A, Bienertova-Vasku J, Sachlova M, Redova M, et al. (2012) Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. WORLD JOURNAL OF GASTROENTEROLOGY 18: 2827–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catucci I, Verderio P, Pizzamiglio S, Bernard L, Dall’Olio V, et al. (2012) The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. BREAST CANCER RESEARCH AND TREATMENT 133: 805–807. [DOI] [PubMed] [Google Scholar]
- 29. Woolf B (1955) On Estimating the Relation between Blood Group and Disease. ANNALS OF HUMAN GENETICS 19: 251–253. [DOI] [PubMed] [Google Scholar]
- 30. Cochran Wg (1954) The Combination of Estimates from Different Experiments. BIOMETRICS 10: 101–129. [Google Scholar]
- 31. DerSimonian R, Kacker R (2007) Random-effects model for meta-analysis of clinical trials: An update. CONTEMPORARY CLINICAL TRIALS 28: 105–114. [DOI] [PubMed] [Google Scholar]
- 32. Mantel N, Haenszel W (1959) Statistical Aspects of the Analysis of Data from Retrospective Studies of Disease. JOURNAL OF THE NATIONAL CANCER INSTITUTE 22: 719–748. [PubMed] [Google Scholar]
- 33.A T (1999) Assessing the influence of a single study in the meta-analysis estimate. 15–17.
- 34. Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BRITISH MEDICAL JOURNAL 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orsos Z, Szanyi I, Csejtei A, Gerlinger I, Ember I, et al. (2013) Association of pre-miR-146a rs2910164 Polymorphism with the Risk of Head and Neck Cancer. ANTICANCER RESEARCH 33: 341–346. [PubMed] [Google Scholar]
- 36. Yamashita J, Iwakiri T, Fukushima S, Jinnin M, Miyashita A, et al. (2013) The rs2910164 G>C polymorphism in microRNA-146a is associated with the incidence of malignant melanoma. MELANOMA RESEARCH 23: 13–20. [DOI] [PubMed] [Google Scholar]
- 37. Wei WJ, Wang YL, Li DS, Wang Y, Wang XF, et al. (2013) Association between the rs2910164 Polymorphism in Pre-Mir-146a Sequence and Thyroid Carcinogenesis. PLOS ONE 8: e56638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma F, Zhang P, Lin DX, Yu DK, Yuan P, et al. (2013) There Is No Association between MicroRNA Gene Polymorphisms and Risk of Triple Negative Breast Cancer in a Chinese Han Population. PLOS ONE 8: e60195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, et al. (2013) Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. METHODS 59: 138–146. [DOI] [PubMed] [Google Scholar]
- 40. Ma L, Zhu LJ, Gu DY, Chu HY, Tong N, et al. (2013) A genetic variant in miR-146a modifies colorectal cancer susceptibility in a Chinese population. ARCHIVES OF TOXICOLOGY 87: 825–833. [DOI] [PubMed] [Google Scholar]
- 41. Song XC, Sturgis EM, Liu J, Jin L, Wang ZQ, et al. (2013) MicroRNA Variants Increase the Risk of HPV-Associated Squamous Cell Carcinoma of the Oropharynx in Never Smokers. PLOS ONE 8: e56622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou FY, Zhu HX, Luo DW, Wang ML, Dong X, et al. (2012) A Functional Polymorphism in Pre-miR-146a Is Associated with Susceptibility to Gastric Cancer in a Chinese Population. DNA AND CELL BIOLOGY 31: 1290–1295. [DOI] [PubMed] [Google Scholar]
- 43. Alshatwi AA, Shafi G, Hasan TN, Syed NA, Al-Hazzani AA, et al. (2012) Differential Expression Profile and Genetic Variants of MicroRNAs Sequences in Breast Cancer Patients. PLOS ONE 7: e30049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang ML, Chu HY, Li P, Yuan L, Fu GB, et al. (2012) Genetic Variants in miRNAs Predict Bladder Cancer Risk and Recurrence. CANCER RESEARCH 72: 6173–6182. [DOI] [PubMed] [Google Scholar]
- 45. Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK, et al. (2012) Impacts of MicroRNA Gene Polymorphisms on the Susceptibility of Environmental Factors Leading to Carcinogenesis in Oral Cancer. PLOS ONE 7: e39777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B, et al. (2012) Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. MOLECULAR CARCINOGENESIS 511: E21–E31. [DOI] [PubMed] [Google Scholar]
- 47. Zhang M, Jin M, Yu Y, Zhang S, Wu Y, et al. (2012) Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. EUROPEAN JOURNAL OF CANCER CARE 21: 274–280. [DOI] [PubMed] [Google Scholar]
- 48. Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, et al. (2012) Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. GENE 504: 92–97. [DOI] [PubMed] [Google Scholar]
- 49. Mihalache F, Hoblinger A, Acalovschi M, Sauerbruch T, Lammert F, et al. (2012) A common variant in the precursor miR-146a sequence does not predispose to cholangiocarcinoma in a large European cohort. HEPATOBILIARY & PANCREATIC DISEASES INTERNATIONAL 11: 412–417. [DOI] [PubMed] [Google Scholar]
- 50. Zhu LJ, Chu HY, Gu DY, Ma L, Shi DN, et al. (2012) A Functional Polymorphism in miRNA-196a2 Is Associated with Colorectal Cancer Risk in a Chinese Population. DNA AND CELL BIOLOGY 31: 349–353. [DOI] [PubMed] [Google Scholar]
- 51. Chen H, Sun LY, Chen LL, Zheng HQ, Zhang QF (2012) A variant in microRNA-196a2 is not associated with susceptibility to and progression of colorectal cancer in Chinese. INTERNAL MEDICINE JOURNAL 42: E115–E119. [DOI] [PubMed] [Google Scholar]
- 52. Zhou Y, Du WD, Chen G, Ruan J, Xu S, et al. (2012) Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. JOURNAL OF CANCER RESEARCH AND CLINICAL ONCOLOGY 138: 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linhares JJ, Azevedo M, Siufi AA, de Carvalho CV, Wolgien M, et al.. (2012) Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC MEDICAL GENETICS 13. [DOI] [PMC free article] [PubMed]
- 54.Lung RW, Wang X, Tong JH, Chau SL, Lau KM, et al.. (2012) A single nucleotide polymorphism in microRNA-146a is associated with the risk for nasopharyngeal carcinoma. Mol Carcinog. [DOI] [PubMed]
- 55. Permuth-Wey J, Thompson RC, Nabors LB, Olson JJ, Browning JE, et al. (2011) A functional polymorphism in the pre-miR-146a gene is associated with risk and prognosis in adult glioma. JOURNAL OF NEURO-ONCOLOGY 105: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garcia AI, Cox DG, Barjhoux L, Verny-Pierre C, Barnes D, et al. (2011) The rs2910164:G>C SNP in the MIR146A Gene is Not Associated with Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. HUMAN MUTATION 32: 1004–1007. [DOI] [PubMed] [Google Scholar]
- 57. Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, et al. (2011) A Functional Variant in MicroRNA-196a2 Is Associated with Susceptibility of Colorectal Cancer in a Chinese Population. ARCHIVES OF MEDICAL RESEARCH 42: 144–148. [DOI] [PubMed] [Google Scholar]
- 58. Zhang XW, Pan SD, Feng YL, Liu JB, Dong J, et al. (2011) [Relationship between genetic polymorphism in microRNAs precursor and genetic predisposition of hepatocellular carcinoma]. Zhonghua Yu Fang Yi Xue Za Zhi 45: 239–243. [PubMed] [Google Scholar]
- 59. Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y (2011) A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. JOURNAL OF VIRAL HEPATITIS 18: E399–E407. [DOI] [PubMed] [Google Scholar]
- 60. Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, et al. (2011) Combined Effect of miR-146a rs2910164 G/C Polymorphism and Toll-like Receptor 4+3725 G/C Polymorphism on the Risk of Severe Gastric Atrophy in Japanese. DIGESTIVE DISEASES AND SCIENCES 56: 1131–1137. [DOI] [PubMed] [Google Scholar]
- 61. Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F, et al. (2011) Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. CLINICAL CHEMISTRY AND LABORATORY MEDICINE 49: 2073–2080. [DOI] [PubMed] [Google Scholar]
- 62. George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD (2011) Genetic variation in microRNA genes and prostate cancer risk in North Indian population. MOLECULAR BIOLOGY REPORTS 38: 1609–1615. [DOI] [PubMed] [Google Scholar]
- 63. Jedlinski DJ, Gabrovska PN, Weinstein SR, Smith RA, Griffiths LR (2011) Single Nucleotide Polymorphism in hsa-mir-196a-2 and Breast Cancer Risk: A Case Control Study. TWIN RESEARCH AND HUMAN GENETICS 14: 417–421. [DOI] [PubMed] [Google Scholar]
- 64. Hong YS, Kang HJ, Kwak JY, Park BL, You CH, et al. (2011) Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. Journal of preventive medicine and public health = Yebang Uihakhoe chi 44: 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu B, Feng NH, Li PC, Tao J, Wu DY, et al. (2010) A Functional Polymorphism in Pre-miR-146a Gene Is Associated With Prostate Cancer Risk and Mature miR-146a Expression In Vivo. PROSTATE 70: 467–472. [DOI] [PubMed] [Google Scholar]
- 66. Guo H, Wang K, Xiong G, Hu HM, Wang DM, et al. (2010) A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. FAMILIAL CANCER 9: 599–603. [DOI] [PubMed] [Google Scholar]
- 67. Li XD, Li ZG, Song XX, Liu CF (2010) A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. PATHOLOGY 42: 669–673. [DOI] [PubMed] [Google Scholar]
- 68. Pastrello C, Polesel J, Della Puppa L, Viel A, Maestro R (2010) Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. CARCINOGENESIS 31: 2124–2126. [DOI] [PubMed] [Google Scholar]
- 69. Zeng Y, Sun QM, Liu NN, Dong GH, Chen J, et al. (2010) Correlation between pre-miR-146a C/G polymorphism and gastric cancer risk in Chinese population. WORLD JOURNAL OF GASTROENTEROLOGY 16: 3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Catucci I, Yang RX, Verderio P, Pizzamiglio S, Heesen L, et al. (2010) Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as Low-penetrance Alleles in German and Italian Familial Breast Cancer Cases. HUMAN MUTATION 31: E1052–E1057. [DOI] [PubMed] [Google Scholar]
- 71. Liu ZS, Li GJ, Wei S, Niu JG, El-Naggar AK, et al. (2010) Genetic Variants in Selected Pre-MicroRNA Genes and the Risk of Squamous Cell Carcinoma of the Head and Neck. CANCER 116: 4753–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qi P, Dou TH, Geng L, Zhou FG, Gu X, et al. (2010) Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. HUMAN IMMUNOLOGY 71: 621–626. [DOI] [PubMed] [Google Scholar]
- 73. Sun QM, Gu HJ, Zeng Y, Xia Y, Wang Y, et al. (2010) Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. CANCER SCIENCE 101: 2241–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang RX, Schlehe B, Hemminki K, Sutter C, Bugert P, et al. (2010) A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. BREAST CANCER RESEARCH AND TREATMENT 121: 693–702. [DOI] [PubMed] [Google Scholar]
- 75. Kim MJ, Yoo SS, Choi YY, Park JY (2010) A functional polymorphism in the pre-microRNA-196a2 and the risk of lung cancer in a Korean population. LUNG CANCER 69: 127–129. [DOI] [PubMed] [Google Scholar]
- 76. Wang K, Guo H, Hu HM, Xiong G, Guan XY, et al. (2010) A functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese Han. BIOMARKERS 15: 614–618. [DOI] [PubMed] [Google Scholar]
- 77. Dou TH, Wu QH, Chen X, Ribas J, Ni XH, et al. (2010) A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. JOURNAL OF CANCER RESEARCH AND CLINICAL ONCOLOGY 136: 1853–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Peng S, Kuang ZS, Sheng CY, Zhang Y, Xu H, et al. (2010) Association of MicroRNA-196a-2 Gene Polymorphism with Gastric Cancer Risk in a Chinese Population. DIGESTIVE DISEASES AND SCIENCES 55: 2288–2293. [DOI] [PubMed] [Google Scholar]
- 79. Srivastava K, Srivastava A, Mittal B (2010) Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. JOURNAL OF HUMAN GENETICS 55: 495–499. [DOI] [PubMed] [Google Scholar]
- 80. Christensen BC, Avissar-Whiting M, Ouellet LG, Butler RA, Nelson HH, et al. (2010) Mature MicroRNA Sequence Polymorphism in MIR196A2 Is Associated with Risk and Prognosis of Head and Neck Cancer. CLINICAL CANCER RESEARCH 16: 3713–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hu ZB, Liang J, Wang ZW, Tian T, Zhou XY, et al. (2009) Common Genetic Variants in Pre-MicroRNAs Were Associated With Increased Risk of Breast Cancer in Chinese Women. HUMAN MUTATION 30: 79–84. [DOI] [PubMed] [Google Scholar]
- 82. Tian T, Shu YQ, Chen JP, Hu ZB, Xu L, et al. (2009) A Functional Genetic Variant in microRNA-196a2 Is Associated with Increased Susceptiblility of Lung Cancer in Chinese. CANCER EPIDEMIOLOGY BIOMARKERS & PREVENTION 18: 1183–1187. [DOI] [PubMed] [Google Scholar]
- 83. Hoffman AE, Zheng TZ, Yi CH, Leaderer D, Weidhaas J, et al. (2009) microRNA miR-196a-2 and Breast Cancer: A Genetic and Epigenetic Association Study and Functional Analysis. CANCER RESEARCH 69: 5970–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xu T, Zhu Y, Wei QK, Yuan YF, Zhou F, et al. (2008) A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. CARCINOGENESIS 29: 2126–2131. [DOI] [PubMed] [Google Scholar]
- 85. Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, et al. (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA 105: 7269–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yang HS, Dinney CP, Ye YQ, Zhu Y, Grossman HB, et al. (2008) Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. CANCER RESEARCH 68: 2530–2537. [DOI] [PubMed] [Google Scholar]
- 87. Wei J, Zheng L, Liu S, Yin J, Wang L, et al. (2013) MiR-196a2 rs11614913 T>C polymorphism and risk of esophageal cancer in a Chinese population. Human Immunology 7: 1199–1205. [DOI] [PubMed] [Google Scholar]
- 88. Wang AX, Xu B, Tong N, Chen SQ, Yang Y, et al. (2012) Meta-analysis confirms that a common G/C variant in the pre-miR-146a gene contributes to cancer susceptibility and that ethnicity, gender and smoking status are risk factors. GENETICS AND MOLECULAR RESEARCH 11: 3051–3062. [DOI] [PubMed] [Google Scholar]
- 89. Guo J, Jin MJ, Zhang MW, Chen K (2012) A Genetic Variant in miR-196a2 Increased Digestive System Cancer Risks: A Meta-Analysis of 15 Case-Control Studies. PLOS ONE 7: e30585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang J, Liu YF, Gan Y (2012) Lack of association between miR-149 C>T polymorphism and cancer susceptibility: a meta-analysis based on 4,677 cases and 4,830 controls. MOLECULAR BIOLOGY REPORTS 39: 8749–8753. [DOI] [PubMed] [Google Scholar]
- 91. Chu H, Wang M, Shi D, Ma L, Zhang Z, et al. (2011) Hsa-miR-196a2 Rs11614913 polymorphism contributes to cancer susceptibility: evidence from 15 case-control studies. PLOS One 6: e18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang F, Ma Y, Zhang P, Yang J, Chen H, et al. (2012) A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Molecular biology reports 39: 269–275. [DOI] [PubMed] [Google Scholar]
- 93. Wang P, Xie S, Cui A, Zhang Y, Jiang B (2012) miR-196a2 polymorphisms and susceptibility to cancer: A meta-analysis involving 24,697 subjects. Experimental and Therapeutic Medicine 3: 324–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhong S, Chen Z, Xu J, Li W, Zhao J (2013) Pre-mir-27a rs895819 polymorphism and cancer risk: a meta-analysis. Mol Biol Rep 40: 3181–3186. [DOI] [PubMed] [Google Scholar]
- 95. Xu Q, He C, Liu J, Yuan Y (2013) Pre-miR-27a rs895819A/G Polymorphisms in Cancer: A Meta-Analysis. PLOS One 8: e65208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Qiu LX, Wang Y, Xia ZG, Xi B, Mao C, et al. (2011) miR-196a2 C allele is a low-penetrant risk factor for cancer development. Cytokine 56: 589–592. [DOI] [PubMed] [Google Scholar]
- 97. Qiu LX, He J, Wang MY, Zhang RX, Shi TY, et al. (2011) The association between common genetic variant of microRNA-146a and cancer susceptibility. Cytokine 56: 695–698. [DOI] [PubMed] [Google Scholar]
- 98. Zou P, Zhao L, Xu H, Chen P, Gu A, et al. (2012) Hsa-mir-499 rs3746444 polymorphism and cancer risk: a meta-analysis. Journal of Biomedical Research 26: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lian H, Wang L, Zhang JM (2012) Increased Risk of Breast Cancer Associated with CC Genotype of Has-miR-146a Rs2910164 Polymorphism in Europeans. PLOS One 7(2): e31615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang F, Sun GP, Zou YF, Li YY, Hao L, et al. (2012) Association of microRNA-499 rs3746444 Polymorphism with Cancer Risk: Evidence from 7188 Cases and 8548 Controls. PLOS One 7(9): e45042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang J, Bi JW, Liu X, Li KN, Di JS, et al. (2012) Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Molecular Biology Reports 39: 4571–4579. [DOI] [PubMed] [Google Scholar]
- 102. Wang Y, Yang BH, Ren XB (2012) Hsa-miR-499 polymorphism (rs3746444) and cancer risk: A meta-analysis of 17 case-control studies. Gene 509: 267–272. [DOI] [PubMed] [Google Scholar]
- 103. Qiu MT, Hu JW, Ding XX, Yang X, Zhang Z, et al. (2012) Hsa-miR-499 rs3746444 Polymorphism Contributes to Cancer Risk: A Meta-Analysis of 12 Studies. PLOS One 7(12): e50887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang F, Sun GP, Zou YF, Fan LL, Song B (2012) Lack of Association of miR-146a rs2910164 Polymorphism with Gastrointestinal Cancers: Evidence from 10206 Subjects. PLOS One 7(6): e39623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chen P, Zhang J, Zhou F (2012) miR-499 rs3746444 polymorphism is associated with cancer development among Asians and related to breast cancer susceptibility. Molecular Biology Reports 39: 10433–10438. [DOI] [PubMed] [Google Scholar]
- 106. Wang LN, Qian SS, Zhi H, Zhang Y, Wang B, et al. (2012) The association between hsa-miR-499 T>C polymorphism and cancer risk: A meta-analysis. Gene 508: 9–14. [DOI] [PubMed] [Google Scholar]
- 107. Yuan ZR, Zeng X, Yang D, Wang WL, Liu ZH (2013) Effects of Common Polymorphism rs11614913 in Hsa-miR-196a2 on Lung Cancer Risk. PLOS One, 2013 8(4): e61047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang F, Sun GP, Zou YF, Fan LL, Song B (2013) Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and gastrointestinal cancer risk. Molecular Biology Reports 40(1): 109–116. [DOI] [PubMed] [Google Scholar]
- 109. Fan CJ, Chen CY, Wu DQ (2013) The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Molecular Biology Reports 40: 3389–3394. [DOI] [PubMed] [Google Scholar]
- 110. Wang ZX, Lai J, Wang YR, Nie WW, Guan XX (2013) The Hsa-miR-27a rs895819 (A>G) polymorphism and cancer susceptibility. Gene 521: 87–90. [DOI] [PubMed] [Google Scholar]
- 111. Wang ZX, Cao Y, Jiang CP, Yang G, Wu JH, et al. (2012) Lack of Association of Two Common Polymorphisms rs2910164 and rs11614913 with Susceptibility to Hepatocellular Carcinoma: A Meta-Analysis. PLOS ONE 7: e40039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. He BS, Pan YQ, Cho WC, Xu YQ, Gu L, et al. (2012) The Association between Four Genetic Variants in MicroRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and Cancer Risk: Evidence from Published Studies. PLOS ONE 7: e49032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang JB, Wang QW, Liu H, Shao N, Tan BX, et al. (2012) The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. MUTAGENESIS 27: 779–788. [DOI] [PubMed] [Google Scholar]
- 114. Srivastava K, Srivastava A (2012) Comprehensive Review of Genetic Association Studies and Meta-Analyses on miRNA Polymorphisms and Cancer Risk. PLOS ONE 7: e50966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tian T, Xu Y, Dai J, Wu J, Shen H, et al. (2010) Functional polymorphisms in two pre-microRNAs and cancer risk: A meta-analysis. International Journal of Molecular Epidemiology and Genetics 1: 358–366. [PMC free article] [PubMed] [Google Scholar]
- 116. Gao LB, Bai P, Pan XM, Jia J, Li LJ, et al. (2011) The association between two polymorphisms in pre-miRNAs and breast cancer risk: a meta-analysis. Breast Cancer Research and Treatment 125: 571–574. [DOI] [PubMed] [Google Scholar]
- 117. Yin ZH, Yan L, Cui ZG, Li XL, Ren YW, et al. (2013) Effects of common polymorphisms rs2910164 in miR-146a and rs3746444 in miR-499 on cancer susceptibility: a meta-analysis. Molecular Biology Reports 40: 3003–3013. [DOI] [PubMed] [Google Scholar]
- 118. Hu M, Zhao LY, Hu SR, Yang JT (2013) The Association between Two Common Polymorphisms in MicroRNAs and Hepatocellular Carcinoma Risk in Asian Population. PLOS One 8(2): e57012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen ZW, Xu L, Ye XY, Shen SP, Li ZM, et al. (2013) Polymorphisms of microRNA Sequences or Binding Sites and Lung Cancer: A Meta-Analysis and Systematic Review. PLOS One 8(4): e61008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xu Y, Li L, Xiang X, Wang H, Cai W, et al. (2013) Three common functional polymorphisms in microRNA encoding genes in the susceptibility to hepatocellular carcinoma: A systematic review and meta-analysis. Gene 527: 584–593. [DOI] [PubMed] [Google Scholar]
- 121. Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, et al. (2008) Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 27: 5643–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li L, Chen XP, Li YJ (2010) MicroRNA-146a and human disease. Scand J Immunol 71: 227–231. [DOI] [PubMed] [Google Scholar]
- 123. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, et al. (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739. [DOI] [PubMed] [Google Scholar]
- 124. Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4: 45–61. [DOI] [PubMed] [Google Scholar]
- 125. Chen C, Zhang Y, Zhang L, Weakley SM, Yao Q (2011) MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med 15: 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Miller GJ, Miller HL, van Bokhoven A, Lambert JR, Werahera PN, et al. (2003) Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res 63: 5879–5888. [PubMed] [Google Scholar]
- 127. Calvo R, West J, Franklin W, Erickson P, Bemis L, et al. (2000) Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci U S A 97: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ying J, Srivastava G, Hsieh WS, Gao Z, Murray P, et al. (2005) The stress-responsive gene GADD45G is a functional tumor suppressor, with its response to environmental stresses frequently disrupted epigenetically in multiple tumors. Clin Cancer Res 11: 6442–6449. [DOI] [PubMed] [Google Scholar]
- 129. Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, et al. (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584: 4575–4580. [DOI] [PubMed] [Google Scholar]
- 130. Ma Y, Yu S, Zhao W, Lu Z, Chen J (2010) miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett 298: 150–158. [DOI] [PubMed] [Google Scholar]
- 131. Liu T, Tang H, Lang Y, Liu M, Li X (2009) MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett 273: 233–242. [DOI] [PubMed] [Google Scholar]
- 132. Zhang Z, Liu S, Shi R, Zhao G (2009) miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet 204: 486–491. [DOI] [PubMed] [Google Scholar]
- 133. Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, et al. (2011) Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene 30(36): 3875–3886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Begg’s funnel plot of publication bias for miR-146a rs2910164 G>C: C vs. G. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-196a2 rs11614913 C>T: T vs. C. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-499 rs3746444 T>C: C vs. T. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-149 rs2292832 C>T: T vs. C. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)
Begg’s funnel plot of publication bias for miR-27a rs895919A>G: G vs. A. Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
(TIF)